Abstract
Adipose tissue, once viewed as an inert organ of energy storage, is now appreciated to be a central node for the dynamic regulation of systemic metabolism. There are three general types of adipose tissue: white, brown, and brown-in-white or “beige” fat. All three types of adipose tissue communicate extensively with other organs in the body including skin, liver, pancreas, muscle, and brain to maintain energy homeostasis. When energy intake chronically exceeds energy expenditure, obesity and its comorbidities can develop. Thus, understanding the molecular mechanisms by which different types of adipose tissues develop and function could uncover new therapies for combating disorders of energy imbalance. In this article, we highlight recent findings on the transcriptional and chromatin-mediated regulation of brown and beige adipose tissue activity.
Keywords: thermogenesis, brown fat, beige fat, transcription, metabolism
I. WHITE, BROWN, AND BEIGE ADIPOSE TISSUE
Adipocytes, skeletal muscle cells, chondrocytes, and bone cells descend from mesenchymal progenitor cells. Adipogenesis is initiated when mesenchymal progenitors undergo progressive fate restrictions to become committed preadipocytes. This is followed by differentiation, as preadipocytes acquire the morphological and functional characteristics of mature adipocytes (Figure 1A). White adipose tissue (WAT) stores energy in the form of triglyceride, secretes hormones, provides cushioning to protect organs and bones, and insulates the body (1). The predominant function of WAT is to balance lipid storage (lipogenesis) and breakdown (lipolysis) in response to the nutritional state of the organism. The ability of white adipocytes to store energy in the form of lipids (triglycerides) protects other tissues such as muscle, pancreas, and liver against the harmful effects of lipid overload, called lipotoxicity. When energy demand increases, such as during fasting or exercise, triglycerides undergo lipolysis in adipocytes. This generates glycerol and free fatty acids for use as an energy substrate in other tissues. Sympathetic nerves play a major role in promoting lipolysis via the secretion of the β-adrenergic agonist norepinephrine (NE) in adipose tissue. This triggers a cAMP/Protein Kinase A (PKA)-dependent signaling cascade leading to the breakdown of lipid droplets (2–5). Conversely, in response to feeding, insulin induces energy storage in adipocytes, while simultaneously suppressing lipolysis (2).
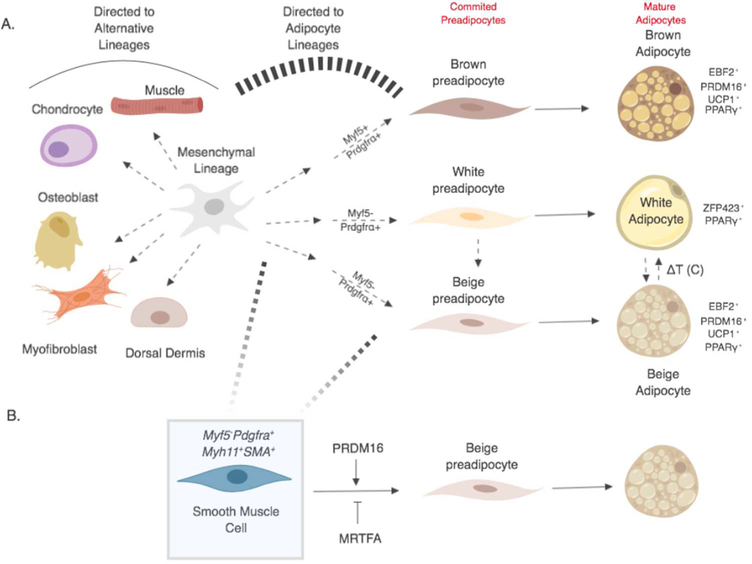
Figure 1. Adipocyte lineage commitment and differentiation.
(A) Mesenchymal precursors (gray) undergo progressive cell fate restrictions and commit to the adipocyte lineage. Brown adipocytes develop from a Myf5+Pdgfrα+ cells, whereas white and beige preadipocytes arise from a Myf5-Pdgfrα+ lineage (multiple steps indicated by dashed arrows). Upon terminal differentiation signals, preadipocytes accumulate lipid and become mature PPARγ+ adipocytes. Cold exposure and re-warming can provoke chromatin state shifts between beige and white adipocytes. Mesenchymal precursors also give rise to alternative cell types including muscle, bone, cartilage, and dermis. (B) Beige adipocytes have been proposed to develop from smooth muscle-like precursor cells (blue shaded box).
WAT is remarkably flexible and can respond to energy surplus through adipocyte hypertrophy and hyperplasia. How are new adipocytes made during periods of energetic overload? Genetic tracing of adipocyte fate in mice demonstrated that initially, both subcutaneous and visceral white adipose undergo hypertrophy to accommodate the increased demand for lipid storage. However, following prolonged high-fat diet and even before existing adipocytes reach capacity, visceral adipose tissues undergo a wave of new adipogenesis when adipocyte progenitors proliferate and differentiate, a phenomenon not observed in subcutaneous depots (6, 7). Thus, different WAT depots adapt to changing systemic energy levels via distinct mechanisms.
The adipose tissue extracellular matrix also undergoes extensive remodeling in response to changing energy flux. Obesity induces the differentiation of fibrogenic precursor cells into myofibroblast-like cells, which deposit extracellular matrix and contribute to fibrosis (8). Activation of the protein Myocardin-related transcription factor A (MRTFA) induces myofibroblast-like differentiation and blocks adipocyte development through stimulating the transcription of extracellular matrix and pro-fibrogenic genes (9). Mice lacking MRFTA have reduced levels of adipose fibrosis and increased thermogenic fat development, leading to improved glucose tolerance and higher energy expenditure. Upon HFD feeding, MRTFA knockout mice are further protected from obesity-induced fibrosis and show increased adipose hyperplasia compared to control animals (10). Together, these results implicate MRTFA in a fibro-adipogenic branch point and thus targeting this pathway may hold promise for combating obesity-induced adipose tissue fibrosis.
WAT also secretes many endocrine factors (termed “adipokines”) that communicate the nutritional status of adipose tissue to the central nervous system and peripheral tissues. A significant number of adipokines have been identified including leptin, adiponectin, resistin, chimerin, inflammatory cytokines such as TNFα and IL-6. The critical function of leptin is underscored by the finding that loss-of-function mutations in leptin or the leptin receptor lead to monogenic forms of obesity in mice and humans (11–13). Of note, adipocyte-derived lipid species with signaling activity (“lipokines”) such as C16:1n7-palmitoleate and fatty acid– hydroxy–fatty acids (FAHFAs) have also been identified (14, 15).
