Abstract
This review summarizes the proceedings of a symposium presented at the “Alcoholism and Stress: A Framework for Future Treatment Strategies” conference held in Volterra, Italy on May 9–12, 2017. Psychiatric diseases, including alcohol use disorders (AUD), are influenced through complex interactions of genes, neurobiological pathways, and environmental influences. A better understanding of the common neurobiological mechanisms underlying an AUD necessitates an integrative approach, involving a systematic assessment of diverse species and phenotype measures. As part of the World Congress on Stress and Alcoholism, this symposium provided a detailed account of current strategies to identify mechanisms underlying the development and progression of AUD. Dr. Sean Farris discussed the integration and organization of transcriptome and postmortem human brain data to identify brain regional- and cell type-specific differences related to excessive alcohol consumption that are conserved across species. Dr. Brien Riley presented the results of a genomewide association study of DSM-IV alcohol dependence; although replication of genetic associations with alcohol phenotypes in humans remains challenging, model organism studies show that COL6A3, KLF12, and RYR3 affect behavioral responses to ethanol, and provide substantial evidence for their role in human alcohol-related traits. Dr. Rob Williams expanded upon the systematic characterization of extensive genetic-genomic resources for quantifying and clarifying phenotypes across species that are relevant to precision medicine in human disease. The symposium concluded with Dr. Robert Hitzemann’s description of transcriptome studies in a mouse model selectively bred for high alcohol (“binge-like”) consumption and a non-human primate model of long-term alcohol consumption. Together, the different components of this session provided an overview of systems-based approaches that are pioneering the experimental prioritization and validation of novel genes and gene networks linked with a range of behavioral phenotypes associated with stress and AUD.
Keywords: alcohol use disorder, species conservation, co-expression networks, systems biology, RNA-seq, genome wide association study, recombinant inbred mice, transcriptome
Introduction
Chronic and uncontrolled consumption of alcohol is a defining trait underlying the development of an alcohol use disorder (AUD). The degree of alcohol consumption can differ substantially among individuals because of variation in genetic and environmental factors. The interaction of chronic alcohol abuse with these factors shape the molecular dynamics of complex systems. Understanding the overall structure and preservation of these biological networks is critical for determining the etiology of AUD, as well as potential points of medical intervention.
The human brain is composed of diverse cell types, consisting of approximately eighty billion neurons and a similar number of glial cells; however, the cellular composition of the human brain is shared across many species (Herculano-Houzel, 2014). Compared to other species, the human frontal cortex accounts for a larger proportion of the overall brain volume. Despite the evolutionary expansion of the human frontal lobe, many brain structures and the circuits connecting them are deeply conserved across species (Janak & Tye, 2015). Studying the similarities and dissimilarities among humans and model organisms provides a rational framework for inferring fundamental molecular systems that affect maladaptive behaviors.
Strong evidence from family and twin studies demonstrates that alcohol dependence and AUD are phenotypically complex with heritability estimated at approximately 50–60% of total phenotypic variability (Reilly, Noronha, Goldman & Koob, 2017). This degree of heritability is consistent with the polygenic nature of AUD, which includes numerous loci with small effect size. Such heterogeneity has been a significant challenge in genetic mapping and identification of specific genes that influence AUD. Genome-wide association studies (GWAS) that are designed to identify single genes in polygenic traits have been hampered by high false-positive discovery rates (Ioannidis, Ntzani, Trikalinos & Contopoulos-Ioannidis, 2001; Ioannidis, Trikalinos & Khoury, 2006; Ioannidis, Trikalinos, Ntzani & Contopoulos-Ioannidis, 2003; Lohmueller, Pearce, Pike, Lander & Hirschhorn, 2003), which are due in part by small sample sizes, resulting in statistically weak genetic associations. Current GWAS methods utilizing very large sample sizes and rigorous statistical analyses have overcome many of these limitations.
There is a rapidly growing number of publically available datasets that include genetic and transcriptional features relevant to alcohol-related risk, consumption, and withdrawal. These data have been collected from diverse species including worms, flies, mice, rats, macaques, and humans. Model organisms represent an alternative source to identify genes involved in alcohol-related phenotypes. New insights into the molecular mechanisms underlying AUD can be gained by taking advantage of the cross-species conservation of genomic features identified in these datasets.
Considering the volume and complexity of alcohol-related data, strategies must be developed to extract meaningful information across genetic, epigenetic, genomic, and phenotypic features. New analytical tools, as well as existing tools such as the NIH Common Fund’s Library of Integrated Network-based Cellular Signatures (LINCS) program and GeneNetwork (University of Tennessee) and many others, are critical for the integration and interpretation of complex datasets. The following sections summarize the findings and approaches presented at the Fourth International Congress on Alcoholism and Stress in Volterra, Italy, detailing the systems-based approaches that are being used to pioneer methods for prioritization and validation of novel genes and gene networks in conjunction with quantifiable behavioral phenotypes associated with stress and alcoholism.
Integrative Analysis of Human and Non-Human Primate Brain Tissue for Alcohol Consumption
Identification of Molecular Systems Involved in Human Disorders.
Transcriptome profiling provides a systematic and unbiased assessment of protein-coding and non-coding transcripts expressed within a given cellular environment. RNA-Sequencing (RNA-Seq) of multiple tissues across species has demonstrated the preservation of tissue-dependent transcriptome architecture (Sudmant, Alexis & Burge, 2015), supporting the utility of model organisms for investigating human disorders. Similar patterns or gene expression profiles are preserved across discrete brain regions; however, variation in expression mirrors neuroanatomical separation of brain regions across species, with the greatest overall separation occurring in the frontal cortex. Many individual protein-coding genes are deeply conserved across phylogenetic kingdoms, demonstrating marked sequence conservation and mechanistic direction. The individual contribution of any single gene or protein is likely inconsequential in comparison to the complete assembly and exchange of information among expressed molecules. Examining the universal layout of all expressed elements within the genome shows the exquisite distribution of gene networks in different tissues and species. By de-emphasizing the role of any single candidate gene or particular transcript, this brings into focus how the total transcriptome contributes to the broader biological pathways involved in behavioral phenotypes.
Modeling the complete profile of expressed genes using bioinformatics tools helps identify distinctive sets of features with shared biological function. Due to the cooperative roles of DNA and RNA for long-term cellular functioning, the collective substructure is intimately linked. GWAS have shown the widespread impact of genetic variation across biological pathways for psychiatric disorders (Breen et al., 2016). Brain-specific transcriptome networks are significantly enriched for GWAS derived gene candidates involved in disease (Farris, Arasappan, Hunicke-Smith, Harris & Mayfield, 2015) and are generally clustered into a discrete number of definable and coherent properties. Overlaying these human demarcated components with evidence from rodent studies indicates the cooperative causal role of multiple candidate genes in particular alcohol behavioral domains (Mayfield, Arends, Harris & Blednov, 2016). Combining cross-species datasets substantiates the involvement of a series of molecular building blocks in the manifestation and continuation of disease.
Conservation of Human Networks in Model Systems.
Dr. Sean Farris discussed how transcriptome data can be integrated with human postmortem brain data to isolate those brain regional- and cell type-specific changes associated with alcohol consumption that are conserved across species. Due to the practical and ethical limitations of studying human brain tissue, identifying the underlying molecular components is a challenging hurdle. Human postmortem brain research is restricted to retrospective observations across a limited set of available phenotypes, wherein the molecular changes found in brain are representative of the cumulative effects at the end-stage of disease. Methodically identifying and linking changes in brain processes to human behaviors thus requires an integrative cross-species based approach. Selection of an appropriate model system will supplement existing human evidence and facilitate translational research.
Non-human primates are the closest living evolutionary relatives of Homo sapiens, sharing more than 90% of DNA sequence identity (Chimpanzee Sequencing Analysis Consortium, 2005; Gibbs et al., 2007). In line with these evolutionary origins, non-human primate brains have the greatest human resemblance among all the model organisms used in biomedical research (Roelfsema & Treue, 2014). Cortical layers have shared expression patterns of co-regulated genes that may be associated with particular cellular subtypes and compartments (Bernard et al., 2012). Consistent with these findings, we found that the frontal cortex and central amygdala shared several corresponding transcriptional signatures. Although the transcriptome profiles were not an exact match between species, the constituent parts represented were remarkably consistent. Non-human primates are a valuable resource for conducting long-term studies on individual variation in alcohol drinking behavior (Jimenez & Grant, 2017). RNA-Seq of non-human primate brain tissue has highlighted the relationship between alcohol self-administration and dysregulation of synaptic signaling elements (Hitzemann et al., 2013). Circulating hormone levels, governed by the endocrine system, can simultaneously impinge upon these synaptic-related mechanisms and influence allostatic load. The transcriptional response to hormones involves specific nuclear hormones that may be essential mediators of alcohol consumption across species (Aoun et al., 2017). Regulation of gene expression and downstream molecular pathways by nuclear receptors involves an intricate interaction of multiple transcriptional proteins. The assembly and functions of multi-protein complexes guiding the production of RNA from DNA may be distinctly different across species. Additionally, there is a paucity of evidence regarding transcriptional regulation in different tissues and cell types.
Linking Affected Networks to Effective and Novel Medications.
