Summary
We examined nevi and melanomas in 10 xeroderma pigmentosum (XP) patients with defective DNA repair. The lesions had a lentiginous appearance with markedly increased numbers of melanocytes. Using laser capture microdissection, we performed DNA sequencing of 18 benign and atypical nevi and 75 melanomas (melanoma in situ and invasive melanomas). The nevi had a similar high frequency of PTEN mutations as melanomas [61% (11/18) versus 53% (39/73)]. Both had a very high proportion of UV-type mutations (occurring at adjacent pyrimidines) [91% (10/11) versus 92% (36/39)]. In contrast to melanomas in the general population, the frequency of BRAF mutations (11%, 7/61), NRAS mutations (21%, 13/62), and KIT mutations (21%, 6/28) in XP melanomas was lower than for PTEN. Phospho-S6 immunostaining indicated activation of the mTOR pathway in the atypical nevi and melanomas. Thus, the clinical and histological appearances and the molecular pathology of these UV-related XP nevi and melanomas were different from nevi and melanomas in the general population.
Keywords: xeroderma pigmentosum, melanoma, PTEN, nevi, mTOR, DNA repair, UV carcinogenesis
Introduction
Melanoma is a highly aggressive malignant tumor of melanocytes. In the United Sates, in 2013, melanoma was diagnosed in about 80 000 people with more than 9000 deaths. The frequency of melanoma has been increasing for many years (Howlader, 2013). Pigmented nevi can be present at birth but more frequently arise as benign tumors of melanocytes. Although the link between melanoma and nevi is disputed (Ackerman, 1980; Whiteman et al., 2011), the Clark model indicates that some nevi may progress to melanomas (Clark et al., 1984).
Xeroderma pigmentosum (XP) is an autosomal recessive cancer-prone disease with photosensitivity and defective DNA repair of UV-induced DNA damage such as cyclobutane pyrimidine dimers and 6–4 pyrimidine-pyrimidone photoproducts (DiGiovanna and Kraemer, 2012). Unrepaired post-UV DNA damage may lead to UV signature mutations (transitions at dipyrimidine sites) (Wang et al., 2009; Ziegler et al., 1993). XP patients have a greater than 1000-fold – increase in non-melanoma and melanoma skin cancers under the age of 20 yr (Bradford et al., 2011). These skin cancers in XP patients have been reported to show molecular evidence of UV damage (D’Errico et al., 2000; Daya-Grosjean and Sarasin, 2005; Wang et al., 2009).
In recent years, high frequencies of mutations in different genes have been identified in melanomas in the general population (Miller and Mihm, 2006; Nikolaou et al., 2012). For example, in the US and European general population, 60% of melanomas have BRAF (v-raf murine sarcoma viral oncogene homolog B) mutations and more than 80% of these have V600E change (Davies et al., 2002). This c.1799T > C transversion mutation at an isolated pyrimidine does not have the UV signature, and these melanomas are considered to be unrelated to chronic sun damage of the skin (Curtin et al., 2005; Liu et al., 2007). PTEN (phosphatase and tension homologue), a tumor suppressor gene in the PIP3/AKT/mTOR pathway, has been shown to be mutated in many forms of human cancers (Baker, 2007; Bonneau and Longy, 2000). PTEN is also recognized as a key regulator of melanoma (Marsh et al., 2013). Approximately 16% of melanomas in the general population carry PTEN mutations (Sanger Institute, 2012). Previously, we reported that 56% of the melanomas in XP patients have PTEN mutations and 91% of these were UV-type base substitution mutations (Wang et al., 2009). In the general population, mutations in the NRAS oncogene have been found in 15–20% of melanomas with 90% of the mutations localized in codon 61 (Davies et al., 2002; Tsao et al., 2000). In addition, KIT mutations are reported to be frequently found in melanomas appearing in chronic sun-damaged skin (Carvajal et al., 2011; Curtin et al., 2006) especially those of lentigo melanoma type resulting in altered melanocyte migration (Whiteman et al., 2011).
To examine the relationship between pigmented lesions and melanomas in XP patients, we studied biopsies of pigmented skin lesions from XP patients. These lesions were sampled because they had an atypical appearance clinically or by dermatoscopy. We classified these pigmented lesions into five histopathologic groups (lentigo, benign XP nevi, atypical XP nevi, XP melanoma in situ, and XP invasive melanoma). We hypothesized that UV may play a major role in induction of XP benign and malignant pigmented lesions and looked for molecular evidence of UV mutagenesis in four candidate melanoma genes, PTEN, BRAF, NRAS, and KIT and for activation of the mTOR pathway.
Results
Characteristic clinical and histological appearance of premalignant pigmented lesions and malignant melanomas in XP patients
XP patients typically have numerous pigmented lesions on sun-exposed skin surfaces (DiGiovanna and Kraemer, 2012; Figure S1A,B). These vary in size, shape, and color ranging from light brown, to dark brown or black. They differ from common freckles (ephiledes) in the general population in that they are larger, more variable in color and shape within an individual, and have a lentiginous clinical and histological appearance (Stern et al., 1993). There often are small interspersed areas of hypopigmentation and telangiectasia along with atrophy resulting in poikiloderma. We frequently used dermatoscopy to attempt to distinguish benign pigmented lesions from malignant melanomas (Figure S1C). All of the XP pigmented lesions biopsied were clinically suspicious for melanoma.