In addition to white adipose tissue, placental mammals are equipped with brown adipose tissue (BAT), which serves as a critical site of non-shivering thermogenesis (16). Brown adipocytes are specialized to burn energy, including lipid and glucose, for the production of heat. Brown fat cells have a high density of mitochondria and display high rates of fatty acid oxidation. BAT-mediated thermogenesis is activated by cold and protects mammals against hypothermia. Cold is sensed through the central nervous system, which rapidly elicits sympathetic outflow to brown and white adipose depots (17, 18). NE activates a coordinated program of glucose and lipid uptake, lipolysis and thermogenesis in brown adipocytes (16, 19, 20). Sustained cold/NE also induces the differentiation of new brown adipocytes to increase BAT mass and augment thermogenic capacity (21, 22). Further, NE-mediated activation of the cAMP/PKA signaling pathway increases the expression of genes that support thermogenesis, including the hallmark brown fat-specific protein uncoupling protein 1 (UCP1) (23, 24). Brown fat mitochondria contain a high concentration of UCP1 within the inner mitochondrial membrane. When activated, UCP1 partially dissipates the proton gradient generated by oxidative phosphorylation (25, 26). The UCP1-mediated decrease in membrane potential drives very high levels of substrate oxidation with the inherent inefficiency of all the metabolic reactions generating heat as a byproduct. Accordingly, brown fat is highly vascularized, allowing for efficient delivery of oxygen and substrate to brown fat cells and resulting heat to be distributed to the rest of the organism.
Recent work challenged the dogma that lipolysis in brown adipocytes is required for non-shivering thermogenesis. Brown adipocyte-specific deletion of ATGL or CGI-58, which control lipolysis, does not affect cold-stimulated brown fat activity (27, 28). Thus, circulating nutrients obtained from diet or generated by lipolysis in WAT can fuel brown fat thermogenesis. Of note, long chain fatty acids released from WAT are converted to acylcarnitines by liver, released into circulation and taken up by activated BAT to support thermogenesis (29). A recent study also demonstrates that BAT avidly takes up succinate; this stimulates ROS production to activate UCP1 and thermogenesis. Striking, feeding mice a succinate-supplemented diet increases energy expenditure and reverses diet-induced obesity in a UCP1-dependent manner (30).
Brown-like (“beige”) adipocytes that express UCP1 and other thermogenic components develop in WAT in response to cold and other stimuli. Despite their differences in developmental history and location, beige and brown adipocytes can achieve similar levels of uncoupling on a per cell basis and express comparable levels of UCP1 protein, when maximally stimulated (31, 32). Ribosomal profiling studies showed that cold-recruited beige adipocytes express smooth muscle-specific genes such as Smooth muscle actin (Acta2) and Myh11. Lineage tracing studies with Myh11Cre and Acta2Cre drivers indicate that beige adipocytes develop from smooth muscle-like cells (33, 34). These results suggest a developmental relationship between smooth muscle cells and beige adipocytes (Figure 1B).
UCP1 is the defining marker gene/protein for thermogenic brown and beige adipocytes. However, beige adipose tissue also employs UCP1-independent mechanisms that enable futile cycling and metabolic inefficiency. Even before the field had been reinvigorated by the discovery of adult human BAT depots, Kozak and colleagues identified UCP1-independent nonshivering mechanisms (35, 36). When wildtype and Ucp1−/− animals were adapted to long-term cold exposure, Ucp1−/− animals responded by elevating fatty acid oxidation and enhancing AMP kinase activity in inguinal WAT. Interestingly, inguinal fat from Ucp1−/− animals had increased levels of phospholamban, a key regulatory component of calcium cycling (36). More recently, SERCA2b-mediated futile calcium signaling has been further shown to mediate UCP1-independent thermogenesis in beige adipose depots (37). In this model, adrenergic signaling promotes calcium release from the sarco/endoplasmic reticulum that is taken back up by the SER Ca2+ ATPase 2b pump, thus consuming ATP and influencing whole-body energy expenditure.
Beige adipocytes can also expend energy through futile creatine cycling, enabled by coordinated upregulation of creatine metabolism genes and creatine kinase activity (38). Reducing creatine synthesis in adipocytes of mice decreases energy expenditure and sensitizes animals to high fat diet-induced obesity (39). These defects can be rescued by supplementation with dietary creatine, pointing to a key mechanism by which creatine regulates adaptive thermogenesis in vivo. The secreted enzyme PM20D1 also promotes UCP1-independent uncoupling by regulating the production of N-acyl amino acids such as N-oleoyl phenylalanine (40). These N-lipidated amino acids directly uncouple mitochondrial respiration and can increase energy expenditure in obese animals.
II. TRANSCRIPTIONAL NETWORKS AND ADIPOGENIC PROGRAMMING
The capacity for adipose tissue to acquire new white or brown adipocytes improves metabolic disease by reducing lipotoxicity. Thus, understanding the transcriptional networks that control adipocyte differentiation is crucial for future translational efforts. The coordinated recruitment of transcription factors defines cell-type specific gene expression during development. Transcription factors are broadly defined as DNA binding proteins that activate or repress RNA polymerase II (Pol II)-mediated transcription. These proteins bind to cognate DNA sequences (motifs) in core promoter or regulatory sequences, namely enhancers, silencers and insulators (Figure 2). These regulatory elements far outnumber the approximately 20,000 coding genes in the human genome and are often mutated in the setting of disease (41, 42). Over the past few decades, the field has gained a fundamental understanding of the transcriptional hierarchies that govern adipogenesis using both in vivo mouse models and in vitro systems. In particular, the terminal differentiation cascade has been studied in detail using preadipocyte cell lines that undergo differentiation into mature adipocytes upon treatment with a standard induction cocktail. Both white and brown adipogenesis can be modeled in cell culture using immortalized cell lines, such as 3T3-L1 (white) adipocytes or HIB-1B (brown) adipocytes (43, 44). Further, primary preadipocytes can be isolated from the stromal vascular fraction of white or brown adipose tissue and then differentiated in cell culture (45).
