Abstract
With the extraordinary rise in available biological data, biologists and clinicians need unbiased tools for data integration in order to reach accurate, succinct conclusions. Network biology provides one such method for high-throughput data integration, but comes with its own set of algorithmic problems and needed expertise. We provide a step-by-step guide for using Omics Integrator, a software package designed for the integration of transcriptomic, epigenomic, and proteomic data. Omics Integrator can be found at http://fraenkel.mit.edu/omicsintegrator.
Keywords: Data Integration, Network Biology, Computational Biology, High-throughput Data
1. INTRODUCTION
As biologists gain access to increasing amounts of data, the challenges associated with interpreting those data have increased. Biologists and clinicians can obtain high-throughput information about a cell’s genome, transcriptome, epigenome, and proteome with reasonable effort and constantly decreasing costs. Indeed, much of those data are freely available to scientists through resources such as The Cancer Genome Atlas[1] and ENCODE[2]. The challenge remains, however, in knowing how to interpret those rich datasets. These “omic” data can be extraordinarily valuable. However, this value can only be extracted if data are properly analyzed using methods that account for the relatively high error rate of high-throughput experiments[3], and then condensed into understandable and actionable hypotheses about the underlying biology. This process can be especially difficult, and especially rewarding, when attempting to integrate several kinds of high-throughput data. Our group and others have shown that integrating data from several sources can lead to novel discoveries that each assay could have missed on its own[4–6].
Network biology is a fast-growing category of methods for this type of analysis [7]. Network models provide a valuable resource for biologists looking to analyze their high-throughput data in a systems context. By mapping ‘hits’ from high-throughput assays onto interaction networks, the mechanistic connections between the hits become obvious, and investigators can focus on pathways, or series of interactions in the cell that are related to a certain function, that may be perturbed in the system.
Network methods typically involve modeling the molecules within a cell – which can for example be DNA, mRNAs, proteins, or metabolites – as nodes in a graph. Edges between these nodes connect molecules that are functionally or physically connected [7]. For example, a protein-protein interaction network (PPI) would represent the binding of protein A to protein B by drawing an edge between the ‘A’ node and ‘B’ node in the network. Several publicly available databases have been created to translate experimentally discovered protein interactions into PPIs, such as iRefIndex [8], BioGRID [9], and STRING [10]. There are also databases that store interactions of proteins with other molecules, such as metabolites [11–13]. In other types of networks, the edges can represent more abstract relationships. For example, in a correlation-based network, edges between nodes might represent probable co-regulation, rather than physical interactions, based on covariance between the concentration of molecule A and molecule B [14, 15].
Mapping high-throughput hits onto networks in search of affected pathways has several advantages. Hits that are close to each other in a network might function in the same pathway. Focusing on subnetworks of functionally related nodes can produce a more tractable number of targets, rather than the potentially hundreds of individual factors identified in high-throughput experiments. In addition, this type of pathway identification reduces the chance of devoting resources to the analysis of false positives from the high-throughput screen. Although the confidence for each hit in a screen may be low, the confidence in a pathway that contains many hits is much higher. Finally, pathway analysis can help to find novel nodes that may not have appeared in a high-throughput screen. These “hidden nodes” can be false negatives in a screen, or true negatives that are nonetheless important players in the investigated biological system. Our work has shown that these hidden nodes can often be important to a system under study, despite the lack of direct experimental evidence [4, 16, 17]. Using the PPI to discover these pathways de novo, rather than relying on pre-determined pathway databases like KEGG [18], expands our ability to find novel information, and avoids biasing the results towards well-studied pathways.
However, network analysis is not as simple as just mapping high-throughput assay hits onto PPIs and finding all possible connections through them. Because of the large and highly connected nature of most biological networks [7], this “brute force” method results in extremely dense, uninterpretable “hairballs” rather than clear pathways [16]. Moreover, combining several types of experimental assays into a unified analysis can be complex. For example, experiments assessing changes in mRNA levels and protein levels are often not well correlated [19]. It is not trivial to map them onto one protein or RNA interaction network together. This chapter will walk you through the use of Omics Integrator, a software package that proposes a solution to these problems [17].