Studying the basic principles of molecular biology for human disorders across different model organisms is especially prudent when tissues of interest are not readily accessible. AUD, like other psychiatric disorders, usurps control of human brain processes that may only be revealed through samples collected posthumously. Transcriptome-based studies of human postmortem brain from cases and appropriately matched control subjects can pinpoint some of the perturbed systems. Medications are useful adjunctive agents to behavioral therapy for mental health disorders, and drug discovery of safe and effective agents is of paramount importance. Large-scale studies of gene expression signatures for small molecules have aided the discovery-based efforts for neoteric uses of existing therapeutics (Lamb et al., 2006). Taking advantage of a constantly increasing volume of data and available resources, several studies have predicted the efficacy of existing compounds for previously non-indicated conditions, which can be further validated in a suitable laboratory model (Duan et al., 2016; Dudley et al., 2011; Liu, Lee, Salazar Hernandez, Mazitschek & Ozcan, 2015). Repurposing or repositioning of medications broadens their respective clinical utility and provides opportunities to examine previously unknown mechanisms of action. Compounds have a myriad of on- and off-target effects, which can lead to unwanted side effects and interfere with long-term treatment. Screening genomic signatures for available compounds and pursuing unreported mechanisms expedites the future development of more selective compounds with mitigated side effects.
Thousands of compounds have been tested and medically approved by governments and regulatory agencies throughout various industrialized countries. Psychiatric conditions, including substance use disorders, are a significant source of the global disease burden; however, few effective medications are currently available. Similar to many other human diseases, psychiatric disorders are not caused by any singular determinant. Treating the illness will not only require a comprehensive understanding of genetic, molecular, and behavioral imbalances, but also a source of medications aimed at restoring the affected biological networks. Utilizing data across multiple biological systems and species will be instrumental in the discovery of new medications without prior indications for a given disorder. A number of symptoms, and associated endophenotypes, may underlie a specific psychiatric diagnosis. Devising an appropriate research strategy for each diagnostic or observable criterion may be important for the nomination and classification of therapeutic options. Symptomology for individuals can vary dramatically; however, these differences likely converge on a common set of genes and molecular pathways in the human genome. Alterations in human postmortem gene expression related to the core disease state are a composite of genetic- and environmental-induced adaptations. In spite of the many similarities present across evolutionary distant taxonomies, some of these changes are conceivably human-specific and may be pivotal factors in disease, as well as biological avenues of therapeutic intervention.
Cross-Species Convergence in the Genetics of Ethanol Response and Alcohol Dependence: a Genomewide Association Study of Alcohol Dependence
Genetic Studies of Alcohol Use Disorder.
AUD show robust evidence for genetic risk (Cotton, 1979; Prescott, Caldwell, Carey, Vogler, Trumbetta & Gottesman, 2005; Sigvardsson, Bohman & Cloninger, 1996; Zhou, Colombo, Gessa & Kreek, 2013) with heritability estimated at ~50% for DSM-IV defined alcohol dependence (Ystrom, Reichborn-Kjennerud, Aggen & Kendler, 2011), consistent with a polygenic architecture and many loci of small effect. Variation in these loci may influence a number of different domains of risk including 1) alcohol-specific physiological measures like initial sensitivity and tolerance (Schuckit, Tipp, Smith, Wiesbeck & Kalmijn, 1997), 2) substance-independent activation of brain reward circuitry (Leeman & Potenza, 2012; Volkow, Wang, Fowler & Tomasi, 2012) common to all addictions, and 3) personality traits like internalizing and externalizing behaviors (Harford, Chen, Saha, Smith, Ruan & Grant, 2013). These features all suggest AUD will require large sample sizes for robust signal detection.
Genetic association studies of single genes in complex traits have a well-documented high false-positive rate (Ioannidis, Ntzani, Trikalinos & Contopoulos-Ioannidis, 2001; Ioannidis, Trikalinos & Khoury, 2006; Ioannidis, Trikalinos, Ntzani & Contopoulos-Ioannidis, 2003; Lohmueller, Pearce, Pike, Lander & Hirschhorn, 2003) due to small sample sizes, weak candidate selection (yielding low prior probability of genetic association), and lax statistical frameworks (Crowe, 1993). Many of these problems are overcome by applying rigorous, unbiased GWAS methods to large samples. In current GWAS, ~1 million single nucleotide polymorphisms (SNPs) across the genome are directly genotyped in every subject on microarrays. In most GWAS, these directly observed genotypes form the backbone for using linkage disequilibrium (the correlation between alleles at different polymorphic positions due to the limited number of specific haplotypes in the population) to infer the genotype probabilities at ~34 million ungenotyped polymorphic positions through the process of imputation (Marchini, Howie, Myers, McVean & Donnelly, 2007). In samples of several thousand cases and controls of European descent, ~8 million of these SNPs have sufficiently high minor allele frequencies for individual SNP analyses to have reasonable power, and this number rises as sample sizes increase and the minor allele frequency threshold for analytic power falls. Bonferroni correction for 1 million independent tests yields rigorous genomewide significance levels of 5 × 10-8.
Prior GWAS of AUD and alcohol-related phenotypes in European samples detected novel signals in the PECR (Treutlein et al., 2009) and AUTS2 (Schumann et al., 2011) genes. Neither of these novel signals were replicated in human samples, but Drosophila studies showed that reduced expression of the AUTS2 ortholog reduced ethanol sensitivity. Two independent signals were detected and replicated around the long-standing candidate gene ADH1B (Frank et al., 2012; Gelernter et al., 2014). A meta-analysis of >105,000 subjects using alcohol consumption as a phenotype identified a novel signal in the beta-Klotho (KLB) gene; brain-specific Klb knockout mice showed increased ethanol preference compared to control animals (Schumann et al., 2016). The UK Biobank GWAS of ethanol consumption in >112,000 subjects detected significant signals in 8 loci including ADH1B and KLB, and provided additional support for AUTS2 (Clarke et al., 2017). The UK Biobank study provides a particularly sobering picture of the challenges of robust identification of loci influencing alcohol-related traits when compared to the identification of 108 associated loci for schizophrenia in a meta-analysis of >150,000 subjects (Schizophrenia Working Group of the Psychiatric Genomics, 2014). Although the first robust genetic associations for alcohol-related phenotypes are emerging, power issues and the marked differences in sample type, ascertainment criteria, and phenotype tested between studies add to the challenges of replication in subsequent human cohorts.
Model organisms offer an alternative source of support for a gene’s involvement in alcohol-related phenotypes. Well-developed experimental approaches can test directly whether perturbation of a candidate gene impacts behavioral response to ethanol. Vertebrate approaches have been extensively described (Crabbe, 2002) while invertebrate approaches (Grotewiel & Bettinger, 2015) are less familiar to many investigators. A single, continuous acute exposure of C. elegans to 400 mM exogenous ethanol in the agar on a test plate yields (through cuticle absorption) an internal concentration of 40–50 mM (Alaimo et al., 2012) or ~200 mg/dL, within the range observed in humans after heavy drinking (Bond et al., 2010). A concentration-dependent slowing of locomotion at 10 minutes exposure (measuring initial sensitivity) is followed at ~30 minutes by an increase in speed of locomotion (measuring acute functional tolerance, AFT) (Davies, Bettinger, Thiele, Judy & McIntire, 2004; Davies et al., 2003) despite an increase in the internal tissue concentration of ethanol (Alaimo et al., 2012). Both measures can be independently affected by knockout or RNA interference (RNAi) downregulation of individual genes (Bettinger, Leung, Bolling, Goldsmith & Davies, 2012; Bhandari et al., 2012; Davies, Bettinger, Thiele, Judy & McIntire, 2004; Davies et al., 2003; Kapfhamer et al., 2008; Mathies et al., 2015).
In Drosophila studies, negative geotaxis assays are widely used. Adult flies 2–5 days-old are exposed to ethanol vapor in stoppered vials. At regular intervals, the vials are gently tapped on a table to knock flies to the bottom. The number of flies that are unable to climb or otherwise move in a typical, coordinated fashion is counted 30 seconds after agitation, and the amount of time required for 50% of the flies in each vial to become sedated (sedation time 50, ST50) is then calculated, either during the initial exposure (to determine sedation sensitivity) or during a second exposure four hours after the first (to determine rapid ethanol tolerance, measured as the ratio of ST502/ST501).
The choice between C. elegans and Drosophila for invertebrate studies depends on the presence of orthologous genes and the availability of genetic reagents (knockout or transposon insertion strains and/or RNAi vectors for the relevant ortholog). Some difference in the phenotypes assessed in invertebrate models is unavoidable because AFT has not been observed in Drosophila despite direct attempts to elicit this response (Chan et al., 2014). Orthologs of genes that affect simple ethanol responses in invertebrates also affect more complex ethanol responses in mammals, including measures of voluntary drinking and sensitivity (Bhandari et al., 2012; Kapfhamer et al., 2008; Liu, Vaithianathan, Manivannan, Parrill & Dopico, 2008). Both vertebrate and invertebrate models have previously demonstrated functional relevance of genes implicated by GWAS in ethanol response behaviors, as noted above.
Dr. Brien Riley discussed the results of a GWAS of DSM-IV defined alcohol dependence in a sample of cases and controls from Ireland conducted by the Virginia Commonwealth University Alcohol Research Center. To provide functional support for GWAS candidates, Drs. Jill Bettinger and Mike Grotewiel tested whether perturbation of orthologous genes altered behavioral response to ethanol in C. elegans or Drosophila, respectively. Dr. Michael Miles took advantage of existing, curated mouse experimental data to query candidate genes bioinformatically for 1) localization to ethanol behavioral QTL intervals using the Mouse Genome Informatics (MGI) tool set and/or 2) evidence that basal candidate gene expression correlated with measured ethanol behavioral phenotypes in the gene expression and behavioral response datasets within the GeneNetwork resource of genetic, phenotypic, and genomic data from C57BL/6J x DBA/2J recombinant inbred (BXD) mouse lines. Dr. M. Scott Bowers tested the effect of pharmacological antagonism of one candidate gene product on motivation to self-administer ethanol in rats after chronic ethanol exposure.