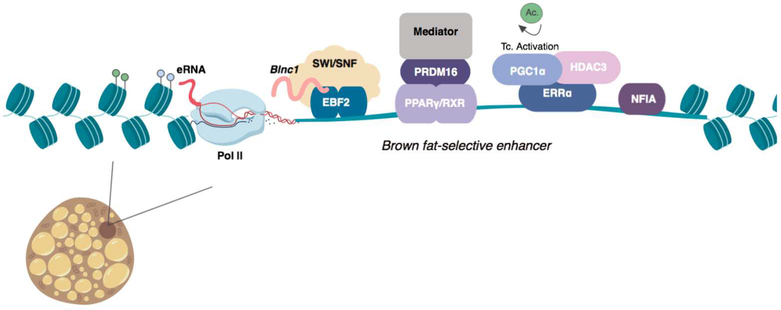
Figure 2. Chromatin state dynamics at brown fat-specific enhancers.
A model brown fat specific enhancer (ex. Ucp1, Cidea, etc.) has characteristic enhancer marks including H3K27ac (blue) and H3K4me1 (green). Enhancer RNAs are transcribed by RNA polymerase II at active enhancers. The transcription factors EBF2 interacts with the SWI/SNF chromatin remodeling complex and the lncRNA Blnc1 to activate brown fat genes. PRDM16 interacts with PPARγ/RXR heterodimers and recruits the Mediator complex to promote enhancer looping and activation. The histone deactylase HDAC3 deactylates PGC1α to coactivate the nuclear receptor ERRα and promote transcription of Pgc1α as well as other brown fat-selective genes. NFIA binds to brown fat selective enhancers to facilitate chromatin accessibility early in differentiation.
The identity and in vivo profile of adipose precursor cells is not well defined, due to a paucity of specific marker genes. Two recent studies employed single cell RNA-sequencing (scRNA-seq) to provide unbiased insight into the cellular populations that are capable of undergoing adipocyte differentiation. scRNA-seq analysis of the subcutaneous adipose tissue revealed three population clusters stratified by gene expression profiles. Interestingly, these populations showed different tendencies towards adipocyte differentiation, including a novel “Areg” population shown to inhibit adipocyte differentiation in a paracrine manner in both mice and humans (46). An independent experiment utilized the β3-adrenergic agonist CL316, 243 (CL) to stimulate de novo adipogenesis in adipose tissues. scRNA-seq analysis resolved distinct adipose precursor populations along the differentiation trajectory, and further identified tissueresident immune cells in the adipogenic niche (47). Together, these studies provide a valuable transcriptomic resource and lay the groundwork for future studies aimed at elucidating the mechanisms controlling adipocyte stem/precursor cell activity in different depots.
The nuclear receptor PPARγ is the master regulator of adipocyte differentiation and function. Ectopic expression of PPARγ drives adipose differentiation in fibroblasts and muscle cells. Conversely, PPARγ is required for adipocyte differentiation in vivo and in vitro (48, 49). At a mechanistic level, PPARγ cooperates extensively with the basic leucine-zipper factor C/EBPα to regulate adipocyte-specific genes (50). Genome-wide ChIP studies show that PPARγ binds to most, if not all, adipocyte-selective genes and that C/EBPα is bound to the majority of these sites in adipocytes (51, 52). PPARγ and C/EBPα function predominantly in the second of two defined “waves” of adipogenesis. In the first wave, expression of the pro-adipogenic factors C/EBPβ and C/EBPδ is induced. Interestingly, C/EBPβ/δ can bind to DNA/chromatin within just a few hours of adipogenic stimulus, and profiling of C/EBPβ in preadipocytes demonstrates some level of chromatin occupancy even before hormonal cocktail induction (53). C/EBPβ both facilitates the binding of and cooperates with the adipogenic transcription factor glucocorticoid receptor (GR) early in 3T3-L1 adipocyte differentiation (54). GR also interacts with phosphorylated STAT5A, which is required for efficient adipogenic differentiation. Indeed, shRNA-mediated depletion of C/EBPβ, GR, or STAT5A reduces the recruitment of other earlyacting adipogenic factors, implying that these early transcription factors facilitate cooperative binding. ChIP-seq studies revealed that C/EBPβ/δ, GR, RXR and STAT5A bind transiently at transcriptional “hotspots” (4h post-induction) during differentiation and cooperate to regulate cell cycle and cell growth genes (55).
The transcriptional cascade set in motion in the first wave of adipogenesis activates the second wave, which centers on PPARγ and C/EBPα. Indeed, many of the early transcriptional hotspots occupied by C/EBPβ/δ, GR, RXR, and STAT5A become bound by PPARγ during the second wave (“inherited” hotspots) (56). PPARγ and C/EBPα sustain each other’s expression through a positive feedback loop. These factors also cooperate to establish late transcriptional hotpots at genes linked to the adipocyte phenotype, including lipid and glucose metabolism genes.
III. TRANSCRIPTION FACTORS IN BROWN ADIPOGENESIS
In addition to activating the general adipocyte gene program, brown adipocytes express a broad set of so-called “thermogenic genes” that support uncoupled respiration and high levels of fatty acid/glucose oxidation. Transcriptomic studies have identified a core brown vs. white fatselective gene signature. These genes include: Ucp1, Pparα (fatty acid metabolism), Cidea (lipid droplet remodeling), Dio2 (thyroid hormone metabolism), Elovl3 (fatty acid elongase), Pgc1α (mitochondrial biogenesis), Cpt1b (fatty acid transport), and genes encoding respiratory chain components (both nuclear and mitochondrial-encoded).
Differentiation of brown fat cells, as well as the acute response to stimulation are regulated via different pathways. The transcriptional factor PR Domain Zinc Finger 16 (PRDM16) is selectively expressed in brown/beige vs. white fat cells and plays an important role in controlling the differentiation-linked brown fat gene program (57). Ectopic PRDM16 expression is able to activate the expression of thermogenic genes in a number of cell systems, including white preadipocytes, muscle precursors, and NIH-3T3 fibroblasts. Mechanistically, PRDM16 binds to brown fat gene enhancers and cooperates with the Mediator complex to help establish enhancer-promoter contacts at genes such as Pparα and Pgc1α (58, 59). PRDM16 also represses white fat and muscle gene expression though interactions with co-repressor complexes (60, 61). Loss of PRDM16 in cultured brown adipocytes abrogates brown fat gene expression and increases the expression of muscle genes under certain conditions (62). PRDM16 protein itself is post-translationally modified by the Polycomb group protein CBX4, which sumoylates PRDM16 at lysine 917, thus blocking ubiquitination-mediated degradation of PRDM16 (63).