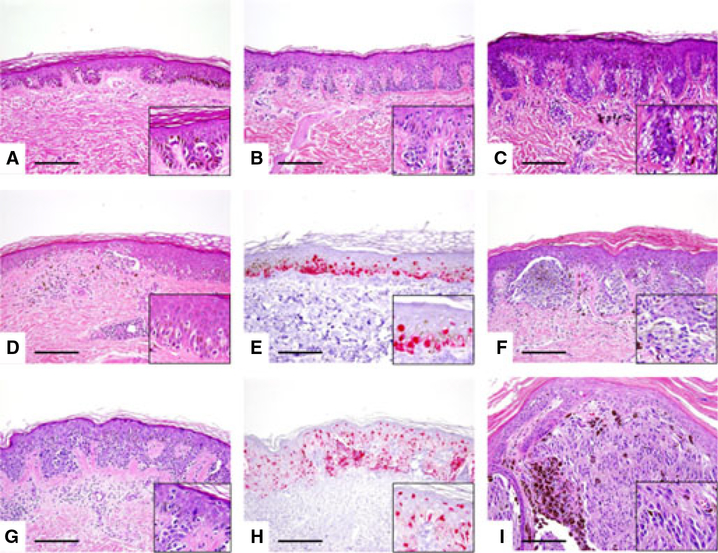
The histological features of these lesions differ from those in the general population. XP nevi are characterized by a lentiginous proliferation of single unit or small nests of melanocytes along the dermal–epidermal junction and often show attenuation of rete ridges; this is in contrast to the junctional nevi seen in the general population which characteristically show elongated rete ridges with small nests of melanocytes along the tips and sides of the rete ridges. Figure 1A shows a benign XP lentigo with hyper-pigmentation of basal epidermal layer keratinocytes and slightly increased density of single unit melanocytes along the dermal–epidermal junction. A unique feature of XP lesions is the attenuation of rete ridges over a relatively broad area of the lesion. Lentiginous XP junctional nevus (Figure 1B) and junctional XP nevus with architectural disorder and cytologic atypia or so-called ‘dysplastic nevus’ (Figure 1C) show similar histologic features to those in the general population. We have classified these lesions as benign XP nevi. We defined atypical XP nevi as junctional melanocytic proliferation with increased density of predominantly single cells and occasional nests of junctional melanocytes, with focal pagetoid melanocytes. The lesional melanocytes show random cytological atypia. The rete ridges are attenuated to flat (Figure 1D,E). These lesions are relatively common in XP patients.
Figure 1.
Histopathology of XP melanocytic lesions (photographs taken at 200× original magnification). (A) Benign XP lentigo: There is hyperpigmentation of basal epidermal layer keratinocytes, attenuated rete ridges, and slightly increased density of single unit melanocytes along the base of epidermis. There are no melanocytic nests seen (H&E taken at 200× original magnification; inset shows higher magnification of single unit melanocytes). (B) Lentiginous XP junctional nevus: There is regular elongation of rete ridges, slight hyperpigmentation of basal epidermal layer keratinocytes, and predominantly single unit and a few small nests of junctional melanocytes (H&E taken at 200× original magnification; inset shows higher magnification of single unit melanocytes and small melanocytic nests along the tips of rete ridges). (C) Junctional XP nevus with architectural disorder and cytologic atypia or so-called ‘dysplastic nevus’: There are single unit and nests of melanocytes along the tips and sides of rete, ‘bridging’ of rete ridges, and fibrotic changes of papillary dermal collagen surrounding rete ridges. The lesional melanocytes show random cytologic atypia. Perivascular chronic inflammation and pigmentary incontinence is present in the subjacent dermis (H&E taken at 200× original magnification; inset shows random cytologic atypia within the lesional melanocytes). (D) Atypical XP junctional melanocytic proliferation with cytologic atypia and focal pagetoid melanocytosis: There is attenuation of rete ridges, increased density of predominantly single unit and occasional nests of junctional melanocytes, and focal pagetoid melanocytes. The pagetoid melanocytosis is focal and mostly confined to the medial portion of the lesion; high-level or extensive pagetoid melanocytosis is not present. The lesional melanocytes show random cytologic atypia (H&E taken at 200× original magnification; inset shows single unit melanocytes permeating above the basal epidermal layer). (E) Melan-A immunohistochemical staining depicts the lesional melanocytes in (D). (F) In situ XP melanoma: The lesion is characterized by confluent nests of uniformly atypical melanocytes with conspicuous nucleoli and enlarged nuclei. Lesional melanocytes show abundant cytoplasm containing finely granular melanin pigments at the dermal–epidermal junction (H&E taken at 200× original magnification; inset shows irregular nests of cytologically atypical melanocytes with moderate to abundant dusky cytoplasm). (G) Another example of in situ XP melanoma: There is high-level and extensive pagetoid melanocytes over a broad area and focal confluency of junctional melanocytes (H&E taken at 200× original magnification; inset shows high-level and extensive pagetoid melanocytosis). (H) Melan-A immunohistochemical staining depicts the lesional melanocytes in (G). (I) Invasive XP melanoma: Sheets of uniformly atypical melanocytes invade into the reticular dermis. There are foci of darkly pigmented melanophages within the lesion (H&E taken at 200× original magnification; inset shows sheets of melanoma cells with prominent nuclear atypia).
In contrast, in XP melanoma, various histologic patterns were seen except for acral lentiginous type. Multiple primaries were relatively common in the XP patients. But, solar elastosis was absent. In situ XP melanomas were characterized by confluent nests of uniformly atypical melanocytes (Figure 1F) or high-level and extensive pagetoid melanocytes over a broad area and focal confluency of junctional melanocytes (Figure 1G,H). Invasive XP melanomas were characterized by sheets of uniformly atypical melanocytes that invade into the reticular dermis (Figure 1J).
We analyzed 18 premalignant pigmented lesions (11 benign nevi and seven atypical nevi) and 75 malignant melanomas (54 melanoma in situ and 21 invasive melanomas) from 10 XP patients (six females and four males) with mean age of 44.1 yr (range 21–63 yr). The patient’s cells were in complementation group C (seven patients), D (two patients), and XP variant (one patient) (Tables 1 and S1).
Table 1.