Irish Affected Sib-Pair GWAS Discovery Study.
Detailed descriptions of the subjects, GWAS genotyping, data cleaning, analysis, and results have been published (Adkins et al., 2017). Briefly, participants were ascertained in alcoholism treatment facilities in Ireland and Northern Ireland. Probands were eligible for inclusion if they met DSM-IV criteria for lifetime alcohol dependence, reported a sibling affected, and if all four grandparents had been born in Ireland or the United Kingdom. Probands, siblings, and parents were interviewed by clinically trained research interviewers using a modified version of the Semi-Structured Assessment of the Genetics of Alcoholism (SSAGA) interview, version II (Bucholz et al., 1994). All participants provided informed consent. DNA samples from healthy, unpaid volunteers donating blood at the Irish Blood Transfusion Service were obtained from the Trinity Biobank at Trinity College Dublin for use as controls, and were eligible for inclusion if they denied any problems with alcohol or history of mental illness and if all four grandparents had been born in Ireland or the United Kingdom. Information about age and sex was available, but because of the sample source, controls were not formally screened for alcohol dependence. Both the relatedness of cases and the lack of formal screening of controls were addressed analytically (see below). After genotype calling, imputation and all sample and genotype quality control procedures, 8,344,348 SNPs were available for analysis in 706 probands and affected siblings and 1748 population controls.
Individual SNPs were tested for association by Modified Quasi-Likelihood Score (MQLS) (Thornton & McPeek, 2007) because MQLS accepts genotypes in post-imputation dosage format and can account for subject relatedness by using a kinship matrix calculated from pedigree data. Unscreened Biobank controls were coded as phenotype unknown with an estimated sex-weighted 8.9% population alcohol dependence prevalence derived from population (Hasin, Stinson, Ogburn & Grant, 2007) and unpaid Dutch blood donor (Atsma, Veldhuizen, de Vegt, Doggen & de Kort, 2011) data to account for lack of control screening. Replication meta-analyses were undertaken in N = 15,496 European subjects, first in three alcohol dependence case-control samples (Edenberg et al., 2010; Frank et al., 2012; Gelernter et al., 2014) and then adding one population sample, for which alcohol dependence diagnoses were derived rather than directly assessed (Heath et al., 2011), using METAL (Willer, Li & Abecasis, 2010). Similar to many prior studies, no significant evidence of replication for discovery signals was detected.
Variation in human COL6A3 is associated with alcohol dependence.
In single marker analyses, 13 SNPs within the collagen VI A3 (COL6A3) gene on chromosome 2q37.2 achieved genome-wide significance, including the most significant SNP in this study, rs2256485, p=6.17×10−9 (Figure 1A). COL6A3 encodes an extracellular matrix (ECM) protein expressed in brain. Although there is no prior human association evidence for this gene, there is mounting evidence (Lubbers, Smit, Spijker & van den Oever, 2014) that multiple substances of abuse increase ECM remodeling, and that remodeling is required for the expected behavioral changes following exposure. Ethanol dose-dependently induces tissue plasminogen activator (tPA), required for ECM remodeling, which enhances ethanol reward (Bahi & Dreyer, 2012). Withdrawal seizures are reduced in tPA-deficient mice following chronic ethanol administration (Pawlak, Melchor, Matys, Skrzypiec & Strickland, 2005). Inhibition of proteolytic enzymes that degrade the ECM block escalated responding during acute withdrawal in dependent animals (Smith, Nealey, Wright & Walker, 2011). Collectively, these results indicate that ECM structural components (like COL6A3) and remodeling enzymes (like TPA) are important determinants of ethanol-induced neuroadaptations.
Figure 1.
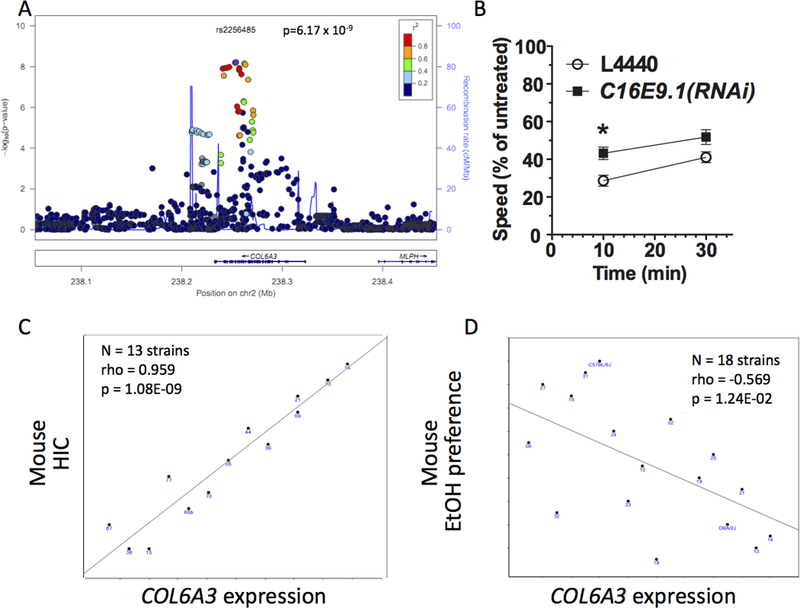
COL6A3 results. A) LocusZoom plot of human COL6A3 GWAS signal. B) RNAi-induced gene knockdown of C16E9.1 reduced sensitivity to ethanol in C. elegans. C) Col6a3 mouse whole brain expression is significantly correlated with total handling induced convulsion (HIC) score (sum of baseline subtracted HIC at 4, 6 and 7 hrs after 4 g/kg intraperitoneal ethanol). D) Col6a3 mouse whole brain expression is negatively correlated with ethanol 2-bottle choice voluntary consumption.
COL6A3 orthologs influence ethanol sensitivity in C. elegans and handling-induced convulsions in mice.
Three C. elegans genes with the highest orthology to human COL6A3 were tested for effects on initial sensitivity and AFT. RNAi knockdown of C16E9.1 decreased initial sensitivity compared to control RNAi animals (p < 0.05, Figure 1B) but did not affect the development of AFT. RNAi knockdown of the other COL6A3 orthologs (C18H7.1 and cutl-23) produced no significant differences in either measure.
In mice, Col6a3 is located within the Alcw5 QTL interval (MGI:3037048) for handling-induced convulsions (HIC) following 72-hour ethanol vapor exposure (Bergeson, Kyle Warren, Crabbe, Metten, Gene Erwin & Belknap, 2003). In GeneNetwork, the strongest correlation observed for mouse Col6a3 basal whole brain expression (GN113, probeset 1424131_at_A) is with total HIC score (sum of baseline subtracted HIC at 4, 6 and 7 hours) after 4 g/kg ethanol injected intraperitoneally (IP) in males (Philip et al., 2010) (trait 11382, correlation rank = 1, rho = 0.959, p = 1.05×10−9, N = 13 strains, Figure 1C). More than 5000 traits are present in GeneNetwork, but they are not all independent because of the multiple related measures made within studies and the partial overlap of BXD lines used between studies. GeneNetwork developers suggest that Bonferroni correction for 200 independent traits approximates an FDR of 0.2. To increase stringency, GeneNetwork results were corrected for 2000 independent tests, yielding a corrected significance threshold of 2.5 × 10-5. Col6a3 expression correlated negatively with 2-bottle choice ethanol preference (Phillips, Crabbe, Metten & Belknap, 1994) (trait 10479, correlation rank = 67, rho = −0.569, p = 0.0124, N = 18 strains, Figure 1D). While not significant after multiple test correction, this is consistent with the expectation that factors increasing HIC will decrease voluntary consumption (Metten et al., 1998). During discussion of this paper, Dr. Hitzemann noted that Col6a3 and numerous other collagen genes are differentially expressed in RNA-Seq data from the ventral striatum between male HDID2 mice, selected for high drinking in the dark, and the HS/NPT progenitor strain.
This study detected suggestive evidence of association for two loci with substantial prior support from both human and model organism (MO) alcohol studies (the human Krueppel-like factor 12 (KLF12) gene, chr 13q22.1, rs117695261, p = 6.63×10−8, (Figure 2A) and the human ryanodine receptor 3 (RYR3) gene, chr 15q14, rs4780153, p = 1.47×10−7 (Figure 3A). Although the signal in KLF12 is with a single imputed SNP of low minor allele frequency, in 1000 Genomes data from the population of Great Britain, closely related to the Irish, the associated SNP rs117695261 (MAF 0.03) in KLF12 has no r2 > 0.2 with any other SNP, consistent with the lack of correlated signals in Irish subjects.
Figure 2.