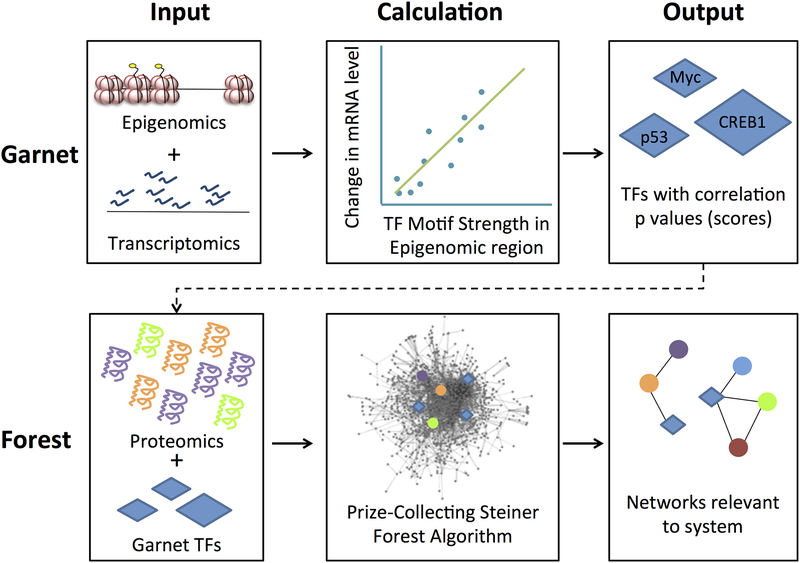
Omics Integrator is a new software tool designed to help biologists analyze and synthesize several kinds of high throughput omics data, and reduce it to a few important, high-confidence pathways. Omics Integrator is designed for ease of use by biologists with basic computer skills (comfort with using the Unix command line is helpful). Omics Integrator first uses transcriptomic and epigenomic data to reconstruct transcriptional regulatory networks, and then integrates those with proteomic data by mapping them onto a protein interaction network [17]. It uses two modules – Garnet and Forest, which are designed to run sequentially, but can also be run individually. Garnet mines transcriptomic and epigenomic information in order to predict transcription factors that may be responsible for gene expression changes in the studied system. Forest maps these transcription factors and protein-level experimental information onto a PPI. Forest then implements the Prize-Collecting Steiner Forest algorithm [16] to predict high-confidence low-density protein interaction pathways that are important to the studied system (See Figure 1).
Figure 1.
Outline of the Omics Integrator workflow. Epigenomic data (open chromatin regions or histone marks) and transcriptomic data are used to predict influential transcription factors (TFs). Transcription factors and proteomic data are then mapped onto an interactome, and the Prize Collecting Steiner Forest algorithm is used to produce small pathways and sub-networks predicted to be relevant to the experimental system.
2. Materials
2.1. Finding transcriptional regulators with Garnet
Transcriptomics data, i.e. differential gene expression between different conditions in your study (i.e. tumor vs. control).
Epigenomic data from a source such as TCGA [1], ENCODE [2], Roadmap [20], Omics Integrator example data, or experimentally derived epigenomic data (in a BED formatted file).
Transcription factor sequence binding motif predictions, from a source such as TRANSFAC [21], and/or Neph et. al. [22]. Omics Integrator provides a file derived from the TRANSFAC database.
2.2. Network integration with Forrest
Prize-collecting Steiner tree algorithm executable (msgsteiner can be downloaded from http://areeweb.polito.it/ricerca/cmp/code/bpsteiner).
Interactome file indicating all known interactions between proteins. Omics Integrator provides an interactome for mouse and human proteins derived from iRefIndex [8].