Age, complementation group, and number of nevi and melanomas in 10 XP patients
| Total Number per Patient of XP Nevi | Total Number of XP Patient Melanomas per | ||||||
|---|---|---|---|---|---|---|---|
| Patient | Age (yr)/Sex | XP Complementation Group | Benign XP Nevi (BN) | Atypical XP Nevi (AN) | XP Melanoma in situ (MIS) | XP Invasive Melanoma (IM) | |
| 1 | XP295BE | 49/F | XPC | 1 | 3 | 8 | 0 |
| 2 | XP86BE | 52/F | XPC | 1 | 0 | 2 | 0 |
| 3 | XP376BE | 44/F | XPC | 0 | 0 | 3 | 0 |
| 4 | XP21BE | 28/F | XPC | 0 | 0 | 9 | 10 |
| 5 | XP24BE | 35/F | XPC | 0 | 0 | 6 | 0 |
| 6 | XP1BE | 49/F | XPC | 2 | 1 | 12 | 2 |
| 7 | XP377BE | 63/M | XPC | 0 | 0 | 0 | 2 |
| 8 | XP29BE | 37/M | XPD | 7 | 3 | 13 | 5 |
| 9 | XP400BE | 21/M | XPD | 0 | 0 | 0 | 1 |
| 10 | XP31BE | 63/M | XPV | 0 | 0 | 1 | 1 |
| Total | 10 patients | 44.1 (Av.) | 11 | 7 | 54a | 21a | |
Including 59 melanomas from Wang et al. (2009).
PTEN mutations in XP nevi
PTEN mutations were present in high frequencies in the benign and the atypical XP nevi [55% (6/11) and 71% (5/7), respectively]. These frequencies were not significantly different (Table 2). Of the 21 PTEN mutations in the XP nevi, 15 (71%) were UV-type involving adjacent pyrimidines (Table 2). Multiple PTEN mutations were found in 4 (22%) of the nevi (one benign nevus and three atypical nevi).
Table 2.
Similar frequency of PTEN mutations in premalignant XP nevi and melanomas from 10 XP patients
| Total lesions sequenced | Number of lesions with mutations (% of lesions) | Number of lesions with UV-type mutations (% of lesions with mutations) | Total number of mutations | Total number of UV-type mutations (% of mutations) | Total number of non-UV-type mutations (% of mutations) | The number of amino acid substitutions (% of mutations) | Multiple mutations (% of lesions) | Loss of Heterozygosity (% Total mutations) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No mutations | 1 mutation | >1 mutation | |||||||||
| Benign XP nevi | 11 | 6 (55) | 6 (100) | 7 | 7 (100) | 0 (0) | 3 (43) | 5 (45) | 5 (45) | 1 (9) | 3 (43) |
| Atypical XP nevi | 7 | 5 (71) | 4 (80) | 14 | 8 (57) | 6 (43) | 4 (29) | 2 (29) | 2 (29) | 3 (43) | 5 (36) |
| Total XP nevi | 18 | 11 (61) | 10 (91) | 21 | 15 (71) | 6 (29) | 7 (33)a,b | 7 (39) | 7 (39) | 4 (22) | 8 (38) |
| XP melanoma in situ | 52 | 29 (56) | 26 (90) | 46 | 40 (87) | 6 (13) | 29 (63) | 23 (44) | 1 7 (33) | 12 (23) | 18 (39) |
| XP invasive melanoma | 21 | 10 (48) | 10 (100) | 16 | 15 (94) | 1 (6) | 12 (75) | 11 (52) | 6 (29) | 4 (19) | 7 (44) |
| Total XP | 73 | 39 (53) | 36 (92) | 62 | 55 (89) | 7 (11) | 41 (66)a,c | 34 (47) | 23 (32) | 16 (22) | 25 (40) |
| Melanoma | |||||||||||
| in situ + | |||||||||||
| Invasive | |||||||||||
| melanoma | |||||||||||
| Total XP nevi + melanomas |
91 | 50 (55) | 46 (92) | 83 | 70 (84) | 13 (16) | 48 (58) | 41 (45) | 30 (33) | 20 (22) | 33 (40) |
Fisher’s exact test P < 0.05 XP nevi versus XP melanoma in situ and invasive melanoma;
Including 6 missense and 1 splicing mutations.
Including 37 missense, 2 nonsense, and 2 splicing mutations.
Two intronic mutations were found, including one splice site mutation. Seven mutations (33%) were amino acid substitutions. Loss of heterozygosity (LOH) was found in eight of the mutations (38%).
PTEN mutations in melanoma
We added 16 additional melanomas from two additional XP patients to the 59 previously reported XP melanomas in eight patients (Wang et al., 2009; Tables 2 and S1). Twenty-nine melanoma in situ (MIS) lesions had PTEN mutations (56%), while 10 invasive melanomas (IM) had PTEN mutations (48%), a similar frequency. The 53% of XP melanomas with PTEN mutations was not significantly different from the frequency of benign and atypical XP nevi PTEN mutations (61%) or the frequency of lesions with multiple PTEN mutations (22% in melanomas and nevi). However, the frequency of amino acid substitutions in the XP melanomas (66%) was significantly greater than in the XP nevi (33%, P < 0.05).
Anatomic distribution of nevi and melanomas with PTEN mutations
In the general population, the anatomic distribution of melanomas with preponderance of trunk and lower extremities is different from that of non-melanoma skin cancers which are primarily found on the face, head and neck (Kraemer et al., 1994). These same differences in anatomic site are also seen in melanomas and nonmelanoma skin cancers in XP patients (Kraemer et al., 1994). In the present study, we found a preponderance of melanomas and nevi on the lower extremities and truck (Table S2). There was a similar site distribution between the lesions with and without PTEN mutations for the melanomas and for the nevi (Table S2).