Genetic experiments in mice show that PRDM16 is required for the maintenance of brown adipocyte identity, though the related factor PRDM3 can compensate for loss of PRDM16 in young animals (58). PRDM16 also plays a critical role in regulating the browning of WAT. Transgenic mice overexpressing PRDM16 in adipose tissue develop beige adipocytes in WAT, display increased energy expenditure, and resist high-fat diet induced obesity (64). PRDM16 expression in vascular smooth muscle cells induces their conversion to beige adipocytes (33). Conversely, Prdm16-deficiency in adipocytes inhibits beige adipocyte development (65). These animals develop obesity accompanied by insulin resistance and hepatic steatosis. PRDM16 also represses type 1 interferon response genes in brown and beige adipocytes through its DNA-binding domain. Ectopic interferon signaling causes mitochondrial dysfunction in adipocytes and decreases Ucp1 expression (66). Thus, PRDM16 regulates brown and beige fat fate through multiple mechanisms.
Members of “Early B-Cell Factor” (EBF) protein family have also emerged as key regulators of general adipogenesis and thermogenic programing. EBF1 is expressed in 3T3-L1 adipocytes early in differentiation, and ectopic EBF1 expression is sufficient to stimulate adipocyte differentiation in embryonic fibroblasts. Conversely, fusing EBF1 to the Engrailed repressor domain blocks adipocyte differentiation (67), and whole body Ebf1−/− mice are lipodystrophic (68). Ebf2 and Ebf3 are also expressed at lower levels in differentiated 3T3-L1 adipocytes, and all EBF isoforms (1,2,3) are able to induce adipogenesis in NIH-3T3 fibroblasts (69). shRNA-mediated depletion of Ebf1 or Ebf2 (but not Ebf3) blocks adipogenesis (69).
Genome-wide analysis of the PPARγ cistrome in white and brown adipose tissue identified high enrichment of the EBF motif at brown adipocyte-selective PPARγ binding sites (70). Among the EBF isoforms, Ebf2 was the most highly enriched in brown relative to white adipose tissue. Exogenous EBF2 expression in myoblasts or white preadipocytes induces brown fat differentiation program and uncoupled respiration. Mechanistically, EBF2 forms a ribonucleoprotein complex with the brown fat-selective lncRNA Blnc1 and cooperates with PPARγ at brown fat enhancers to activate transcription (70, 71). Ebf2-deficiency ablates the brown fat-specific characteristics of BAT or isolated brown adipocytes (70). In a separate line of investigation, RNA-seq analysis identified Ebf2 as a marker of committed brown adipocyte precursors during mouse development. Ebf2 expression is activated in the dermomyotome of somites starting from E11, and only Ebf2+ cells isolated from embryos displayed brown adipogenic potential in vitro (72).
EBF2 is also required and sufficient for the beiging of WAT. Overexpression of EBF2 in primary white adipocytes or WAT induces the expression of thermogenic genes, increases oxygen consumption and suppresses high-fat diet induced weight gain (73). EBF2 does not induce the expression of identified “beige adipose specific genes” such as Cited1 or Tmem26. Thus, EBF2 may reprogram white adipocytes into bona fide brown adipocytes. The transcriptional activity of EBF2 (and possibly other EBFs) is normally inhibited in white adipocytes by the zinc finger protein ZFP423. In mature adipocytes, ZFP423 binds and represses EBF2 activity to maintain white fat cell identity. Deletion of ZFP423 in adipocytes leads to unrestrained EBF2-activity and induction of thermogenic genes (74).
Genome-wide chromatin immunoprecipitation (ChIP) and RNA-seq analyses show that EBF2 binds directly to many brown fat-specific enhancer regions. The transcription factor NFIA also binds to brown fat-selective active enhancer marks along with PPARγ, C/EBPβ, and EBF2. Notably, NFIA is uniquely present at a number of brown fat enhancers prior to induction of differentiation and before PPARγ recruitment to these sites. This suggests that NFIA may play an early role in establishing a permissive chromatin landscape at lineage-specific cis regulatory elements (75, 76).
PPARγ co-activator 1A (PGC1α) was first discovered as an interacting partner of PPARγ in brown adipocytes (77). Pgc1α expression is highly induced by cold exposure and is further activated following phosphorylation by the cAMP-PKA-p38/MAPK signaling pathway. Upon interaction with its binding partners, PGC1α recruits histone acetyltransferases such as CBP/p300 and GCN5 to augment transcription (78). PGC1α binds to Nuclear Respiratory Factors 1 and 2 to promote the activation of many mitochondrial genes. PGC1α also co-activates a number of nuclear hormone receptors, including PPARγ, PPARα, and ERRα/β/γ, all of which participate in the transcription of brown fat genes (79). Overexpression of PGC1α in adipocytes, myotubes, or cardiomyocytes promotes mitochondrial biogenesis and increases oxygen consumption (80, 81). Pgc1α-deficient BAT displays mildly increased lipid droplet accumulation but expresses normal levels of Ucp1 and other brown fat-selective genes (82). Pgc1α-deficient brown fat cells in culture fail to efficiently activate the thermogenic machinery in response to adrenergic stimulation (83). These results demonstrate that PGC1α is required for the acute transcriptional activation of thermogenesis but not BAT development per se. Interestingly, deletion of Pgc1α in adipocytes severely impairs the development of beige adipocytes in WAT (84).
Peroxisome proliferator activated receptor alpha (PPARα) is a nuclear hormone receptor that regulates lipid metabolism, including mitochondrial and peroxisomal β-oxidation, fatty acid uptake, and lipoprotein transport. Pparα itself is a transcriptional target of EBF2, PRDM16, PPARγ, and C/EBPβ. It can also bind and directly activate Ucp1 expression (85). PPARα further cooperates with PRDM16 to activate Pgc1α expression, thus linking coordinated induction of fatty acid oxidation and mitochondrial metabolism (86). Surprisingly, mice lacking Pparα in brown adipocytes have normal Ucp1 expression and are able to defend their body temperature against cold stress (87). However, fasted Pparα−/− mice become hypothermic, likely because dampened hepatic fatty acid oxidation reduces the amount of fuel available for thermogenesis (88).
To date, many other transcriptional regulators, nuclear receptors, and long non-coding RNAs have been implicated in activating or repressing brown fat-specific gene expression. ZFP516, KLF11, IRF4, TAF7L, ZBTB16, EWS, PLAC8, the lncRNA Blnc1, ERRα, ΕRRγ and others have all been shown in different contexts to promote activation of brown fat genetic programming (89–91). Conversely, FOXO1, TWIST1, p107, LXRα, pRB, RIP140, TLE3, REVERBα and ZFP423 repress brown fat gene expression either directly or by inhibiting activators such as PRDM16, EBF2, or PGC1α (89–91).