Input prize file, indicating the proteins you would like to include in the final solution (See Note 1).
(Optional) Output from Garnet to include transcription factors implicated by transcriptomic data in the final solution.
Cytoscape to visualize the final network solution.
3. METHODS
3.1. Installation of Omics Integrator
To install Omics Integrator, follow the instructions on our website: http://fraenkel.mit.edu/omicsintegrator/. You should make sure you have all dependencies (see Note 2) installed and that you have the most updated version of Omics Integrator from our GitHub page (see Note 3).
3.2. Finding transcriptional regulators with Garnet
Garnet uses differentially expressed genes from your transcriptomic assays (i.e. RNA-seq) to predict transcription factors (TFs) that are likely to be responsible for the altered gene expression. It uses epigenomic data to find regions of the genome to look for differential TF binding. For example, this could be ATAC-seq data that points out accessible regions of the genome in your cell type. The algorithm will search for transcription factor binding motifs within regions implicated by your epigenomic data. The strength of these motifs is then correlated with the magnitude of change of nearby differentially expressed genes to give each TF a score.
Obtain epigenomic data for cell lines related to your samples from one of the sources listed under 2.1. Alternatively, if you have epigenomic data for your own samples, you can use this as well. These data can be in the form of histone marks ChIP-seq, or DNase-seq or ATAC-seq, all of which indicate accessible chromatin regions where a TF might be bound. Collect these data in a BED-formatted file.
Go to the Galaxy webserver [23] (see Note 4) to extract the DNA sequences for your epigenomic regions. Upload your BED file to Galaxy under the “Get Data” tool, specify which genome you are using, and then use the “Fetch Alignments/Sequences” > “Extract Genomic DNA” tool to download a FASTA-formatted file.
Format your experimentally-derived gene expression data in a tab-delimited file with two columns. The first should be the name of the gene, and the second should be the log-fold-change of that gene in the study conditions (i.e. tumor vs. control). We recommend only including genes with a statistically significant change in expression (see Note 5).
Create the Garnet configuration file. For an example configuration file, see the README on the Omics Integrator GitHub page, or the comment on the top of scripts/garnet.py. Your configuration file should be formatted similarly, but you should replace the paths to the bedfile, fastafile, and expressionFile with the paths to the files you created in Steps 3.2.1–3.2.3. Make sure the annotation files referenced by genefile, xreffile, and genome are using the correct genome for your sample (files for mm9 and hg19 are provided with Omics Integrator).
You can change the parameters to your liking (Table 1).
-
Run Garnet on the command line by navigating to the directory with garnet.py and running python garnet.py yourconfigfile.cfg. You can also add a --outdir directoryname flag if you would like to put the output from garnet into a different directory.
Garnet will run through several steps, informing you on the command line where it is in the process. These steps include:- Mapping the genes to nearby epigenetic regions
- Scanning those regions for tf binding motifs
- Building a matrix of gene expression changes and binding motif scores for each TF
- Running a regression to check the correlation of TF binding score with differential gene expression
Garnet will print results into several tab-delimited files. These files are described in the README file on the Omics Integrator GitHub page. The file which ends in regression_results.tsv shows all TFs, clustered by similar binding sites, along with their p- and q-values from the regression. The file which ends in FOREST_INPUT.tsv contains only significant results and will be used in future steps.
Table 1.
An explanation of the parameters used by Garnet.