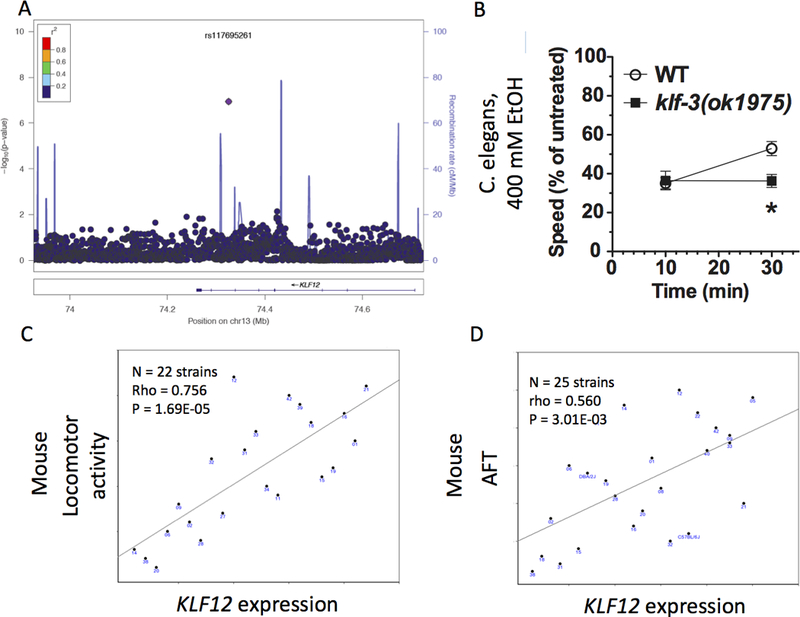
KLF12 results. A) LocusZoom plot of human KLF12 GWAS signal. B) Knockout of klf3, the C. elegans ortholog of KLF12, blocks the development of acute functional tolerance (AFT). C) Mouse basal Klf12 expression in prefrontal cortex (PFC) is significantly correlated with locomotor activity 0–5 minutes after 2.25 g/kg intraperitoneal ethanol. D) Mouse basal Klf12 expression in nucleus accumbens (Nac) is suggestively correlated with the development of acute functional tolerance (AFT).
Figure 3.
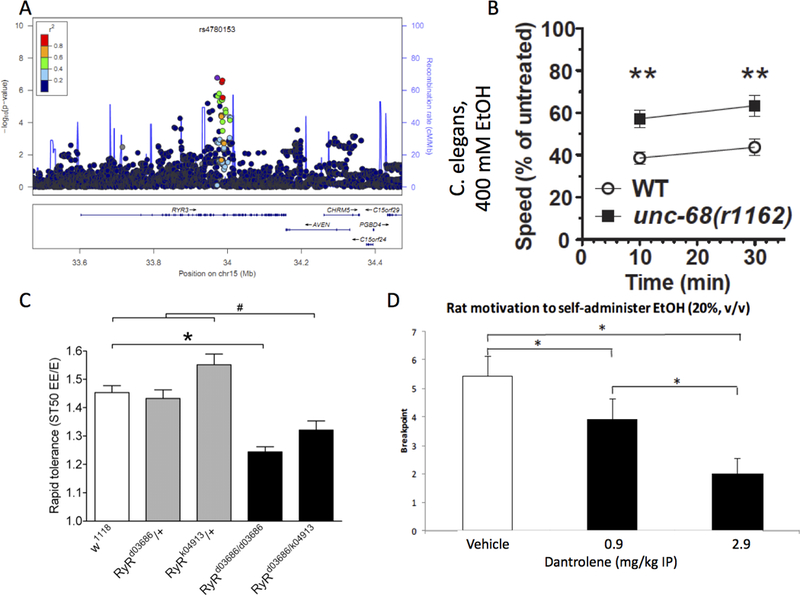
RYR3 results. A) LocusZoom plot of human RYR3 GWAS signal. B) Knockout of unc-1168, the single C. elegans ryanodine receptor gene, reduces initial sensitivity. C) Rapid tolerance was significantly reduced in RyRd03686 homozygous flies and in RyRd03686/k04913 transheterozygous flies compared to w1118 controls and RyR heterozygous animals (#, Bonferroni multiple comparison, p<0.05). D) Dantrolene treatment (0, 0.9, 2.9 mg/kg IP) dose-dependently reduced the motivation to self-administer ethanol. Breakpoint was defined as the maximum number of presses completed on an exponential progressive ratio schedule for ethanol reinforcement. Data represent mean ± SEM, * p < 0.05, n = 12.
There is substantial prior evidence for a role of KLF12 in ethanol response behaviors across species. In BXD mice, Klf12 is regulated by acute ethanol in prefrontal cortex (PFC), nucleus accumbens (NAc), and ventral tegmental area (VTA), and is a hub in a network of ethanol responsive genes (Wolen et al., 2012). In humans, KLF12 is known to act in combination with the co-repressor CTBP1 (Schuierer et al., 2001), and in C. elegans, the ctbp-1 gene is required for the development of AFT (Bettinger, Leung, Bolling, Goldsmith & Davies, 2012). The closest C. elegans ortholog to human KLF12 is klf-3, and this evidence collectively suggests KLF-3 is likely to act together with CTBP-1 to regulate AFT in worms. A strong loss-of-function allele in klf-3 showed no difference in initial sensitivity between wild-type and klf-3(ok1975) mutants, but while wild-type worms demonstrated normal AFT, klf-3 mutants showed no development of AFT at 30 minutes (Figure 2B, t-test of AFT, wild-type vs. klf-3 mutants, t3 = 8.99, p < 0.001). These data strongly suggest that the transcriptional regulation provided by KLF-3 is required for the development of AFT in worms.
In GeneNetwork, basal Klf12 expression in PFC (GN135, probeset 1455521_at) is significantly correlated with locomotor activity 0–5 minutes after 2.25 g/kg IP ethanol (Philip et al., 2010) trait 11708, correlation rank = 1, rho = 0.756, p = 1.69×10−5, N = 22 strains, Figure 2C). Basal Klf12 expression in mouse NAc (GN156, probeset 1439847_s_at) was suggestively correlated with AFT (Kirstein, Davidson, Ehringer, Sikela, Erwin & Tabakoff, 2002) (trait 10348, correlation rank = 29, rho = 0.560, p = 0.003, N = 25 strains, Figure 2D). While not significant after Bonferroni correction, this is consistent with the failure to develop AFT in C. elegans klf-3 mutants.
Previous studies have also implicated ryanodine receptors (RyR) in ethanol phenotypes; in humans, RYR3 was implicated in a GWAS of alcohol responses (Joslyn, Ravindranathan, Brush, Schuckit & White, 2010). Ryr1 and Ryr2 upregulation in mouse brain is observed following acute exposure to multiple drugs including alcohol (Kurokawa, Mizuno, Shibasaki & Ohkuma, 2010) and behavioral changes like conditioned place preference and withdrawal following acute exposure are blocked by the RyR antagonist dantrolene (Kurokawa, Mizuno & Ohkuma, 2013; Kurokawa, Mizuno, Shibasaki & Ohkuma, 2010). Dr. Riley discussed findings demonstrating that RYR3 orthologs influence initial sensitivity in C. elegans, rapid tolerance in Drosophila, and motivation to self-administer ethanol in rats.
C. elegans has one RyR gene, unc-68. Loss of unc-68 confers reduced sensitivity to ethanol (p < 0.001, Figure 3B). There is a single RYR3 ortholog in Drosophila, RyR. Two insertional mutations that cause partial loss of function in the Drosophila RyR gene reduce the development of rapid tolerance to ethanol in homozygotes and transheterozygotes (p < 0.05, Figure 3C). The mouse Ryr3 gene is localized to the support intervals for a complex group of ethanol behavioral QTL mapped to Chr. 2, but Ryr3 basal whole brain expression (GN113; probeset 1427427_at) is not strongly correlated with ethanol-related phenotypes.
The availability of dantrolene allowed us to assess the effect of antagonism of RyRs on the complex behavior of ethanol self-administration in rats. Male Han Wistar rats (p60 on arrival) were trained to self-administer ethanol (20% v/v) on a fixed ratio 3 reinforcement schedule, as previously described (Bowers et al., 2008; Bull et al., 2014; Bull, Syed, Minter & Bowers, 2015; Hopf et al., 2010). After 50 contiguous days of self-administration, the effect of three different doses (0, 0.9, and 2.9 mg/kg, IP) of dantrolene on the motivation to self-administer ethanol was measured, as previously reported (Bowers et al., 2008; Bull et al., 2014; Hopf et al., 2010) using a within-subjects, counterbalanced design where all doses were administered on all days and all animals received all doses. Between tests, rats were returned to daily fixed ratio 3 ethanol self-administration until responding was stable at pre-treatment baseline (~1 week). On test day, breakpoint served as a proxy for motivation to self-administer ethanol via ethanol-maintained responding on a progressive ratio reinforcement schedule (Richardson & Roberts, 1996). Dantrolene dose-dependently reduced motivation to self-administer ethanol in this experimental paradigm (Figure 3D). This effect was mirrored in responding on the ethanol-paired (p < 0.05), but not on the control (p = 0.12) lever. There was no effect of dosing order (p = 0.36) and no interaction of injection order with dantrolene (p = 0.88). Previous studies have shown that dantrolene has no effect on sucrose self-administration following a 48-hour drinking in the dark paradigm (Tarragon, Balino & Aragon, 2012).