Characteristic UV-type PTEN mutations in nevi and malignant pigmented lesions
All six benign nevi (100%) and five of the atypical nevi (83%) had UV-type mutations (Table 2). Thus, 92% of the XP nevi and melanomas with mutations had UV-type mutations. This frequency is significantly greater (P < 0.0001) than that expected compared with the frequencies of adjacent pyrimidines (54%) in the PTEN gene (Wang et al., 2009). This finding is consistent with the hypothesis that UV exposure plays a central role in induction of these mutations.
The type of PTEN mutations was similar between XP nevi and XP melanoma in situ (Figure S2). Most of the mutations (89%) were transitions. It is intriguing that the proportion of C to T and T to C mutations were almost same. The frequencies of T to C mutations in XP melanoma in situ (48%) were much higher than melanoma in the Sanger Catalog of Somatic Mutations in Cancer (COSMIC) Database (13%: P < 0.001; Sanger Institute, 2012). Benign nevi (57%) and atypical nevi (44%) also had a high frequency of T to C mutations which was significantly different from the COSMIC (P < 0.05). However, there was no significant difference in the frequency of T to C mutations between the XP invasive melanoma (22%) and the melanomas in the COSMIC Database.
Location of mutations in the PTEN gene
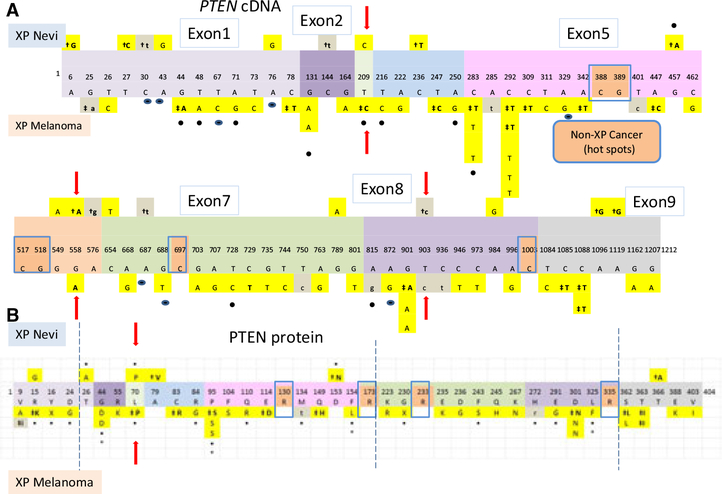
We compared the location of the PTEN base substitution mutations in the cDNA in the nevi to that in the melanomas (Figure 2A). These mutations were scattered in all nine PTEN exons in the nevi and in the melanomas. Most of these mutations were UV type (Figure 2A – yellow boxes). However, only three mutations(c.209T > C, c.558G > A, c.903T > C; Figure 2A – arrows) were the same in the nevi and the melanomas. Interestingly, the c.209T > C mutation was found in an invasive XP melanoma and has also been reported in the Sanger COSMIC database (Sanger Institute, 2012). The other two mutations were associated with XP atypical nevi as well as with XP melanomas. There were three transition hotspots (c.283C > T, c.292C > T, andc.901G > A) in exons 5 and 8 in the melanomas. These were different from the non-XP cancer PTEN mutation hotspots in exons 5, 6, 7, and 8 (Figure 2A – brown boxes).
Figure 2.
Schematic diagram of PTEN mutations in nevi and melanomas. (A) The nine exons of the 1212 nucleotide PTEN cDNA are indicated. PTEN mutations in XP nevi are located above the exons, while PTEN mutations in melanomas are below. (B) Sites of 43 non-synonymous amino acid substitution mutations and 2 nonsense mutations are shown above and below the 404 amino acids of PTEN protein (colored boxes indicate same exons and symbols as in Figure 2A). Arrows shows same altered amino acid in both nevus and melanoma. Dual specificity protein phosphatase domain is from amino acids 25–179. C2 calcium-/lipid-binding region are from amino acids 190–347. Key for (A and B) yellow boxes indicateS UV-type mutations, and gray boxes indicate non-UV-type mutations. † indicates mutations in atypical XP nevi. ‡ indicates mutations in invasive malignant melanomas. (•) indicates mutations listed in Sanger COSMIC database (Sanger Institute, 2012). Arrows show same mutations in nevi and melanomas. Brown boxes indicate hot spots in non-XP melanoma in Sanger Data base. * indicates reduced PTEN protein activity as reported previously (Wang et al., 2009).
The location of missense and nonsense mutations in PTEN protein in XP nevi and melanomas is shown in Figure 2B. All six of the missense mutations in the XP nevi were at UV-induced sites, and 37 of the 39 missense and nonsense mutations in the XP melanomas were at UV-induced sites. Only one of these mutations was the same in the XP nevi and in invasive melanomas (p.L70P) (Figure 2B – arrow) and was reported in the Catalog of Somatic Mutations in Cancer COSMIC database (Sanger Institute, 2012). The two nonsense mutations (p.Y16X and p.G230X) were found in MIS and reported in the COSMIC database.
Loss of heterozygosity in XP pigmented lesions
Loss of heterozygosity (LOH) was found in nevi and melanomas (Table 2). There was a similar frequency of LOH in benign XP nevi (43%), atypical XP nevi (31%), XP MIS (41%), and XP IM (36%). This result resembles haplo-insufficiency which subtle variations of PTEN gene expression may cause tumor and progression (Alimonti et al., 2010; Berger and Pandolfi, 2011). This may be an indication of genetic instability in all XP pigmented lesions reflecting the greatly increased risk of melanoma in XP patients (Bradford et al., 2011).