The growing number of whole-genome transcriptional and epigenomic studies continues to strengthen our understanding of how brown and beige adipogenesis is transcriptionally regulated (75, 76, 92, 93). All of the transcription factors discussed above cooperate with epigenetic factors such as histone modification readers, writers, and erasers as well as chromatin remodeling enzymes (89) to establish a permissive chromatin environment for brown fat gene activation. Identification of brown fat gene enhancers has been enabled by a number of genomewide interrogations of chromatin state. For example, lineage-specific enhancers are marked by high levels of activating histone marks such as H3K27ac and H3K4me1/2 in brown, but not white adipose tissue (93). Functional brown fat enhancers have been further defined by nascent enhancer RNA transcription (92).
Most available studies on genome regulation have focused on brown fat, with less known about chromatin regulation during beige fat induction. A recent study addressed this question using the NuTRAP mouse, which allows for isolation of nuclei from specific cell types within a heterogeneous tissue (94). Using this system, brown and beige adipocyte nuclei were purified and analyzed for changes in chromatin state in response to temperature shifts (95). Interestingly, the chromatin state in classic brown adipocytes remains fairly stable during temperature changes, while beige adipocytes were found to show extreme plasticity in their chromatin state. During cold exposure, beige adipocytes adopt a chromatin signature that was almost indistinguishable from that of classical brown adipocytes. After re-warming to 30°C, the H3K27ac pattern of “previously beige adipocytes” became identical to that of warm white adipocytes. However, “rewarmed” beige adipocytes displayed a primed chromatin state characterized by H3K4me1+/H3K27ac- histones at certain brown fat genes, including Ucp1 and Cpt1b. These poised enhancers presumably enable rapid re-engagement of the thermogenic program upon β-adrenergic stimulation. These studies thus point to a novel mechanism of epigenomic memory utilized by beige adipocytes for rapid adaptation to cold exposure.
Several histone methyltransferases and demethylases have been implicated in regulating the chromatin state at brown fat-selective genes. PRDM16 cooperates with the H3K9 methyltransferase EHMT1 to repress muscle and white adipogenic gene expression. Genetic deletion of Ehmt1 in brown fat precursors causes ectopic activation of skeletal muscle genes and impaired brown adipocyte differentiation (61). EHMT2 (G9a) has a broader role in regulating general adipogenesis by catalyzing H3K9me2 deposition at the PPARγ promoter in pre-adipocytes. EHMT2 and H3K9me2 levels decrease during adipogenesis thereby facilitating the differentiation process (96). The H3K4 methyltransferase MLL4 is essential for brown fat and muscle development (97). In addition, the H3K27 demethylases UTX and JMJD3 promote brown adipogenesis through direct binding and demethylation at brown fat gene promoters (98, 99), although the mechanism by which these proteins specifically target brown fat genes is unknown. LSD1, an H3K4/K9 demethylase, cooperates with NRF1 to promote expression of genes involved in oxidative phosphorylation and transgenic LSD1 expression induces beiging of inguinal white adipose tissue (100, 101). LSD1 also interacts with PRDM16 at white fat-specific cis-regulatory elements to remove H3K4me1/2 and repress transcription (102).
Histone acetylation is correlated with gene activation, as negatively charged acetyl groups neutralize the interactions between the DNA phosphate backbone and basic histone lysine residues. A number of histone deacetylase enzymes (HDACs) have been implicated in control of brown adipose physiology. Pan-inhibition of class I HDACs (HDACs 1,2,3, and 8) promotes mitochondrial respiration and biogenesis in vitro through augmented PGC1α expression. Further, administration of HDAC inhibitors in vivo promotes beige adipogenesis and augments energy expenditure in the db/db mouse model of obesity (103). In support of this result, HDAC1 silencing enhances brown fat-specific gene activation through coordinate removal of repressive H3K27me3 and increased H3K27ac deposition at promoters (104).
Histone acetyltransferases and deacetylases can also modify non-histone substrates (105). In this regard, the histone deacetylase HDAC3, typically considered as a transcriptional corepressor, was shown to co-activate ERRα in BAT via deacetylation of PGC1α (92). Mice lacking HDAC3 in brown fat do not express UCP1, fail to activate non-shivering thermogenesis in cold, and show markedly diminished mitochondrial respiration (92). Thus, HDACs likely have context-dependent roles in priming and activation of thermogenic gene expression.
Histone modifying enzymes are often found in large complexes with chromatin remodelers (106). Chromatin remodeling complexes use energy derived from ATP hydrolysis to alter nucleosome-DNA contacts in a non-covalent manner. The four classes of chromatin remodelers (CHD, INO80, ISWI, and SWI/SNF) are classified based on their varied ATPaseflanking domains. In particular, SWI/SNF family remodelers, which are characterized by Cterminal bromodomains that interpret histone acetylation modifications, play important roles in both general and brown fat gene activation. The mammalian SWI/SNF (mSWI/SNF or BAF) complex is built around two related ATPases, either Brahma homolog (BRM) or BRM-related gene 1 (BRG1), which interact with distinct classes of transcription factors and have unique functional outputs (107). BRG1/BRM cooperate with non-catalytic subunits to alter nucleosome positioning or expose occluded cis regulatory elements (108). Early studies focusing on the role of the BAF complex in adipogenesis found that BRG1 facilitates stable preinitiation complex assembly at the Pparγ2 promoter during white adipogenesis (53). Recent studies indicate that BRG1 is required for the induction of thermogenic genes, but dispensable for general adipocyte differentiation in brown adipocytes (109). Upon β-adrenergic stimulation, BRG1/BAF forms a complex with phosphorylated JMJD1A and PPARγ. This complex promotes enhancer-promoter chromatin looping at the Ucp1 locus to increase transcription (109). EBF2 also physically interacts with BRG1 and the BAF chromatin remodeling complex. Interestingly, the histone reader Double PHD Fingers 3 (DPF3) is specifically incorporated into the BAF complex of brown fat cells. DPF3 is required downstream of EBF2 for the activation of thermogenic genes and mitochondrial function in brown adipocytes (110). Importantly, many epigenetic modules such as bromodomains or enzyme active sites are druggable and thus could be targeted for therapeutic purposes. Thus, understanding how transcription factors interact with epigenetic factors may provide new avenues to pharmacologically regulate brown fat transcriptional programming.