In summary, although human replication remains challenging in genetic studies of alcohol-related traits, as shown by multiple GWAS of these phenotypes, model organisms provide substantial convergent evidence that discovery loci are involved in behavioral response to alcohol. Model organism studies can also provide independent evidence regarding the potential function of these genes in relation to ethanol, although the degree of phenotypic consilience in response to manipulation of candidate orthologs can vary considerably between species. Mammalian and invertebrate nervous systems show extensive molecular and functional conservation (Bargmann, 1998; Brownlee & Fairweather, 1999) and many drugs mediate their behavioral effects through orthologous target proteins (Kaletta & Hengartner, 2006; Matthews & Kopczynski, 2001). In the case of manipulation of othologs of human KLF12, strong phenotypic consilience in effects on AFT and consistent direction of those effects are observed. In contrast, orthologs of RYR3 affect quite different alcohol-related phenotypes across species, similar to prior reports of the effects of manipulations of chloride intracellular channel 4 (Clic4) orthologs, which altered sensitivity in flies and mice but in different directions (Bhandari et al., 2012). Where phenotypic consilience is observed, it can provide strong evidence for a gene’s role in a specific function. Where consilience is not observed, this may be due to differences in ethanol-response measures available for different species (such as the apparent lack of AFT in flies) or to the phenotypic measures favored by a specific laboratory, or to real differences between species. Despite this variable phenotypic consilience, model organism studies remain of critical importance in the elucidation of genes and mechanisms underlying AUD and alcohol-related traits.
Systems Genetics of Substance Use Disorders: Resources, Methods, and Challenges
Effective treatment of alcoholism and other substance use disorders will have a complexity that reflects numerous causes—intrinsic and extrinsic, genetic, epigenetic, and environmental. While simple systems that exploit single strains of rodents raised in single environments have their place, even the most statistically significant findings may not extrapolate to real-world complexity (Williams, 2009). Genetic and environmental differences contribute to the serious replication problem highlighted by Crabbe, Wahlsten and Dudek (1999) and more recently by Collins and Tabak (2014).
To address this problem, teams are building up systematic data across diverse and large cohorts. The NIAAA Collaborative Studies on Genetics of Alcoholism (COGA) is the most prominent example in our field, and this program has demonstrated key findings that were achieved by increasing the scale of science and level of collaboration by an order of magnitude. The NIAAA Integrative Neuroscience Initiative on Alcoholism (INIA) is a second example, encompassing efforts to jointly assemble, analyze, and share datasets on alcoholism across many genotypes, many scales, and several species, including mouse, rat, macaque, and human. In particular, the presentation by Dr. Rob Williams focused on the use of new and old mouse cohorts to dissect gene-by-ethanol-and-environmental interactions (GXEE) that influence alcohol consumption before and after stress. The group summarized their recent progress using both the expanded set of BXD strains of mice (Wang et al., 2016) and a significantly upgraded version of the GeneNetwork web service (Mulligan et al., 2017)( Figure 4).
Figure 4.
Two massive CIE gene expression data sets are accessible at www.genenetwork.org. Left: Published Phase 1 CIE and control data includes RNA-seq assays of mRNA levels in the mesocorticolimbic region. Right: Phase 2 (unpublished but accessible data by Mulligan and colleagues) includes CIE and control data generated using exon arrays (Affymetrix MTA/Clariom D arrays) for midbrain and (soon) for several other brain regions.
For the last 35 years many teams have been systematically generating genotypes, drug-related phenotypes, and omics data for the BXD family (Chesler et al., 2005; Crabbe, Kosobud, Young & Janowsky, 1983; Hitzemann et al., 2003; Matthews et al., 2005; Mozhui, Ciobanu, Schikorski, Wang, Lu & Williams, 2008; Mulligan, Wang, Adler, Mozhui, Lu & Williams, 2012; Philip et al., 2010; Rodriguez, Plomin, Blizard, Jones & McClearn, 1994). For the past 15 years, the INIA program has supported the construction of a massive compendium of phenome data for these strains in different environments and exposed to different levels of stress, ethanol, and many other drugs of abuse. Almost all of these datasets are embedded with GeneNetwork, a system built with support from INIA. The first tranche of findings of the INIA teams are summarized well in set of companion papers in a recent issue of Alcohol (Lopez, Miles, Williams & Becker, 2017; Mulligan et al., 2017; Porcu et al., 2017; Rinker et al., 2017; van der Vaart et al., 2017; Williams & Holmes, 2017).
The recent expansion of the BXD strains has revitalized their use and they are now a pre-eminent model with which to study GXEE. While the early phase of work-the first 20 years-was limited to 25 BXD strains, the expanded family now includes 150 lines. Limitations of power and precision have given way to new limitations of protocol throughput and budget. Hence the need to collaborate. The great benefit of the BXDs is being able to replicate each of over 100 genomes across multiple treatments and both sexes. It is possible to study the correlation structure of many different types of stressors and their effects on alcohol use. Once these complex molecular and behavioral responses to ethanol have been replicated for each genotype and sex in different environments, it becomes possible to map loci and genes and to test molecular targets and therapeutic interventions. In essence, INIA-Stress has helped build an experimental murine branch of precision medicine relevant to alcoholism and other addictive disorders.
Dr. Williams summarized a large-scale analysis of gene expression in the mesocorticolimbic system of the parent strains of the large BXD family-C57BL/6J (also known as B6 or simply as B; the mother of the family) and DBA/2J (D2 or D, the father of the family). As has been known for nearly 60 years (McClearn, 1959), these strains differ greatly in willingness to consume alcohol. B6 is a heavy drinker by murine standards and is also resilient to many stressors (Graybeal et al., 2014; McClearn, 1959; Mozhui et al., 2010). The D2 strain is the flip-side—minimal voluntary drinking and easily stressed. What are the underlying genetic and molecular causes of this heritable behavioral difference, and how does chronic intermittent ethanol (CIE) treatment impact long-term ethanol consumption in these parents and in their many BXD progeny? Can we extract the persistent imprints of allostatic load at the transcriptional level? Given enough data, can we model and predict behavioral outcomes from genetics and a well-defined treatment? While it would be premature to claim that we have answers to these important questions, the presentation demonstrated that we are now on terra firma, and have the resources with which to discover the answers. The more powerful mouse models also provide the link between genetically engineered mouse models on single background strains to highly complex and admixed human cohorts. They are a translational intermediate for precision medicine.
In phase 1 of the study, RNA-Seq data (N = 190 data sets) was acquired from both parents under three conditions: baseline, air control, and CIE treatment in a vapor chamber) and using precisely defined regions (laser capture microdissection, see Mulligan et al. (2017). CIE is a complex model, but one that has been studied and carefully parameterized in over 160 publications (Lopez, Anderson & Becker, 2016; Rodberg, den Hartog, Anderson, Becker, Moorman & Vazey, 2017). This protocol models both binge drinking and the acquisition of a heavy allostaic load on body and brain. In phase 2 of the study (work that is still in progress), four brain regions are being profiled using new exon arrays (Affymetrix MTA 1.0, Clariom D) across the parents, but now with many more replicates and at time points up to 14 days after the last CIE treatment. Michael Miles and colleagues are extending this analysis to many of the BXD strains themselves, with data on the impact of CIE in the PFC and NAc (see BXD GeneNetwork data sets). The key goal is to uncover the imprints of chronic alcoholism on CNS transcriptomes. These two phases of transcriptome data can be easily analyzed in GeneNetwork (Figure 4) and can be used to evaluate known or suspected genes that modulate core behavioral differences and the impact of alcohol exposure and potential treatments.
Brain Transcription Changes in the Mouse and Macaque Brain Associated with Excessive Ethanol Consumption
Extensive data are now available for the transcriptional features associated with the risk of developing and/or the consequences of excessive ethanol consumption and withdrawal. Data have been collected in fly, mouse, rat, macaque, and human samples. It is assumed that there will be conservation of these transcriptional features and that it is the conserved features that will present the best opportunities for developing new therapeutic targets. However, over the past 20 years of research, there is now a clear recognition that the transcriptional features associated with excessive ethanol consumption are exceedingly complex. Mulligan et al. (2006), the first genome-wide meta-analysis of gene expression data for any behavioral trait, found > 3000 genes that were associated with ethanol preference (2-bottle choice) consumption. As new technologies have emerged to analyze the brain transcriptome (e.g. RNA-Seq), complexity has not decreased. Furthermore, there has been a significant shift in how the transcriptional data are analyzed, moving from an emphasis on differential expression to more network-centric approaches (Iancu, Colville, Oberbeck, Darakjian, McWeeney and Hitzemann (2015). Dr. Hitzemann described some recent studies conducted at Oregon Health and Science University using both mouse and macaque models of excessive consumption. The highlights of each study are noted, followed by a discussion of conceptual overlap between the mouse and macaque analyses. The theme that develops is that excessive ethanol consumption is associated with a marked synaptic reorganization that in some cases involves structural proteins, such as cadherins and protocadherins, and in other cases elements of the extracellular matrix (e.g. collagens).
Iancu et al. (2013) used a microarray-based approach to examine how selection for high drinking in the dark (HDID) affected the ventral striatal transcriptome. The DID phenotype captures some aspects of binge consumption. Data were collected from the two selected lines, (HDID-1 & −2) which routinely reach a blood alcohol level of > 150 mg% (Crabbe, 2014), and the heterogeneous stock (HS/NPT) founders. The HS/NPT founders capture approximately 30% of the genetic variance that is available in Mus musculus (Roberts, Pardo-Manuel de Villena, Wang, McMillan & Threadgill, 2007). The initiation of the two selections were spaced by two years. The data analysis focused on finding QTL and transcriptional features common to both selections. Common QTLs were found on chromosomes 4, 14, and 16; these QTLs were distinct from those detected for preference consumption (Belknap & Atkins, 2001; Hitzemann & Oberbeck, 2008) and thus further support the idea that binge and preference phenotypes are mostly genetically different (Crabbe, Harris & Koob, 2011), even though some genetic similarity is indicated by significant genetic correlations among inbred strain values for preference drinking and DID (Crabbe et al., 2012). Perhaps the most notable preference QTL not detected was the preference QTL found on Chr 9 and centered at approximately 50 Mbp. Ninety-four common genes were differentially expressed (FDR < 0.05) in both selections; however, this list was largely independent of the differentially expressed genes detected for preference consumption (e.g. Mulligan et al. (2006). The data were further analyzed using a network-centric approach. Two coexpression modules were significantly affected in both selections; intra-modular connectivity was significantly disrupted. A number of genes known to be associated with ethanol phenotypes (e.g. Gabarg1, Glra2, Grik1, Npy2r, and Nts) showed significant changes in connectivity. Overall it was found that network based results showed significantly higher concordance across the two selections compared to the QTL and differential gene expression.