BRAF, NRAS, and KIT mutations
In the XP melanomas, we sequenced selected exons in BRAF (exon 15 including codon V600), NRAS (exons 2 and 3 including codons 12, 13 and 61), and KIT gene (exons 11, 13, 17, and 18) which include frequent sites of mutations in the general population (Curtin et al., 2006; Davies et al., 2002; Tsao et al., 2000). We found a similar frequency of mutations in the XP MIS as in the XP IM for each of these oncogenes (Table 3). Unlike the general population, the frequency of XP melanomas with BRAF (11%, P < 0.0001), NRAS (21%, P = 0.0002), or KIT (21%, P = 0.004) mutations was significantly lower than that of PTEN (53%) mutations (Table 3). Similarly, no mutations in BRAF or NRAS were detected in the XP nevi (Table S1B), while 61% had PTEN mutations (Table 2).
Table 3.
Higher frequency of mutations in PTEN than in BRAF, NRAS, or KIT in XP melanomas
| Gene | Melanoma in situ (%) | Invasive Melanoma (%) | Total (number of lesions with mutations/Total lesions sequenced) (%) |
|---|---|---|---|
| PTEN | 29/52 (56) | 10/21 (48) | 39/73 (53) |
| BRAF | 5/43 (12)*** | 2/18 (11)* | 7/61 (11)*** |
| NRAS | 10/43 (23)** | 3/19 (16)* | 13/62 (21)*** |
| KIT | 2/16 (13)** | 4/12 (33) | 6/28 (21)** |
P < 0.05
P < 0.01
P < 0.001 using Fisher’s exact test compared with PTEN mutations of same type (melanoma in situ, invasive melanoma, or total).
There were 11 mutations in the BRAF gene in the XP melanomas (Table S1B and Figure S3A). Ten of these (91%) were UV type. There was a small mutation hotspot at c.1803A > T which was listed in the Sanger database for non-XP cancers. Eight of these BRAF mutations in the XP melanomas were missense and two nonsense (Table S1B and Figure S3B). There was a UV-type mutation hotspot at K601N. However, we found only one XP melanoma which had the (non-UV type) BRAF V600E mutation that is frequently observed in melanomas in the general population (Davies et al., 2002).
We found 13 NRAS mutations in the XP melanomas (Table S1B and Figure S3C) and 11 (92%) of the 12 base substitution mutations were UV type. None of these were at the NRAS mutation hotspots found in melanomas in the general population (Figure S3C). There were seven NRAS missense mutations in the XP melanomas and 6 (86%) were UV type (Table S1B and Figure S3D).
There were seven KIT mutations in the XP melanomas and 5 (71%) of these were UV type (Table S1B and Figure S3E). None of these were at KIT mutation hotspots found in melanomas in the general population (Figure S3E). Three of the mutations were missense, and all were UV type in invasive XP melanomas (Table S1B and Figure S3F). The D820E mutation in the XP melanoma was previously reported in soft issue and thymic cancer (Sanger Institute, 2012).
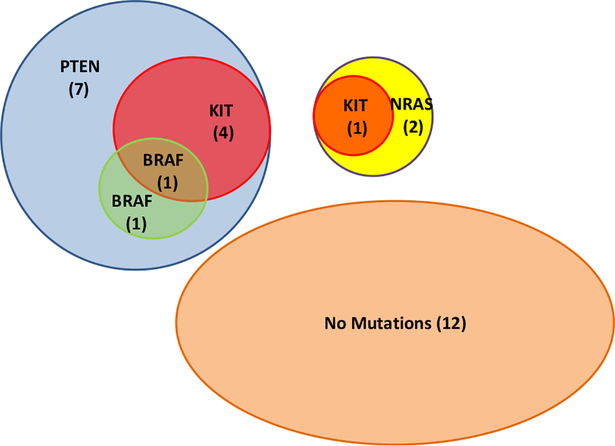
We were able to sequence all four genes (PTEN, BRAF, NRAS, and KIT) in 28 XP melanomas (Figure 3 and Table S2). Twelve (43%) of these did not have mutations detected in any of these genes. Thirteen (81%) of the other 16 XP melanomas had mutations in PTEN. These were associated with KIT mutations in four and BRAF mutations in two. One of these had mutations in PTEN, BRAF, and KIT. The other three XP melanomas had NRAS mutations, and one of these also had a KIT mutation. Thus, it is likely that there are interactions among these genes in the process of melanomagensis.
Figure 3.
Venn diagram of mutations found in 28 XP melanomas. Overlapping circles show mutations in PTEN, KIT, BRAF, and NRAS in each melanoma. Parenthesis shows the number of melanomas with given mutations.
mTOR pathway
We assessed 38 pigmented lesions (BN:10, AN:4, MIS:15, IM:9) with phospho-S6 staining to examine mTOR activity (Table 4 and Figure S3). There was a strong correlation between increasing frequency of phospho-S6 staining and histologic assessment of melanoma progression (P < 0.001). Thus, none of 10 benign XP nevi showed phospho-S6 staining, while 10 (67%) of 15 XP MIS and 8 (89%) of 9 XP IM showed phospho-S6 staining. However, there was no correlation between the frequency of XP lesions with PTEN mutations and phospho-S6 staining (data not shown).
Table 4.
Expression of p-S6 in XP pigmented lesions
| p-S6 staining | ||||
|---|---|---|---|---|
| Histopathology | Positive | Negative | Total | % |
| Benign XP nevi | 0 | 10 | 10 | 0 |
| Atypical XP nevi | 1 | 3 | 4 | 25 |
| XP melanoma in situ | 10 | 5 | 15 | 67 |
| XP invasive melanoma | 8 | 1 | 9 | 89 |
| Total | 19 | 19 | 38 | |
P < 0.001 by use of Cochran-Armitage test comparing increasing frequency of phospho-S6 staining and histologic assessment of melanoma progression.
Discussion
XP, DNA repair, and cancer
The risk of melanoma in XP patients is more than 1000-fold greater than in the general population. The XP melanomas occurred at an average age of 22 yr which is 33 yr earlier than in the general population (Bradford et al., 2011). This increased XP melanoma risk is related to UV exposure and the deficiency of repair of UV-induced DNA damage (Bradford et al., 2011; DiGiovanna and Kraemer, 2012; Wei et al., 2003).