| windowsize | This parameter determines the maximum distance in nucleotides from a gene TSS to a TF binding motif to consider them related. Higher values will find more TFs, but their binding may be farther away from the gene, and thus, less likely to be directly related to expression. Values usually range from 2000 to 20000. |
| pvalThresh | The p value of a correlation measures how likely you are to get this correlation value if the events were not correlated. This threshold determines which transcription factors will be passed to Forest. Only those whose correlation with expression falls below the provided threshold will be included. Recommended values range from 0.01 to 0.05. Leave this value blank to use a q value threshold rather than a p value. |
| qvalThresh | A q value is a False Discovery Rate adjusted p value. This measurement will result in fewer false positives. This threshold determines which transcription factors will be passed to Forest. Only those whose correlation with expression falls below the provided threshold will be included. Recommended values range from 0.01 to 0.05. Leave this value blank if a p value threshold is sufficient. (If you are going on to run Forest, a p value is generally sufficient since the network nature of Forest make false positives less likely to appear in a final network). |
3.4. Network Integration with Forest
Forest integrates proteomic data and the output from Garnet into a network. After mapping the data onto a provided interactome network, it uses the prize-collecting Steiner tree algorithm (solved by the msgsteiner code that you downloaded and installed) to find an optimal set of sub-networks. These sub-networks can then be analyzed for pathway context.
If you are not using the default interactome provided with Omics Integrator, prepare your input interactome file. An interactome file (or “edge file”) contains the large network of all known connections between nodes. The file should be formatted in three tab-delimited columns. Each line should have the form “interactor1 interacter2 weight”. The third column contains an edge weight, between 0 and 0.99, usually representing the confidence in the validity of that edge. Optionally, you can include a fourth column indicating whether that edge is directed (‘D’) or undirected (‘U’). The current default interactome for human or mouse tissue is derived from iRefIndex (version 13) [8] and scored with the MIScore system [24]. You can find it in the data folder, called iref_mitab_miscore_2013_interactome.txt. You should create your own interactome file if you are not running your experiments in mouse or human cell models, or if you have a more updated interactome for your experiments.
Prepare your input prize file. This file contains significant features from your proteomic data (See Note 6). It should have two tab-delimited columns: the protein name (matching the interactome file exactly), and the protein prize. You should assign higher prizes to proteins for which you have stronger evidence that they should be in the final network.
Prepare your configuration file. This file contains input parameters for your run of Forest (Table 2). An example can be found in the example/a549 folder, called tgfb_forest.cfg. At a minimum this file must contain values for the parameters w, b, and D. If you are including results from Garnet, you will also need a garnetBeta parameter. See Section 3.5.1 “Choosing Parameters for Forest” section below for more information.
You can now run forest with the command python forest.py–p yourprizefile.txt–e youredgefile.txt–c yourconfigfile.txt --garnet yourgarnetoutput_FOREST_INPUT.tsv. You can also add a --outlabel yourexperimentname flag to give your output files a prefix and a --outpath directoryname flag if you would like to put the output from forest into a different directory. You may need to add a --msgpath directoryname flag to indicate where you installed the msgsteiner code during the installation step. There are several other optional flags you can add to this command if wanted (See Note 7).
-
Forest will run through several steps, informing you on the command line where it is in the process. These steps include:
- reading in your input files
- running the msgsteiner optimization
- writing the output files
Output files are described in the README file on the Omics Integrator GitHub page (see Note 8).
To visualize the network output, open the Forest output files in Cytoscape [25]. Open Cytoscape and import a network. The Forest output files that end in .sif have been formatted for this purpose. The file ending in optimalForest.sif contains only those edges used in the optimal Steiner forest, while augmentedForest.sif contains all edges in the interactome between the nodes in the final forest, and is recommended for final analysis. You can then import tables to annotate those networks; the nodeattributes.tsv file and the edgeattibutes.tsv file, to view information about the nodes and edges in the network, such as the edge weights and the node prizes. Node attributes also include the node prize type: TF, proteomic, or blank to indicate a hidden node which had no input prize but was chosen by the algorithm to connect prize nodes. Cytoscape has many useful visualization tools that you can use to better represent these values and types [25, 26] (See Note 9).
Table 2.