IV. CONCLUSION AND OUTLOOK FOR HUMAN HEALTH
A major goal in the adipose field is to establish methods for encouraging the differentiation of white and/or brown adipocytes to enhance insulin sensitivity and reduce diabetes in people. Thus, identifying the transcriptional and epigenetic factors that activate or repress adipose differentiation and function will provide new therapeutic targets. However, much of our current knowledge is based on rodent studies. It will thus be important to assess the relevance of these identified pathways in human adipocytes.
While human brown/beige adipose tissue is currently of limited availability, a number of labs have demonstrated that human mesenchymal stem cells or adipose derived stromal vascular cells can be expanded ex vivo and are amenable to genetic and pharmacologic manipulations. For example, treatment of human mesenchymal adipose-derived stem cells with the PPARγ ligand roziglitzone induces the thermogenic program (111, 112). A key rosiglitazone-induced factor is KLF11, which is required downstream of PPARγ for activation of the thermogenic program in human adipocytes (113). Notably, human adipocyte progenitors can also be isolated and expanded ex vivo in 3D Matrigel-based cultures (114). Following stimulation with proangiogenic and pro-adipogenic cocktail, this vascularized human adipose tissue can be implanted into mice, leading to metabolic benefits.
Embryonic stem (ES) or induced pluripotent (iPS) cells hold great promise for human health as they can be induced to differentiate into a variety of differentiated cell types. Human ES derived mesenchymal progenitors can be differentiated into adipocytes via transduction with PPARγ virus and/or treatment with a PPARγ agonist. Addition of C/EBPβ and PRDM16 vectors, along with exogenous PPARγ2, further programs these mesenchymal progenitors into metabolically active UCP1+ brown adipocytes (115). These cells can engraft and uptake glucose when transplanted into immunocompromised mice, demonstrating that human stem cells can be programmed into functional brown adipocytes. A recent protocol established beige adipocytes from human iPS cells without gene transfer. Importantly, these cells can form differentiated and vascularized adipose tissue when injected subcutaneously (116).
Understanding the transcriptional hierarchies that regulate brown adipogenesis will advance these reprogramming-based strategies to augment thermogenic adipose tissue. For example, little is known about how Ebf2 is transcriptionally activated, with only one study in the literature demonstrating that the lateral plate mesoderm-derived BMP4 signaling induces Ebf2 and Ebf3 mRNA expression in chicken somites (117). Identifying the upstream signals that activate Ebf2 and/or other early browning factors in human cells could be used to induce the reprogramming of white adipocytes or ESCs into brown adipocytes. Future studies should prioritize understanding the upstream signals and transcriptional regulators that activate master lineage-determining factors during the commitment of brown precursors. Looking to the future, the field will continue to broaden and deepen our understanding how signaling pathways, environmental factors, and/or metabolites regulate the activity of master transcriptional regulators, potentially leading to clinically useful methods for increasing brown and beige fat thermogenesis.
Acknowledgements
Due to space limitations, we respectfully note that we were unable to comprehensively cite many worthy contributions to the field. We would like to thank members of the Seale lab for helpful comments and discussion. This work was supported by an NIH/NRSA grant to S.N.S (1F31DK108507) and NIDDK grant 5R01DK10300802 to P.S.
Funding: NIH grants 1F31DK108507 and 5T32GM008216 to S.N.S, and 5R01DK103008 to P.S.
Footnotes
Disclosure: The authors declare no conflict of interest.
References
- 1.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan JJ, Greenberg AS, Chang MK, Wek SA, Moos MC Jr., Londos C Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc Natl Acad Sci U S A. 1992;89(18):8537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young SG, Zechner R. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 2013;27(5):459–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadian M, Wang Y, Sul HS. Lipolysis in adipocytes. Int J Biochem Cell Biol. 2010;42(5):555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolditz CI, Langin D. Adipose tissue lipolysis. Curr Opin Clin Nutr Metab Care. 2010;13(4):377–81. [DOI] [PubMed] [Google Scholar]
- 6.Jeffery E, Church CD, Holtrup B, Colman L, Rodeheffer MS. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol. 2015;17(4):376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19(10):1338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcelin G, Ferreira A, Liu Y, Atlan M, Aron-Wisnewsky J, Pelloux V, et al. A PDGFRalpha-Mediated Switch toward CD9high Adipocyte Progenitors Controls Obesity-Induced Adipose Tissue Fibrosis. Cell Metab. 2017;25(3):673–85. [DOI] [PubMed] [Google Scholar]
- 9.McDonald ME, Li C, Bian H, Smith BD, Layne MD, Farmer SR. Myocardin-related transcription factor a regulates conversion of progenitors to beige adipocytes. Cell. 2015;160(12):105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JZ, Rabhi N, Farmer SR. Myocardin-Related Transcription Factor A Promotes Recruitment of ITGA5+ Profibrotic Progenitors during Obesity-Induced Adipose Tissue Fibrosis. Cell reports. 2018;23(7):1977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387(6636):903–8. [DOI] [PubMed] [Google Scholar]
- 12.Ozata M, Ozdemir IC, Licinio J. Human leptin deficiency caused by a missense mutation: multiple endocrine defects, decreased sympathetic tone, and immune system dysfunction indicate new targets for leptin action, greater central than peripheral resistance to the effects of leptin, and spontaneous correction of leptin-mediated defects. J Clin Endocrinol Metab. 1999;84(10):3686–95. [DOI] [PubMed] [Google Scholar]
- 13.Clement K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392(6674):398–401. [DOI] [PubMed] [Google Scholar]
- 14.Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134(6):933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159(2):318–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological reviews. 2004;84(1):277–359. [DOI] [PubMed] [Google Scholar]
- 17.Morrison SF, Madden CJ, Tupone D. Central control of brown adipose tissue thermogenesis. Frontiers in endocrinology. 2012;3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartness TJ, Vaughan CH, Song CK. Sympathetic and sensory innervation of brown adipose tissue. Int J Obes (Lond). 2010;34 Suppl 1:S36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RE, Horwitz BA. Brown fat and thermogenesis. Physiological reviews. 1969;49(2):330–425. [DOI] [PubMed] [Google Scholar]
- 20.Himms-Hagen J Cellular thermogenesis. Annual review of physiology. 1976;38:315–51. [DOI] [PubMed] [Google Scholar]
- 21.Lee YH, Petkova AP, Konkar AA, Granneman JG. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2015;29(1):286–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bukowiecki LJ, Geloen A, Collet AJ. Proliferation and differentiation of brown adipocytes from interstitial cells during cold acclimation. Am J Physiol. 1986;250(6 Pt 1):C880–7. [DOI] [PubMed] [Google Scholar]