Histopathology of XP pigmented lesions
Histopathologically, we classified XP pigmented lesions into five groups (Figure 1): three types of non-malignant pigmented lesions: lentigo, ‘benign XP nevi’, and ‘atypical XP nevi’ plus in situ and invasive melanomas. The XP nevi predominately had a lentiginous junctional pattern, while the melanomas had various histological patterns with predominately lentiginous changes, but not the acral lentiginous type. The nodular pattern was infrequently found in the XP melanomas. Multiple primary melanomas were relatively common along with a striking absence of solar elastosis. Clinically, the XP patients had multiple, larger, irregular lentiginous pigmented lesions rather than common freckles. Thus, both the clinical and histological characteristics of melanoma in XP were different from those in the general population.
PTEN mutations and cancer
Mutations of PTEN have been reported in several kinds of human carcinoma (glioblastomas, breast cancers, endometrial, prostate cancers, and melanoma) (Bonneau and Longy, 2000; Sanger Institute, 2012; Tsao et al., 2004). A comprehensive review of 16 studies of melanoma in the general population reported a 7% frequency of PTEN mutations in primary melanomas (Aguissa-Toure and Li, 2012). Tsao et al. reported that strong expression of PTEN was detected in acquired nevi but loss of expression was present in 37% of primary cutaneous melanomas and lentigo malignas (Tsao et al., 2003). In contrast, we found a similar high frequency of PTEN mutations in premalignant pigmented lesions (63%) as in melanomas (54%) in the XP patients (Table 2). In all of the studies of melanomas in the general population, the PTEN mutations were scattered throughout all exons (Aguissa-Toure and Li, 2012). In the XP melanomas, the two small PTEN mutation hotspots in exon 5 and one in exon 8 were not present in the general population (Figure 2A).
There is evidence that some of the PTEN mutations in the melanomas (G44D, P95S, F154L, L325F) (Figure 2B – marked as *) diminish PTEN activity (Rodriguez-Escudero et al., 2011; Wang et al., 2009). We used the PolyPhen2.0 program (Ramensky et al., 2002) to predict whether the PTEN missense mutations we found would be damaging (Table S3). This program has good agreement with previous site directed mutagenesis studies of the Akt phosphorylation inhibition function of PTEN (Wang et al., 2009; Table S3).
Interestingly, only three mutations (T209C, G558A, T903C) were coincident between the (premalignant) nevi and the malignant pigmented lesions in the XP patients (Figure 2). Two of these (c.558G > A and c.903T > C) are silent mutations that do not change amino acids and thus probably do not contribute to induction of the melanoma. However, a recent study reported that synonymous mutations may play a role in melanomagenesis. (Gartner et al., 2013). The c.209T > C mutation was reported in the COSMIC database in association with uterine carcinoma (Sanger Institute, 2012). c.209T > C results in a L70P amino acid change in the dual specificity protein phosphatase domain. PolyPhen 2.0 program (Ramensky et al., 2002) has a score of 1.000 predicting the L70P to be damaging (Table S3). Overall, the proportion of mutations that resulted in amino acid substitutions in the XP melanomas (66%) was much higher than that in the nevi (33%) (P < 0.05). In addition, most of the PTEN mutations in the XP nevi were scattered throughout PTEN exons and probably were passenger mutations. In contrast, most of the mutations in XP melanomas were non-synonymous and are likely to have a greater contribution to melanoma induction (Table S3).
However, as 40% (20/50) of the melanocytic lesions with PTEN mutations have multiple mutations (Table 2), it is difficult to determine the contribution of each mutation to the overall PTEN function. In addition, the presence or absence of loss of heterozygosity (LOH) (Tables 2 and S1) may play an essential role in the overall effect of a missense mutation. In an attempt to assess the effects of multiple mutations, we stained selected invasive melanomas for PTEN and pAKT (Figure S5). An invasive XP melanoma on the leg had 3 PTEN mutations: L70P predicted to be damaging but without LOH, D301N predicted to be benign but with LOH, and a splice mutation with LOH [Table S3 and (Wang et al., 2009)]. This melanoma was negative for PTEN staining and positive for pAKT indicating inactivation of the normal PTEN suppressor activity (Figure S5 A and B). This complexity does not permit unambiguous assignment of the inhibition of PTEN function to a single PTEN mutation. In contrast, an invasive XP melanoma from the scalp had 3 PTEN mutations: P95S predicted to be damaging, E114D predicted to be possibly damaging, and a synonymous mutation [Table S3 and (Wang et al., 2009)]. LOH was not found for these mutations. Staining showed PTEN staining and lack of staining for pAKT indicating normal PTEN function (Figure S5 C and D). Thus, it is possible that none of these PTEN mutations were driver mutations. A number of other genes have been implicated in melanomas in the general population and could be playing a role in the XP melanomas. These include NF1, CDKN2A, RB1, and p53 or genes in the mTOR pathway (see below).
PTEN mutation types
As commonly observed in ultraviolet mutagenesis, there was a predominance of PTEN transition mutations (Figures. 2 and S2). In the invasive melanomas in the XP patients and in the general population, the proportion of PTEN C to T mutations was greater than T to C mutations (69% and 58%, respectively) which were similar to the previous reports (Berger et al., 2012; Wei et al., 2011). In contrast, we found high frequencies of T to C changes in benign or atypical XP nevi (57% and 50%) and XP MIS (46%) which was significantly greater (P < 0.01) than in the invasive melanomas in the XP patients (19%) or in the general population (13%). After UV radiation, C to T mutations predominate, however, all types of mutations may be found in mammal cells. 6–4PPs isomer1 frequently induces T to C mutations in E. coli (Kamiya et al., 1998; LeClerc et al., 1991). This result is similar to some reports that XP-V cells which lack Pol η show the equal proportions of C to T and T to C mutations (Dumstorf et al., 2006; Wang et al., 2007). These mutations in the XP patients might be related to bypass polymerase iota which tends to induce T to C transitions (Dumstorf et al., 2006; Wang et al., 2007).