An explanation of the parameters used by Forest.
| w | This parameter influences the number of separate trees detected, which can aid in identifying functionally distinct processes. Higher values of w lead to more trees in the optimal forest, while lower values force most prizes to be found in the same tree. Values usually range from 1–10. See Tuncbag et al 2013[14] for a more detailed explanation. |
| b | This parameter linearly scales the prizes, thereby changing the relative weighting of edge weights and node prizes. Higher values lead to larger trees, including some low-confidence edges, while lower values force networks to be small and use only high confidence edges, and lead to the possible exclusion of some prize terminals. Values usually range from 1–20. |
| D | This parameter sets the maximum depth from the dummy node, or root of the tree, to the leaf nodes. Higher values lead to long pathways, while lower values lead to shorter disparate pathways. Values usually range from 5–15. |
| mu | This parameter controls negative prizes in Forest. Negative prizes are explained in detail above. The default value is zero, and if you want to use negative prizes, values usually range from 0.0001 to 0.1. |
| garnetBeta | This parameter controls the relative weighting of TF scores derived from Garnet and prize values on proteomic nodes. Higher values will encourage the inclusion of more TF nodes in the network, while lower values force networks to include only the most significant or pathway-relevant TF nodes. Typically, the value for this parameter is set to the median value of the proteomic prizes divided by the median value of the TF scores |
3.5. Network Quality control
We recommend checking the robustness and specificity of your networks. You can do this by adding flags to the forest.py command. Add --noisyEdges 10 to test robustness of your network to noise in the edge weights. This command will add Gaussian noise to the edgeweights, re-run Forest ten (or your input number of) times, and then merge the results into output files with noisyEdges in the filenames. Add --randomTerminals 10 to test specificity of your network to your input terminals. This command will randomly redistribute your prizes among the interactome, keeping the degree distribution of your original prizes, re-run Forest ten times, and then merge the results into output files with randomTerminals in the filenames. Both of these flags will increase the runtime of forest significantly (See Note 10).
Forest results include an attribute representing the fraction of optimal forests containing each node, which indicates how often that node appeared in the various forest runs with noise or random inputs. A robust network will have high FractionOfOptimalForestsContaining values for most nodes in noisyEdges run, and nodes that are specific to your input data will have low FractionOfOptimalForestsContaining values after randomTerminals runs. These metrics can be especially useful ways to judge the importance of hidden nodes to your system.
3.5.1. Choosing Parameters for Forest
The resulting network from this data integration algorithm is highly dependent on several parameters. These include w, b, D, mu, and garnetBeta (Table 2).
We recommend running Forest over a range of these values to find the best set for your system. To see an example of a script for testing parameters, see OmicsIntegrator/example/GBM/GBM_case_study.py. Once you have several resulting networks, we recommend choosing the best result by
Choosing a set of parameters that maximizes the fraction of input prize nodes that are included in the final network and are robust to noise (as judged by the noisyEdges runs).
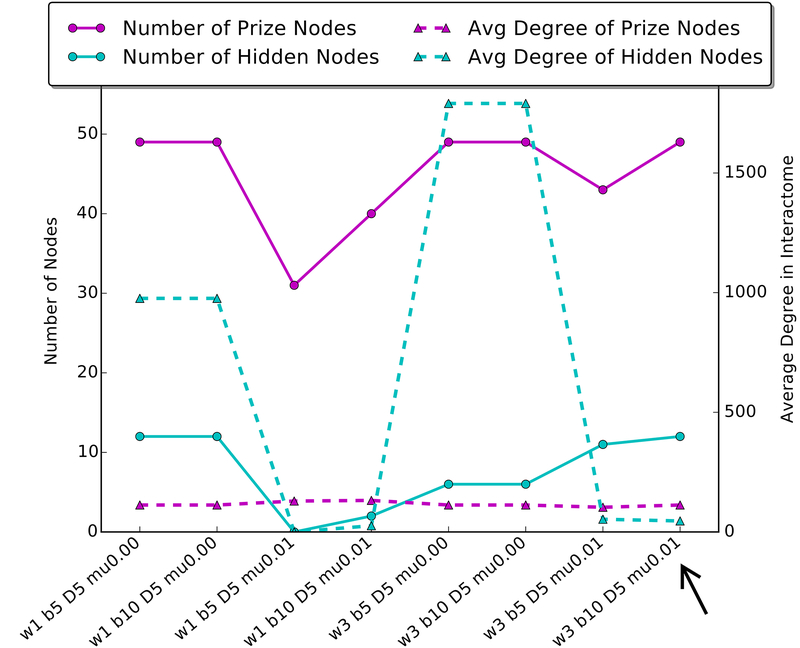
Some parameters will lead to networks with large “hubs”, that is, one hidden protein in the middle connected to several prize nodes with few interactions between these “spokes”. These hubs are usually not informative or very specific to one system. We recommend choosing parameters that minimize this by measuring the average degree of hidden nodes in your network (i.e., the number of edges connecting to those nodes in the interactome) compared to the average degree of prize nodes. A good parameter set will minimize the distance between these metrics. Figure 2 shows an example of this analysis using the data in the example/a549 folder (See Figure 2).