The HDID-2 and HS/NPT samples for Iancu et al. (2013) have been subsequently analyzed using a RNA-Seq approach (Iancu et al. unpublished observations). As noted elsewhere (Iancu, Kawane, Bottomly, Searles, Hitzemann & McWeeney, 2012), RNA-Seq has a number of advantages over microarray-based approaches for constructing and analyzing the coexpression data. A key, new observation extracted from the RNA-Seq data was that selection had a marked effect on genes associated with the extracellular matrix; especially noteworthy were effects on collagen genes. It is well established that excessive ethanol consumption has marked effects on the ECM; however, it is less clear that the ECM has a role in the “risk” for excessive consumption (reviewed in Lasek (2016).
Previous studies on changes in murine brain gene expression associated with the selection for ethanol preference have used F2 intercross or heterogeneous stock founders, derived from standard laboratory strains. However, these populations represent only a small proportion of the genetic variance available in Mus musculus. To investigate a wider range of genetic diversity, Colville et al. (2017) selected mice for ethanol preference using an HS derived from the eight strains of the Collaborative Cross. These heterogeneous stock mice were selectively bred (four generations) for High and Low ethanol preference. The NAc shell of naïve S4 mice was interrogated using RNA-Seq. Gene networks were constructed to assess both coexpression and cosplicing. Selection targeted one of the network coexpression modules that was significantly enriched in genes associated with receptor signaling activity, including Chrna7, Grin2a, Htr2a, and Oprd1. Connectivity in the module as measured by changes in the hub nodes was significantly reduced in the Low preference line. Analysis of the cosplicing network data revealed a significant effect of selection on a large cluster of RasGTPase binding genes, including Cdkl5, Cyfip1, Ndrg1, Sod1, and Stxbp5. These data in part support the earlier observation of Mulligan et al. (2006) that preference is linked to Ras/Mapk pathways.
Iancu et al. (2017) used RNA-Seq to assess the effects of chronic ethanol consumption in 30 rhesus macaques on the transcriptome in the central nucleus of the amygdala (CeA) and Area32 (medial PFC). The macaques had free access to ethanol or water for 12 months, i.e. a preference design. However, it should be noted that patterns of consumption differed markedly among the animals from very light drinkers, to binge consumption, to very heavy consumption. When parsed in this fashion, there were insufficient data per cell for adequate analyses and thus the analysis focused on average daily consumption. Membrane, synaptic, and splicing annotation categories were over-represented in the modules significantly enriched in genes that were positively associated with consumption. One of the modules strongly associated with excessive consumption was enriched in a number of receptor genes, including Chrm3, Chrna4, Chrna7, Glra2, Grm1, and Grm2. Note that Chrna7 was detected in Colville et al. (2017), Glra2 was detected in Iancu et al. (2013), and Grm2 was identified by Zhou, Colombo, Gessa and Kreek (2013) as a key risk gene. Key hub nodes that were significantly correlated with coexpression and cosplicing hub nodes were identified. For the CeA, key hub nodes were Rab6b, Cdk18, and Igs21. For area 32, key hub nodes were Ppr3r1 and Myeov2. As noted in Iancu et al. (2017), each of these genes has an association in mouse and/or human studies with excessive consumption,
Broadly speaking, the data from both the mouse and macaque studies has revealed strong effects on synaptic genes and genes associated with signaling pathways. This is not surprising and continues a theme that has been evident since Mulligan et al. (2006). However, the mouse studies also clearly suggest that the genes associated with the risk for binge and preference consumption are different. Given that we assume excessive consumption, as seen in the macaques, is a blend of the binge and preference phenotypes, there was an expectation that we would detect genes affected in the macaques, reflecting both phenotypes. Indeed, there was some overlap, e.g. for Chrna7 and Glra2. In lieu of independent confirmation, these may be chance observations but the data suggest that there may well be conservation across species. Detecting these genes and the associated gene networks are likely to be key to developing new therapeutic approaches.
Summary
The outcomes of complex systems based approaches in studies of psychiatric disorders are often multi-faceted, making it difficult to discern true biological signals from noise. Combining information across multiple species, especially for conserved phenotypic measures, helps distinguish the reproducible impact on interacting biological systems. The added power gained from combining data across multiple, independent experiments facilitates predictive analytics and adds a degree of confidence for further testing of specific hypotheses. Given the broad spectrum of factors influencing psychiatric disorders, large-scale systems approaches could potentially be used to identify the widespread effects of targeted therapeutic approaches.
The goal of this symposium was to highlight the analytical and integrative data techniques available to increase our understanding of the molecular mechanisms underlying AUDs. The integration of cross-species genetic and genomic information from humans to invertebrates was discussed, confirming the importance of validating current animal models of alcohol consumption. Future studies would benefit from several considerations. First, additional platforms are needed to bridge complex admixed human cohorts (clinical Precision Medicine) to complex admixed animal model cohorts (experimental Precision Medicine). To achieve this, the effects of gene variants on complex genetic backgrounds (knockouts on multiple strain backgrounds) should be studied. Second, improved models are needed to test GXEE interactions. This requires large-scale collaborations because such studies are cost prohibitive, and Consortia such as INIA have been a tremendous benefit to integrative research strategies. Third, large-scale integrative studies would benefit from additional statistical power (driven by sample size), increased replicability and rigor of experiments and conclusions. This session provided an overview of systems-based approaches and tools that are improving the experimental prioritization and validation of novel genes and gene networks linked with multiple behavioral phenotypes associated with stress and AUD.
Acknowledgments
The work presented was supported by NIAAA funding: U01AA020926 (RDM), R01AA012404 (RDM), K99AA024836 (SPF), P20AA017828 (BPR), R37AA011408 (BPR), P50AA022537 (MFM; BPR), U01AA016662 (RWW; MKM) and U01AA013499 (RWW; MKM), U24AA020929 (MFL; MFM), P50AA010761 (MFM), U01AA016667 (MFM), R01AA11034 (RJH), U01AA13484 (RJH), U01AA13510 (KAG; RJH), R24AA19431 (KAG; RJH) and P50AA10760 (RJH). We thank Dr. Rosana Camarini (Universidade de São Paulo, São Paulo, Brazil) for serving as Discussant for the symposium. The authors thank Dr. Jody Mayfield for thoughtful critiques and editing of the manuscript, and Dr. Marisa Roberto and organizers for allowing us to share our work at the 2017 “Alcoholism and Stress: A Framework for Future Treatment Strategies” conference in Volterra, Italy (supported by R13AA017581).