BRAF, NRAS and KIT Oncogenes, and cancer
Mutations in the oncogene, BRAF have been reported in malignant melanomas in the general population (Curtin et al., 2006; Miller and Mihm, 2006; Nikolaou et al., 2012). In addition, BRAF mutations were present in 82% of benign nevi (Poynter et al., 2006). The most frequent BRAF mutation in the general population was V600E. However, in our cases, we found BRAF V600E mutation in only one malignant melanoma (Table S1,B). Instead, we found other UV-type mutations in BRAF exon15 (11%). These findings suggest that melanomas in XP patients should have BRAF V600E mutation assessment before treatment with specific V600E inhibitors.
In addition to BRAF, we analyzed the XP melanomas for mutations in the NRAS and KIT oncogenes. We found mutations in NRAS exon 2 and 3 (21%) and in KIT exons 11, 13, 17, and 18 (21%) (Tables 3 and S1B). In 28 melanoma samples, all four genes were analyzed (PTEN, BRAF, NRAS, and KIT) (Figure 3 and Table S1A,B). In those samples, interestingly, NRAS and PTEN mutations were found in different melanomas, while BRAF mutations were found in association with PTEN mutations. This result is similar to Tsao et al. that melanomas developed through cooperation between BRAF activity and PTEN loss and is independent of NRAS gene (Tsao et al., 2000, 2004).
In the general population, melanomas with BRAF, KIT, and NRAS mutations are associated with different anatomic locations (Whiteman et al., 2011). For example, on non-chronic sun-damaged sites such as the back about 70% of the melanomas have BRAF mutations, while chronic sun-damaged sites such as the face have a lower prevalence of BRAF mutations with 30–40% having mutations in KIT or NRAS. In the XP melanomas, we found a lower frequency of BRAF mutations than KIT or NRAS mutations (Table 3). However, there was a greater frequency of PTEN mutations (Table 3). Further, there was a similar distribution of melanomas and nevi with or without PTEN mutations in the XP patients (Table S2). Acral lentiginous melanomas are located on the UV-protected sites (palms and soles) and have a lower frequency of PTEN mutations than BRAF, NRAS and KIT mutations (Zebary et al., 2013).
Most of the mutations in the XP melanomas were UV type. XP is, in general, likely to have accumulation of mutations by UV because of lack of ability to repair UV-induced pyrimidine dimers. These results also indicate that XP melanomas are related to UV exposure and are consistent with earlier reports (Wang et al., 2009; Wei et al., 2003). As XP patients are hypersensitive to UV, XP melanoma is considered a chronically sun-damaged type of melanoma. Our data showing relative lack of BRAF V600E mutations (a non-UV-type mutation) in the XP melanomas and its absence in the XP nevi is consistent with a previous report that BRAF V600E mutations were found infrequently with melanoma in chronic sun-induced skin (Curtin et al., 2006). These results suggest that PTEN mutations may play a major role in UV-related melanomas such as XP melanoma.
mTOR activation
The mammalian target of rapamycin (mTOR) is a kinase that links growth factor stimulation with protein synthesis and cell growth. Activation of mTOR leads to phosphorylation of several proteins, including p70 ribosomal S6 (phospho-S6) kinase which phosphorylates ribosomal protein S6 thereby resulting in increased protein translation and cell growth. The mTOR pathway is complex and involves regulation by PTEN and involvement of PI3 Kinase, AKT, tuberous sclerosis complex 2, Ras, BRAF, and other proteins. mTOR has been reported to be activated in a high frequency (about 73%) of melanomas in the general population and only a small proportion (about 4%) of benign nevi. Phospho-S6 activation was independent of BRAF and NRAS mutational status (Karbowniczek et al., 2008). mTOR activation in cutaneous melanoma was associated with poorer prognosis characteristics including elevated expression of pAKT Ser473 and low expression of PTEN (Populo et al., 2011). We assessed mTOR pathway using phospho-S6 staining. In agreement with the studies in the general population, we found that the frequency of activation of mTOR pathway was elevated in XP melanomas in situ and invasive melanomas (67% and 80%, respectively) and low in benign nevi (Table 4). The frequency in atypical XP nevi was intermediate (25%).
Increased immunoreactivity for phosphor-S6 in melanoma cells might reflect inactivating mutations in PTEN, mutations elsewhere along the PI3K pathway, activation of pathways that converge on mTOR such as MAPK, or mTOR activation by increased local production of ligands that stimulate this pathway. The lack of correlation of phosphor-S6 with PTEN mutational status suggests these other mechanisms might also be involved.
Dankort et al. (2009) reported that mTOR inhibition with rapamycin or PD325901 prevented the progression of BRAFV600E-/PTEN−/−-induced melanomas in mice. Our data demonstrating a high frequency of PTEN mutations and activation of the mTOR pathway in XP melanomas suggest that treatment of XP patients with mTOR inhibitors could be considered (Populo et al., 2012).
Taken together, our results in XP patients imply that UV plays an important role in induction of PTEN suppressor gene mutations in premalignant and malignant pigmented lesions. There was a high frequency of UV-type mutations, including T to C as well as more common C to T mutations. The greater proportion of mutations resulting in amino acid substitutions of PTEN mutations in the melanomas suggest that some may be driver mutations. The lower frequency of BRAF mutations in the XP melanomas was similar to that in chronic sun-induced damage type melanomas in the general population. These data provide insights into the molecular mechanism resulting in the greater than 1000-fold increase in melanomas in XP patients. The increased frequency of PTEN mutations and activation of the mTOR pathway in the XP melanomas may offer an opportunity for melanoma control by use of PTEN/mTOR pathway inhibitors.