- 23.Jacobsson A, Stadler U, Glotzer MA, Kozak LP. Mitochondrial uncoupling protein from mouse brown fat. Molecular cloning, genetic mapping, and mRNA expression. J Biol Chem. 1985;260(30):16250–4. [PubMed] [Google Scholar]
- 24.Ricquier D, Mory G, Bouillaud F, Thibault J, Weissenbach J. Rapid increase of mitochondrial uncoupling protein and its mRNA in stimulated brown adipose tissue. Use of a cDNA probe. FEBS Lett. 1984;178(2):240–4. [DOI] [PubMed] [Google Scholar]
- 25.Ricquier D Uncoupling protein 1 of brown adipocytes, the only uncoupler: a historical perspective. Frontiers in endocrinology. 2011;2:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arechaga I, Ledesma A, Rial E. The mitochondrial uncoupling protein UCP1: a gated pore. IUBMB Life. 2001;52(3–5):165–73. [DOI] [PubMed] [Google Scholar]
- 27.Schreiber R, Diwoky C, Schoiswohl G, Feiler U, Wongsiriroj N, Abdellatif M, et al. ColdInduced Thermogenesis Depends on ATGL-Mediated Lipolysis in Cardiac Muscle, but Not Brown Adipose Tissue. Cell Metab. 2017;26(5):753–63 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin H, Ma Y, Chanturiya T, Cao Q, Wang Y, Kadegowda AKG, et al. Lipolysis in Brown Adipocytes Is Not Essential for Cold-Induced Thermogenesis in Mice. Cell Metab. 2017;26(5):764–77 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simcox J, Geoghegan G, Maschek JA, Bensard CL, Pasquali M, Miao R, et al. Global Analysis of Plasma Lipids Identifies Liver-Derived Acylcarnitines as a Fuel Source for Brown Fat Thermogenesis. Cell Metab. 2017;26(3):509–22 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills EL, Pierce KA, Jedrychowski MP, Garrity R, Winther S, Vidoni S, et al. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature. 2018;560(7716):102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shabalina IG, Petrovic N, de Jong JM, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell reports. 2013;5(5):1196–203. [DOI] [PubMed] [Google Scholar]
- 33.Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014;19(5):810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry DC, Jiang Y, Graff JM. Mouse strains to study cold-inducible beige progenitors and beige adipocyte formation and function. Nat Commun. 2016;7:10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anunciado-Koza R, Ukropec J, Koza RA, Kozak LP. Inactivation of UCP1 and the glycerol phosphate cycle synergistically increases energy expenditure to resist diet-induced obesity. J Biol Chem. 2008;283(41):27688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ukropec J, Anunciado RP, Ravussin Y, Hulver MW, Kozak LP. UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1−/− mice. J Biol Chem. 2006;281(42):31894–908. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, et al. UCP1independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017;23(12):1454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. 2015;163(3):643–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kazak L, Chouchani ET, Lu GZ, Jedrychowski MP, Bare CJ, Mina AI, et al. Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab. 2017;26(4):660–71 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long JZ, Svensson KJ, Bateman LA, Lin H, Kamenecka T, Lokurkar IA, et al. The Secreted Enzyme PM20D1 Regulates Lipidated Amino Acid Uncouplers of Mitochondria. Cell. 2016;166(2):424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23(22):5866–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissuespecific gene expression. Nat Rev Genet. 2011;12(4):283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3(2):127–33. [DOI] [PubMed] [Google Scholar]
- 44.Klaus S, Choy L, Champigny O, Cassard-Doulcier AM, Ross S, Spiegelman B, et al. Characterization of the novel brown adipocyte cell line HIB 1B. Adrenergic pathways involved in regulation of uncoupling protein gene expression. Journal of cell science. 1994;107 ( Pt 1):3139. [DOI] [PubMed] [Google Scholar]
- 45.Klaus S, Ely M, Encke D, Heldmaier G. Functional assessment of white and brown adipocyte development and energy metabolism in cell culture. Dissociation of terminal differentiation and thermogenesis in brown adipocytes. Journal of cell science. 1995;108 ( Pt 10):3171–80. [DOI] [PubMed] [Google Scholar]
- 46.Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559(7712):103–8. [DOI] [PubMed] [Google Scholar]
- 47.Burl RB, Ramseyer VD, Rondini EA, Pique-Regi R, Lee YH, Granneman JG. Deconstructing Adipogenesis Induced by beta3-Adrenergic Receptor Activation with Single-Cell Expression Profiling. Cell Metab. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Molecular cell. 1999;4(4):585–95. [DOI] [PubMed] [Google Scholar]
- 49.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Molecular cell. 1999;4(4):611–7. [DOI] [PubMed] [Google Scholar]
- 50.Madsen MS, Siersbaek R, Boergesen M, Nielsen R, Mandrup S. Peroxisome proliferator-activated receptor gamma and C/EBPalpha synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol. 2014;34(6):939–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21):2941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22(21):2953–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol Cell Biol. 2004;24(11):4651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24(10):1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, et al. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. Embo J. 2011;30(8):1459–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siersbaek R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab. 2012;23(2):56–64. [DOI] [PubMed] [Google Scholar]
- 57.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 2014;19(4):593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. 2015;29(3):308–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex Genes Dev. 2008;22(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504(7478):163–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Q, Huang L, Pan D, Zhu LJ, Wang YX. Cbx4 Sumoylates Prdm16 to Regulate Adipose Tissue Thermogenesis. Cell reports. 2018;22(11):2860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell. 2014;156(1–2):304–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kissig M, Ishibashi J, Harms MJ, Lim HW, Stine RR, Won KJ, et al. PRDM16 represses the type I interferon response in adipocytes to promote mitochondrial and thermogenic programing. Embo J. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol. 2002;22(22):8015–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fretz JA, Nelson T, Xi Y, Adams DJ, Rosen CJ, Horowitz MC. Altered metabolism and lipodystrophy in the early B-cell factor 1-deficient mouse. Endocrinology. 2010;151(4):1611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27(2):743–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17(4):562–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular cell. 2014;55(3):372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A. 2014;111(40):14466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stine RR, Shapira SN, Lim HW, Ishibashi J, Harms M, Won KJ, et al. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab. 2016;5(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell metabolism. 2016;23(6):1167–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hiraike Y, Waki H, Yu J, Nakamura M, Miyake K, Nagano G, et al. NFIA co-localizes with PPARgamma and transcriptionally controls the brown fat gene program. Nat Cell Biol. 2017;19(9):1081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pradhan RN, Zachara M, Deplancke B. A systems perspective on brown adipogenesis and metabolic activation. Obes Rev. 2017;18 Suppl 1:65–81. [DOI] [PubMed] [Google Scholar]