References:
- Adkins AE, Hack LM, Bigdeli TB, Williamson VS, McMichael GO, Mamdani M, et al. (2017). Genomewide Association Study of Alcohol Dependence Identifies Risk Loci Altering Ethanol-Response Behaviors in Model Organisms. Alcoholism, clinical and experimental research 41: 911–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaimo JT, Davis SJ, Song SS, Burnette CR, Grotewiel M, Shelton KL, et al. (2012). Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcoholism, clinical and experimental research 36: 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoun EG, Jimenez VA, Vendruscolo LF, Walter NAR, Barbier E, Ferrulli A, et al. (2017). A relationship between the aldosterone-mineralocorticoid receptor pathway and alcohol drinking: preliminary translational findings across rats, monkeys and humans. Molecular psychiatry [DOI] [PMC free article] [PubMed]
- Atsma F, Veldhuizen I, de Vegt F, Doggen C, & de Kort W (2011). Cardiovascular and demographic characteristics in whole blood and plasma donors: results from the Donor InSight study. Transfusion 51: 412–420. [DOI] [PubMed] [Google Scholar]
- Bahi A, & Dreyer JL (2012). Involvement of tissue plasminogen activator “tPA” in ethanol-induced locomotor sensitization and conditioned-place preference. Behavioural brain research 226: 250–258. [DOI] [PubMed] [Google Scholar]
- Bargmann CI (1998). Neurobiology of the Caenorhabditis elegans genome. Science (New York, NY) 282: 2028–2033. [DOI] [PubMed] [Google Scholar]
- Belknap JK, & Atkins AL (2001). The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mammalian genome : official journal of the International Mammalian Genome Society 12: 893–899. [DOI] [PubMed] [Google Scholar]
- Bergeson SE, Kyle Warren R, Crabbe JC, Metten P, Gene Erwin V, & Belknap JK (2003). Chromosomal loci influencing chronic alcohol withdrawal severity. Mammalian Genome 14: 454–463. [DOI] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, et al. (2012). Transcriptional architecture of the primate neocortex. Neuron 73: 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger JC, Leung K, Bolling MH, Goldsmith AD, & Davies AG (2012). Lipid Environment Modulates the Development of Acute Tolerance to Ethanol in Caenorhabditis elegans. PLOS ONE 7: e35192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari P, Hill JS, Farris SP, Costin B, Martin I, Chan CL, et al. (2012). Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes, brain, and behavior 11: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Ye Y, Cherpitel CJ, Room R, Rehm J, Borges G, et al. (2010). THE RELATIONSHIP BETWEEN SELF-REPORTED DRINKING AND BAC LEVEL IN EMERGENCY ROOM INJURY CASES: IS IT A STRAIGHT LINE? Alcoholism, clinical and experimental research 34: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers MS, Hopf FW, Chou JK, Guillory AM, Chang SJ, Janak PH, et al. (2008). Nucleus accumbens AGS3 expression drives ethanol seeking through G betagamma. Proceedings of the National Academy of Sciences of the United States of America 105: 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen G, Li Q, Roth BL, O’Donnell P, Didriksen M, Dolmetsch R, et al. (2016). Translating genome-wide association findings into new therapeutics for psychiatry. Nat Neurosci 19: 1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee DJ, & Fairweather I (1999). Exploring the neurotransmitter labyrinth in nematodes. Trends in neurosciences 22: 16–24. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr., et al. (1994). A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of studies on alcohol 55: 149–158. [DOI] [PubMed] [Google Scholar]
- Bull C, Freitas KC, Zou S, Poland RS, Syed WA, Urban DJ, et al. (2014). Rat nucleus accumbens core astrocytes modulate reward and the motivation to self-administer ethanol after abstinence. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 39: 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull C, Syed WA, Minter SC, & Bowers MS (2015). Differential response of glial fibrillary acidic protein-positive astrocytes in the rat prefrontal cortex following ethanol self-administration. Alcoholism, clinical and experimental research 39: 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RF, Lewellyn L, DeLoyht JM, Sennett K, Coffman S, Hewitt M, et al. (2014). Contrasting influences of Drosophila white/mini-white on ethanol sensitivity in two different behavioral assays. Alcoholism, clinical and experimental research 38: 1582–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, et al. (2005). Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nature genetics 37: 233–242. [DOI] [PubMed] [Google Scholar]
- Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. (2017). Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112,117). bioRxiv [DOI] [PMC free article] [PubMed]
- Collins FS, & Tabak LA (2014). Policy: NIH plans to enhance reproducibility. Nature 505: 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colville AM, Iancu OD, Oberbeck DL, Darakjian P, Zheng CL, Walter NAR, et al. (2017). Effects of selection for ethanol preference on gene expression in the nucleus accumbens of HS-CC mice. Genes, Brain and Behavior 16: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing Analysis Consortium (2005). Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. [DOI] [PubMed] [Google Scholar]
- Cotton NS (1979). The familial incidence of alcoholism: a review. Journal of studies on alcohol 40: 89–116. [DOI] [PubMed] [Google Scholar]
- Crabbe JC (2002). Alcohol and genetics: new models. Am J Med Genet 114: 969–974. [DOI] [PubMed] [Google Scholar]
- Crabbe JC (2014). Chapter 18 - The Genetic Complexity of Alcohol Drinking in Rodents. In Neurobiology of Alcohol Dependence Academic Press: San Diego, pp 359–375. [Google Scholar]
- Crabbe JC, Colville AM, Kruse LC, Cameron AJ, Spence SE, Schlumbohm JP, et al. (2012). Ethanol tolerance and withdrawal severity in high drinking in the dark selectively bred mice. Alcoholism, clinical and experimental research 36: 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Harris RA, & Koob GF (2011). Preclinical studies of alcohol binge drinking. Annals of the New York Academy of Sciences 1216: 24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Kosobud A, Young ER, & Janowsky JS (1983). Polygenic and single-gene determination of responses to ethanol in BXD/Ty recombinant inbred mouse strains. Neurobehavioral toxicology and teratology 5: 181–187. [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, & Dudek BC (1999). Genetics of mouse behavior: interactions with laboratory environment. Science (New York, NY) 284: 1670–1672. [DOI] [PubMed] [Google Scholar]
- Crowe RR (1993). Candidate genes in psychiatry: An epidemiological perspective. American Journal of Medical Genetics 48: 74–77. [DOI] [PubMed] [Google Scholar]
- Davies AG, Bettinger JC, Thiele TR, Judy ME, & McIntire SL (2004). Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron 42: 731–743. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, et al. (2003). A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115: 655–666. [DOI] [PubMed] [Google Scholar]
- Duan Q, Reid SP, Clark NR, Wang Z, Fernandez NF, Rouillard AD, et al. (2016). L1000CDS2: LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, et al. (2011). Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med 3: 96ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, et al. (2010). Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcoholism, clinical and experimental research 34: 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, & Mayfield RD (2015). Transcriptome organization for chronic alcohol abuse in human brain. Molecular psychiatry 20: 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, et al. (2012). Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addiction biology 17: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. (2014). Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry 19: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, et al. (2007). Evolutionary and biomedical insights from the rhesus macaque genome. Science (New York, NY) 316: 222–234. [DOI] [PubMed] [Google Scholar]
- Graybeal C, Bachu M, Mozhui K, Saksida LM, Bussey TJ, Sagalyn E, et al. (2014). Strains and stressors: an analysis of touchscreen learning in genetically diverse mouse strains. PLoS One 9: e87745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewiel M, & Bettinger JC (2015). Drosophila and Caenorhabditis elegans as Discovery Platforms for Genes Involved in Human Alcohol Use Disorder. Alcoholism, clinical and experimental research 39: 1292–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harford TC, Chen CM, Saha TD, Smith SM, Ruan WJ, & Grant BF (2013). DSM-IV personality disorders and associations with externalizing and internalizing disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Journal of psychiatric research 47: 1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, & Grant BF (2007). Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Archives of general psychiatry 64: 830–842. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, et al. (2011). A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biological psychiatry 70: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S (2014). The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 62: 1377–1391. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Bottomly D, Darakjian P, Walter N, Iancu O, Searles R, et al. (2013). Genes, behavior and next-generation RNA sequencing. Genes, brain, and behavior 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Malmanger B, Reed C, Lawler M, Hitzemann B, Coulombe S, et al. (2003). A strategy for the integration of QTL, gene expression, and sequence analyses. Mammalian genome : official journal of the International Mammalian Genome Society 14: 733–747. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, & Oberbeck D (2008). Strategies to study the neuroscience of alcoholism: introduction. Alcohol research & health : the journal of the National Institute on Alcohol Abuse and Alcoholism 31: 231–232. [PMC free article] [PubMed] [Google Scholar]
- Hopf FW, Bowers MS, Chang SJ, Chen BT, Martin M, Seif T, et al. (2010). Reduced nucleus accumbens SK channel activity enhances alcohol seeking during abstinence. Neuron 65: 682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Colville A, Oberbeck D, Darakjian P, McWeeney SK, & Hitzemann R (2015). Cosplicing network analysis of mammalian brain RNA-Seq data utilizing WGCNA and Mantel correlations. Frontiers in genetics 6: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Colville A, Walter NA, Darakjian P, Oberbeck DL, Daunais JB, et al. (2017). On the relationships in rhesus macaques between chronic ethanol consumption and the brain transcriptome. Addiction biology [DOI] [PMC free article] [PubMed]
- Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann R, & McWeeney S (2012). Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics (Oxford, England) 28: 1592–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Oberbeck D, Darakjian P, Metten P, McWeeney S, Crabbe JC, et al. (2013). Selection for Drinking in the Dark Alters Brain Gene Coexpression Networks. Alcoholism, clinical and experimental research 37: 1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, & Contopoulos-Ioannidis DG (2001). Replication validity of genetic association studies. Nature genetics 29: 306–309. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Trikalinos TA, & Khoury MJ (2006). Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. American journal of epidemiology 164: 609–614. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Trikalinos TA, Ntzani EE, & Contopoulos-Ioannidis DG (2003). Genetic associations in large versus small studies: an empirical assessment. Lancet (London, England) 361: 567–571. [DOI] [PubMed] [Google Scholar]
- Janak PH, & Tye KM (2015). From circuits to behaviour in the amygdala. Nature 517: 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez VA, & Grant KA (2017). Studies using macaque monkeys to address excessive alcohol drinking and stress interactions. Neuropharmacology 122: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, & White RL (2010). Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcoholism, clinical and experimental research 34: 800–812. [DOI] [PubMed] [Google Scholar]