Once conditions 1 and 2 are satisfied, we prefer larger networks, as those provide the most opportunities for novel discoveries of hidden nodes and pathways enriched in the subnetworks.
Figure 2.
An analysis of several parameter sets when running Forest on the sample A549 data provided with Omics Integrator. A good parameter set will minimize the difference between the average degree of prize nodes and hidden nodes, and will include a large number of prize nodes. A good choice of a parameter set is highlighted by the black arrow. The A549 dataset reflects phosphoproteomic changes in a lung cancer cell line when stimulated with TGF-beta. The black arrow highlights a network that includes relevant nodes such as EGFR, while networks with large average degree of hidden nodes are mostly comprised of a hub centered on ubiquitin-C, which connects to most prize nodes in the interactome, but is not specific to the lung cancer cell system.
3.5.2. Negative Prizes in Forest
One of the more innovative aspects of Omics Integrator is its ability to incorporate negative evidence. There are two settings in which negative prizes can be useful. First, if you have reason to believe certain nodes should not show up in your optimal network you can assign a negative prize to a node and include it in the input prizes file along with positive prizes. Second, negative prizes can be used to avoid bias toward “hub nodes.”
We have found that in many cases, certain nodes are overrepresented in network integration solutions because they have a high “degree”, or number of edges connecting to that node, in the interactome. This could be because they bind with low specificity, e.g. chaperone proteins, or because they are highly studied proteins, causing more of their interactions to be discovered and represented in the literature. Because the optimal solution to the PCSF problem has the lowest cost method of connecting nodes, it will tend to use these nodes regardless of the input data. Simply removing these nodes from the network is not desirable, as there are settings in which they are relevant. To prevent hubs from being over-represented in all networks, Forest adds a penalty to nodes based on their degree. This penalty discourages solutions that include hubs but still allows them to be present when indicated by the data. This has been shown to improve accuracy in certain networks[17]. A positive number of the parameter mu will cause all nodes in the interactome to incur a penalty of mu*degree.
ACKNOWLEDGEMENTS
This work was supported by grants from National Institute of Health (R01-NS089076, T32-GM008334, and U01-CA184898). We thank Tobias Ehrenberger and Renan Escalante-Chong for helpful comments on the manuscript.
4. Notes
Problems in running Omics integrator can originate from spaces in node names, or mismatched node names. Input files to Garnet and Forest should have no spaces in the protein and gene names. In addition, all node names in the input files should match those in the interactome exactly. Forest will try to catch this error by letting you know if a large percentage of your input nodes were not found in the interactome. The provided iRefIndex interactome uses Official Gene Symbols for protein nodes, so when using this interactome, input files should also use this nomenclature.
Currently, Omics Integrator requires Python 2.6 or 2.7, with the python packages numpy, scipy, matplotlib, and Networkx. You will need Cytoscape (http://www.cytoscape.org) [25, 26] for viewing network results. Any updates will be reflected in the “System Requirements” section on our GitHub page (see Note 3).