- 77.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–39. [DOI] [PubMed] [Google Scholar]
- 78.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286(5443):136–871. [DOI] [PubMed] [Google Scholar]
- 79.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–70. [DOI] [PubMed] [Google Scholar]
- 80.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–24. [DOI] [PubMed] [Google Scholar]
- 81.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106(7):847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–35. [DOI] [PubMed] [Google Scholar]
- 83.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3(5):333–41. [DOI] [PubMed] [Google Scholar]
- 84.Kleiner S, Mepani RJ, Laznik D, Ye L, Jurczak MJ, Jornayvaz FR, et al. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proc Natl Acad Sci U S A. 2012;109(24):9635–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barbera MJ, Schluter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J Biol Chem. 2001;276(2):1486–93. [DOI] [PubMed] [Google Scholar]
- 86.Hondares E, Rosell M, Diaz-Delfin J, Olmos Y, Monsalve M, Iglesias R, et al. Peroxisome proliferator-activated receptor alpha (PPARalpha) induces PPARgamma coactivator 1alpha (PGC-1alpha) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J Biol Chem. 2011;286(50):43112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lasar D, Rosenwald M, Kiehlmann E, Balaz M, Tall B, Opitz L, et al. Peroxisome Proliferator Activated Receptor Gamma Controls Mature Brown Adipocyte Inducibility through Glycerol Kinase. Cell reports. 2018;22(3):760–73. [DOI] [PubMed] [Google Scholar]
- 88.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103(11):1489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nature reviews Molecular cell biology. 2016;17(8):480–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang W, Seale P. Control of brown and beige fat development. Nat Rev Mol Cell Biol. 2016;17(11):691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loft A, Forss I, Mandrup S. Genome-Wide Insights into the Development and Function of Thermogenic Adipocytes. Trends Endocrinol Metab. 2017;28(2):104–20. [DOI] [PubMed] [Google Scholar]
- 92.Emmett MJ, Lim HW, Jager J, Richter HJ, Adlanmerini M, Peed LC, et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546(7659):544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic acids research. 2017;45(11):6388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roh HC, Tsai LT, Lyubetskaya A, Tenen D, Kumari M, Rosen ED. Simultaneous Transcriptional and Epigenomic Profiling from Specific Cell Types within Heterogeneous Tissues In Vivo. Cell reports. 2017;18(4):1048–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roh HC, Tsai LTY, Shao M, Tenen D, Shen Y, Kumari M, et al. Warming Induces Significant Reprogramming of Beige, but Not Brown, Adipocyte Cellular Identity. Cell Metab. 2018;27(5):1121–37 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, et al. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. Embo J. 2013;32(1):45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zha L, Li F, Wu R, Artinian L, Rehder V, Yu L, et al. The Histone Demethylase UTX Promotes Brown Adipocyte Thermogenic Program Via Coordinated Regulation of H3K27 Demethylation and Acetylation. J Biol Chem. 2015;290(41):25151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pan D, Huang L, Zhu LJ, Zou T, Ou J, Zhou W, et al. Jmjd3-Mediated H3K27me3 Dynamics Orchestrate Brown Fat Development and Regulate White Fat Plasticity. Developmental cell. 2015;35(5):568–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duteil D, Tosic M, Willmann D, Georgiadi A, Kanouni T, Schule R. Lsd1 prevents ageprogramed loss of beige adipocytes. Proc Natl Acad Sci U S A. 2017;114(20):5265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duteil D, Metzger E, Willmann D, Karagianni P, Friedrichs N, Greschik H, et al. LSD1 promotes oxidative metabolism of white adipose tissue. Nat Commun. 2014;5:4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zeng X, Jedrychowski MP, Chen Y, Serag S, Lavery GG, Gygi SP, et al. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. 2016;30(16):1822–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Galmozzi A, Mitro N, Ferrari A, Gers E, Gilardi F, Godio C, et al. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes. 2013;62(3):732–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li F, Wu R, Cui X, Zha L, Yu L, Shi H, et al. Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation. J Biol Chem. 2016;291(9):4523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. [DOI] [PubMed] [Google Scholar]
- 106.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nature reviews Molecular cell biology. 2017;18(7):407–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Molecular cell. 2003;11(2):377–89. [DOI] [PubMed] [Google Scholar]
- 108.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108(4):475–87. [DOI] [PubMed] [Google Scholar]
- 109.Abe Y, Rozqie R, Matsumura Y, Kawamura T, Nakaki R, Tsurutani Y, et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat Commun. 2015;6:7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shapira SN, Lim HW, Rajakumari S, Sakers AP, Ishibashi J, Harms MJ, et al. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. 2017;31(7):660–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bartesaghi S, Hallen S, Huang L, Svensson PA, Momo RA, Wallin S, et al. Thermogenic Activity of UCP1 in Human White Fat-Derived Beige Adipocytes. Mol Endocrinol. 2015;29(1):130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Elabd C, Chiellini C, Carmona M, Galitzky J, Cochet O, Petersen R, et al. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem cells. 2009;27(11):2753–60. [DOI] [PubMed] [Google Scholar]
- 113.Loft A, Forss I, Siersbaek MS, Schmidt SF, Larsen AS, Madsen JG, et al. Browning of human adipocytes requires KLF11 and reprogramming of PPARgamma superenhancers. Genes Dev. 2015;29(1):7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Min SY, Kady J, Nam M, Rojas-Rodriguez R, Berkenwald A, Kim JH, et al. Human ‘brite/beige’ adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice. Nat Med. 2016;22(3):312–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol. 2012;14(2):209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guenantin AC, Briand N, Capel E, Dumont F, Morichon R, Provost C, et al. Functional Human Beige Adipocytes From Induced Pluripotent Stem Cells. Diabetes. 2017;66(6):1470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.El-Magd MA, Allen S, McGonnell I, Otto A, Patel K. Bmp4 regulates chick Ebf2 and Ebf3 gene expression in somite development. Dev Growth Differ. 2013;55(8):710–22. [DOI] [PubMed] [Google Scholar]