- Kaletta T, & Hengartner MO (2006). Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 5: 387–399. [DOI] [PubMed] [Google Scholar]
- Kapfhamer D, Bettinger JC, Davies AG, Eastman CL, Smail EA, Heberlein U, et al. (2008). Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes, brain, and behavior 7: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein SL, Davidson KL, Ehringer MA, Sikela JM, Erwin VG, & Tabakoff B (2002). Quantitative trait loci affecting initial sensitivity and acute functional tolerance to ethanol-induced ataxia and brain cAMP signaling in BXD recombinant inbred mice. The Journal of pharmacology and experimental therapeutics 302: 1238–1245. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Mizuno K, & Ohkuma S (2013). Dopamine D1 receptor signaling system regulates ryanodine receptor expression in ethanol physical dependence. Alcoholism, clinical and experimental research 37: 771–783. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Mizuno K, Shibasaki M, & Ohkuma S (2010). Regulation of ryanodine receptors by dopamine D1 receptors during methamphetamine-induced place conditioning. Journal of neurochemistry 115: 1206–1214. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. (2006). The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science (New York, NY) 313: 1929–1935. [DOI] [PubMed] [Google Scholar]
- Lasek AW (2016). Effects of Ethanol on Brain Extracellular Matrix: Implications for Alcohol Use Disorder. Alcoholism, clinical and experimental research 40: 2030–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman RF, & Potenza MN (2012). Similarities and differences between pathological gambling and substance use disorders: a focus on impulsivity and compulsivity. Psychopharmacology 219: 469–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee J, Salazar Hernandez MA, Mazitschek R, & Ozcan U (2015). Treatment of obesity with celastrol. Cell 161: 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Vaithianathan T, Manivannan K, Parrill A, & Dopico AM (2008). Ethanol modulates BKCa channels by acting as an adjuvant of calcium. Molecular pharmacology 74: 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, & Hirschhorn JN (2003). Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nature genetics 33: 177–182. [DOI] [PubMed] [Google Scholar]
- Lopez MF, Anderson RI, & Becker HC (2016). Effect of different stressors on voluntary ethanol intake in ethanol-dependent and nondependent C57BL/6J mice. Alcohol (Fayetteville, NY) 51: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MF, Miles MF, Williams RW, & Becker HC (2017). Variable effects of chronic intermittent ethanol exposure on ethanol drinking in a genetically diverse mouse cohort. Alcohol (Fayetteville, NY) 58: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbers BR, Smit AB, Spijker S, & van den Oever MC (2014). Neural ECM in addiction, schizophrenia, and mood disorder. Progress in brain research 214: 263–284. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, & Donnelly P (2007). A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Blackwell GG, Austin MK, Edwards AC, Riley BP, Davies AG, et al. (2015). SWI/SNF chromatin remodeling regulates alcohol response behaviors in Caenorhabditis elegans and is associated with alcohol dependence in humans. Proceedings of the National Academy of Sciences of the United States of America 112: 3032–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DB, Bhave SV, Belknap JK, Brittingham C, Chesler EJ, Hitzemann RJ, et al. (2005). Complex Genetics of Interactions of Alcohol and CNS Function and Behavior. Alcoholism: Clinical and Experimental Research 29: 1706–1719. [DOI] [PubMed] [Google Scholar]
- Matthews DJ, & Kopczynski J (2001). Using model-system genetics for drug-based target discovery. Drug discovery today 6: 141–149. [DOI] [PubMed] [Google Scholar]
- Mayfield J, Arends MA, Harris RA, & Blednov YA (2016). Genes and Alcohol Consumption: Studies with Mutant Mice. Int Rev Neurobiol 126: 293–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClearn GE, Rogers DA, (1959). Differences in alcohol preference among inbred strains of mice. Quart J Studies on Alcohol 20: 691–695. [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, et al. (1998). High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mammalian Genome 9: 983–990. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, & Williams RW (2008). Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS genetics 4: e1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Karlsson RM, Kash TL, Ihne J, Norcross M, Patel S, et al. (2010). Strain differences in stress responsivity are associated with divergent amygdala gene expression and glutamate-mediated neuronal excitability. The Journal of neuroscience : the official journal of the Society for Neuroscience 30: 5357–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Mozhui K, Pandey AK, Smith ML, Gong S, Ingels J, et al. (2017). Genetic divergence in the transcriptional engram of chronic alcohol abuse: A laser-capture RNA-seq study of the mouse mesocorticolimbic system. Alcohol (Fayetteville, NY) 58: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. (2006). Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proceedings of the National Academy of Sciences of the United States of America 103: 6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Wang X, Adler AL, Mozhui K, Lu L, & Williams RW (2012). Complex control of GABA(A) receptor subunit mRNA expression: variation, covariation, and genetic regulation. PLoS One 7: e34586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak R, Melchor JP, Matys T, Skrzypiec AE, & Strickland S (2005). Ethanol-withdrawal seizures are controlled by tissue plasminogen activator via modulation of NR2B-containing NMDA receptors. Proceedings of the National Academy of Sciences of the United States of America 102: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, et al. (2010). High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes, brain, and behavior 9: 129–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, & Belknap JK (1994). Localization of genes affecting alcohol drinking in mice. Alcoholism, clinical and experimental research 18: 931–941. [DOI] [PubMed] [Google Scholar]
- Porcu P, O’Buckley TK, Lopez MF, Becker HC, Miles MF, Williams RW, et al. (2017). Initial genetic dissection of serum neuroactive steroids following chronic intermittent ethanol across BXD mouse strains. Alcohol (Fayetteville, NY) 58: 107–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Caldwell CB, Carey G, Vogler GP, Trumbetta SL, & Gottesman II (2005). The Washington University Twin Study of alcoholism. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 134b: 48–55. [DOI] [PubMed] [Google Scholar]
- Reilly MT, Noronha A, Goldman D, & Koob GF (2017). Genetic studies of alcohol dependence in the context of the addiction cycle. Neuropharmacology 122: 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson NR, & Roberts DC (1996). Progressive ratio schedules in drug self-administration studies in rats: a method to evaluate reinforcing efficacy. Journal of neuroscience methods 66: 1–11. [DOI] [PubMed] [Google Scholar]
- Rinker JA, Fulmer DB, Trantham-Davidson H, Smith ML, Williams RW, Lopez MF, et al. (2017). Differential potassium channel gene regulation in BXD mice reveals novel targets for pharmacogenetic therapies to reduce heavy alcohol drinking. Alcohol (Fayetteville, NY) 58: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, & Threadgill DW (2007). The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mammalian genome : official journal of the International Mammalian Genome Society 18: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodberg EM, den Hartog CR, Anderson RI, Becker HC, Moorman DE, & Vazey EM (2017). Stress Facilitates the Development of Cognitive Dysfunction After Chronic Ethanol Exposure. Alcoholism, clinical and experimental research 41: 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, & McClearn GE (1994). Alcohol acceptance, preference, and sensitivity in mice. I. Quantitative genetic analysis using BXD recombinant inbred strains. Alcoholism, clinical and experimental research 18: 1416–1422. [DOI] [PubMed] [Google Scholar]
- Roelfsema PR, & Treue S (2014). Basic neuroscience research with nonhuman primates: a small but indispensable component of biomedical research. Neuron 82: 1200–1204. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Tipp JE, Smith TL, Wiesbeck GA, & Kalmijn J (1997). The relationship between Self-Rating of the Effects of alcohol and alcohol challenge results in ninety-eight young men. Journal of studies on alcohol 58: 397–404. [DOI] [PubMed] [Google Scholar]
- Schuierer M, Hilger-Eversheim K, Dobner T, Bosserhoff AK, Moser M, Turner J, et al. (2001). Induction of AP-2alpha expression by adenoviral infection involves inactivation of the AP-2rep transcriptional corepressor CtBP1. The Journal of biological chemistry 276: 27944–27949. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. (2011). Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proceedings of the National Academy of Sciences of the United States of America 108: 7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, et al. (2016). KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences of the United States of America 113: 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson S, Bohman M, & Cloninger CR (1996). Replication of the Stockholm Adoption Study of alcoholism. Confirmatory cross-fostering analysis. Archives of general psychiatry 53: 681–687. [DOI] [PubMed] [Google Scholar]
- Smith AW, Nealey KA, Wright JW, & Walker BM (2011). Plasticity associated with escalated operant ethanol self-administration during acute withdrawal in ethanol-dependent rats requires intact matrix metalloproteinase systems. Neurobiology of learning and memory 96: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Alexis MS, & Burge CB (2015). Meta-analysis of RNA-seq expression data across species, tissues and studies. Genome Biol 16: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarragon E, Balino P, & Aragon CM (2012). Dantrolene blockade of ryanodine receptor impairs ethanol-induced behavioral stimulation, ethanol intake and loss of righting reflex. Behavioural brain research 233: 554–562. [DOI] [PubMed] [Google Scholar]
- Thornton T, & McPeek Mary S (2007). Case-Control Association Testing with Related Individuals: A More Powerful Quasi-Likelihood Score Test. American Journal of Human Genetics 81: 321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, et al. (2009). Genome-wide association study of alcohol dependence. Archives of general psychiatry 66: 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart AD, Wolstenholme JT, Smith ML, Harris GM, Lopez MF, Wolen AR, et al. (2017). The allostatic impact of chronic ethanol on gene expression: A genetic analysis of chronic intermittent ethanol treatment in the BXD cohort. Alcohol (Fayetteville, NY) 58: 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang G-J, Fowler JS, & Tomasi D (2012). Addiction Circuitry in the Human Brain. Annual review of pharmacology and toxicology 52: 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, et al. (2016). Joint mouse-human phenome-wide association to test gene function and disease risk. Nature communications 7: 10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, & Abecasis GR (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW (2009). Herding Cats: The Sociology of Data Integration. Frontiers in Neuroscience 3: 154–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, & Holmes A (2017). Preface to a special issue on genetic models of alcoholism and alcohol-stress interactions. Alcohol (Fayetteville, NY) 58: 23–24. [DOI] [PubMed] [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, et al. (2012). Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PLoS One 7: e33575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ystrom E, Reichborn-Kjennerud T, Aggen SH, & Kendler KS (2011). Alcohol dependence in men: reliability and heritability. Alcoholism, clinical and experimental research 35: 1716–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Colombo G, Gessa GL, & Kreek MJ (2013). Effects of voluntary alcohol drinking on corticotropin-releasing factor and preprodynorphin mRNA levels in the central amygdala of Sardinian alcohol-preferring rats. Neuroscience letters 554: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]