GitHub is an online hosting service for repositories of code. It lets the community contribute to improvements of open source projects like Omics Integrator, and keeps track of changes made and bugs reported. The latest version of Omics Integrator, including any future updates, can be found on its GitHub page: https://github.com/fraenkel-lab/OmicsIntegrator
Galaxy is an online platform for computational biologists. In addition to the Extract Genomic Sequences tool described here, Galaxy provides several tools and workflows for analyzing biological data [23].
Genes used in Garnet should be significantly differentially expressed according to your transcriptomic data. For example, RNA-seq data can be analyzed with tools such as DEseq [27] or CuffDiff [28]. Genes that these tools report as differential with a p value less than 0.05 should be used as the input to Garnet.
Similar to transcriptomic data, your proteomics data will indicate which proteins should be used as the input to Forest. A review of tools for differential proteomics can be found here [29]. Many of these tools will provide a metric for determining statistical significance of differential expression of proteins, such as a p value. We generally use all proteins with a (modified) p value of less than 0.05. Prizes for the proteins are then the absolute value of the log of the fold change of protein expression. Be sure to use the absolute value, to avoid assigning a negative prize to downregulated proteins, which would encourage the algorithm to leave that node out of the networks, rather than including it.
There are several other flags available for advanced users, which change the behavior of forest.py. For example, you can change the group of nodes Forest uses to root each resulting tree (by default, this is all nodes which have been assigned a positive prize). There is a knockout option for doing an in silico knockout experiment by removing a protein from the interactome. For details on these and other flags, run python forest.py -h or read our GitHub repository page.
Many problems can lead to the final Forest output being empty (i.e. not containing any nodes). Check the output file ending in “info.txt” for some statistics of the run. One common problem, once formatting and input protein name problems have been ruled out, is a mu parameter set too high or other Forest parameters that lead to an empty optimal solution. Try changing your parameter values.
Cytoscape is a popular open source software for visualizing and analyzing networks [25, 26]. It is highly flexible and there are several available plug-ins for extending its use[30]. Omics Integrator can output results formatted for import into Cytoscape versions 2.8 or 3 by use of a flag for forest.py (it defaults to version 3). Once the networks and node and edge attributes are imported into Cytoscape, you can use options in Cytoscape to create informative figures of your results. For example, we often use the Style tab to change the color of a node to represent its prize, the shape of a node to represent its Terminal Type (TF vs proteomic vs hidden node), and the edge width to represent its confidence. We recommend playing around with Styles and Layouts to best display your network.
Depending on your input data and run set-up, a run of Omics Integrator can take a few hours. We recommend running in a screen session (https://www.gnu.org/software/screen/) or tmux (https://tmux.github.io/), which will allow the program to run continuously in the background, or on a computer that is set not to turn off or interrupt the run. You can also run Omics Integrator on a cloud server. However, if the run is taking more than a day, you should cancel the run and look for errors. In particular, try running Forest without or with a smaller input to noisyEdges or randomTerminals, as these options can lead to large memory and time consumption. High values for the D parameter can also increase runtime.
REFERENCES
- 1.Tomczak K, Czerwińska P, Wiznerowicz M (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Poznań, Poland) 19:A68–77. doi: 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium Encode (2013) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247.An [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malo N, Hanley JA, Cerquozzi S, et al. (2006) Statistical practice in high-throughput screening data analysis. Nat Biotechnol 24:167–75. doi: 10.1038/nbt1186 [DOI] [PubMed] [Google Scholar]
- 4.Huang S-SC, Fraenkel E (2009) Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci Signal 2:ra40. doi: 10.1126/scisignal.2000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ideker T, Thorsson V, Ranish J a, et al. (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934. doi: 10.1126/science.292.5518.929 [DOI] [PubMed] [Google Scholar]
- 6.Huang SSC, Clarke DC, Gosline SJC, et al. (2013) Linking Proteomic and Transcriptional Data through the Interactome and Epigenome Reveals a Map of Oncogene-induced Signaling. PLoS Comput Biol. doi: 10.1371/journal.pcbi.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabási A-L, Oltvai ZN (2004) Network Biology: Understanding the cell’s functional organization. Nat Rev Genet 5:101–113. doi: 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 8.Razick S, Magklaras G, Donaldson IM (2008) iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9:405. doi: 10.1186/1471-2105-9-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyers M, Breitkreutz A, Stark C, et al. (2006) BioGRID: a general repository for interaction datasets. Nucl Acids Res 34:D535–539. doi: 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Franceschini A, Wyder S, et al. (2015) STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart DS, Jewison T, Guo AC, et al. (2013) HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. doi: 10.1093/nar/gks1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiele I, Swainston N, Fleming RMT, et al. (2013) A community-driven global reconstruction of human metabolism. Nat Biotechnol 31:419–425. doi: 10.1038/nbt.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn M, Szklarczyk D, Pletscher-Frankild S, et al. (2014) STITCH 4: Integration of protein-chemical interactions with user data. Nucleic Acids Res. doi: 10.1093/nar/gkt1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valcárcel B, Würtz P, al Basatena NKS, et al. (2011) A differential network approach to exploring differences between biological states: An application to prediabetes. PLoS One. doi: 10.1371/journal.pone.0024702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotze HL, Armitage EG, Sharkey KJ, et al. (2013) A novel untargeted metabolomics correlation-based network analysis incorporating human metabolic reconstructions. BMC Syst Biol 7:107. doi: 10.1186/1752-0509-7-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuncbag N, Braunstein A, Pagnani A, et al. (2013) Simultaneous reconstruction of multiple signaling pathways via the prize-collecting steiner forest problem. J Comput Biol 20:124–36. doi: 10.1089/cmb.2012.0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuncbag N, Gosline SJ, Kedaigle AJ, et al. (2016) Network-based interpretation of diverse high-throughput datasets through the Omics Integrator software package. PLoS Comput. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki-Kinoshita KF, Kanehisa M (2007) Gene annotation and pathway mapping in KEGG. Methods Mol Biol 396:71–91. doi: 10.1007/978-1-59745-515-2_6 [DOI] [PubMed] [Google Scholar]
- 19.Maier T, Güell M, Serrano L (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett 583:3966–3973. doi: 10.1016/j.febslet.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, Stamatoyannopoulos J a, Costello JF, et al. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28:1045–1048. doi: 10.1038/nbt1010-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matys V, Kel-Margoulis OV, Fricke E, et al. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34:D108–10. doi: 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neph S, Vierstra J, Stergachis AB, et al. (2012) An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489:83–90. doi: 10.1038/nature11212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blankenberg D, Von Kuster G, Coraor N, et al. (2010) Galaxy: A web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. doi: 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villaveces JM, Jiménez RC, Porras P, et al. (2015) Merging and scoring molecular interactions utilising existing community standards: Tools, use-cases and a case study. Database. doi: 10.1093/database/bau131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, et al. (2003) Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smoot ME, Ono K, Ruscheinski J, et al. (2011) Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27:431–432. doi: 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Anders S, Huber W (2014) Differential analysis of count data - the DESeq2 package. Genome Biol. doi: 10.1186/s13059-014-0550-8 [DOI] [Google Scholar]
- 28.Trapnell C, Hendrickson DG, Sauvageau M, et al. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bantscheff M, Lemeer S, Savitski MM, Kuster B (2012) Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal Bioanal Chem 404:939–965. doi: 10.1007/s00216-012-6203-4 [DOI] [PubMed] [Google Scholar]
- 30.Saito R, Smoot ME, Ono K, et al. (2012) A travel guide to Cytoscape plugins. Nat Methods 9:1069–76. doi: 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]