Methods
Tissue samples and Histopathology
In this retrospective study, we retrieved blocks of paraffin fixed tissues removed from XP patients who had been examined at the NIH Clinical Center under protocols approved by the National Cancer Institute Institutional Review Board from 1987 to 2012. All of the surgical procedures were performed at NIH except for five melanomas that were excised by the patient’s local physicians. Specimens were recut and processed for histological examination using hematoxylin and eosin staining. Melan-A immunohistochemistry was performed to highlight the lesional melanocytes using a Ventana Benchmark immunostain and a red chromogen to distinguish from other darkly pigmented cells. Lesions were selected that showed distinct histological features of nevi and melanomas with sufficient material to perform laser capture microdissection. Lesions interpreted as lentigos were considered to have insufficient density of melanocytes to perform laser capture. Phospho-S6 immunostaining for activation of the mTOR pathway was performed as described previously (Li et al., 2008). Sections were incubated with rabbit antiphospho-ribosomal protein S6 (Ser235/236) (1:200; Cell Signaling Technology, Danvers, MA, USA) and stained with a Vectastain ABC-alkaline phosphatase with Vector red substrate (Vector Laboratories, Burlingame, CA, USA), using Mayer’s hematoxylin solution (Sigma–Aldrich, St. Louis, MO, USA) for counterstain.
Immunohistochemistry for PTEN and pAKT (Ser473) was performed on an automated immunostainer (Dako Autostainer Plus, Carpinteria, CA, USA). Briefly, after deparaffinization and rehydration, heat-induced antigen retrieval was performed prior to primary antibody incubation for both antibodies. For pAKT, slides were placed in a microwavable pressure cooker (TenderCooker; Nordic-ware, Minneapolis, MN, USA) into preheated buffer (0.01 M citrate buffer, pH 6.0, containing 0.1% Tween 20) and heated in a microwave oven to full pressure/temperature for 10 min. For PTEN, slides were placed into preheated buffer (0.01 M EDTA, pH 8.0) and heated in a microwavable pressure cooker to full pressure/temperature for 2 min. The slides were cooled and immediately incubated with the primary antibody overnight and detection was carried out on the Dako autostainer using Envision-HRP (rabbit) detection (Dako). Positive control cell lines were used to confirm the adequacy of the staining for each antibody. Antibodies were used at the following dilutions: PTEN (#9559 Cell Signaling; dilution 1:500), pAKT Ser473 (Cell Signaling; dilution 1:1000).
Laser capture microdissection and DNA extraction
We prepared 5-μm PEN membrane-coated slides from paraffin-embedded tissue blocks. After staining with hematoxylin and eosin, we collected DNA by using laser microdissection (Leica LMD 6000; Buffalo Grove, IL, USA) as described previously (Wang et al., 2009). To minimize formalin sequencing artifacts (Williams et al., 1999), approximately 700–1000 cells were microdissected from each slide. DNA was extracted using PicoPure DNA Extraction Kit using protocols supplied by the manufacturer (Applied Biosystems, Grand Island, NY, USA) (Vandewoestyne et al., 2012).
DNA analysis
We performed direct Sanger sequencing of all nine exons of PTEN, exon 15 of BRAF, exons 2 and 3 of NRAS, and exons 11, 13, 17, and 18 of KIT using sequencing techniques and primers described previously (Curtin et al., 2005; Wang et al., 2009). To minimize contamination, we used gloves and barrier pipet tips in the laminar hood for PCR reactions. Sequencing reactions were performed on a Prism Model 3700 Capillary Array Sequencer using PCR primers (Figure S1F; Wang et al., 2009). We checked mutations with both forward and reverse primers.
Statistical methods
We used the Fisher’s exact test for comparisons with significance P = 0.05 and Cochran-Armitage exact test for assessment of trends.
Supplementary Material
Significance.
We found that pigmented nevi and melanomas from xeroderma pigmentosum patients had a very high frequency of UV-type mutations in the PTEN tumor suppressor gene and few mutations in BRAF, NRAS, or KIT. These data provide insights into the molecular mechanisms of UV damage resulting in their greater than 1000-fold increase in melanomas. The increased frequency of PTEN mutations and activation of the mTOR pathway in these lesions may offer an opportunity for melanoma prevention by use of PTEN/mTOR pathway inhibitors.
Acknowledgements
We thank Ji-an Wang, who performed immunohistochemical studies, Seth Steinberg Ph.D. for statistical assistance, and Medha Bhagwat PhD for computational predictions of effects of mutations. The authors declare no conflict of interest.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Malignant melanomas in XP patients analyzed using laser capture microdissection.
Figure S2. Different types of PTEN base substitution mutations in XP nevi and melanomas compared to Sanger melanoma database.
Figure S3. Schematic Diagram of mutations and amino acid alterations in BRAF, NRAS and KIT.
Figure S4. Phospo-S6 immunostaining of XP lesions for activation of mTOR pathway.
Figure S5. Immunohistochemical staining of PTEN and pAKT in cutaneous melanomas of XP patients (200× original magnification).
Table S1a. PTEN, BRAF, NRAS and KIT mutations in XP melanomas and nevi.
Table S2. Similar site distribution of melanomas and nevi with or without PTEN mutations in XP patients.
Table S3. PTEN mutations - predicted and measured assessment of function and presence in cancer.
References
- Ackerman AB (1980). Malignant melanoma: a unifying concept.Hum. Pathol. 11, 591–595. [DOI] [PubMed] [Google Scholar]
- Aguissa-Toure AH, and Li G (2012). Genetic alterations of PTEN in human melanoma. Cell. Mol. Life Sci 69, 1475–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimonti A, Carracedo A, Clohessy JG et al. (2010). Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet 42, 454–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ (2007). PTEN enters the nuclear age. Cell 128, 25–28. [DOI] [PubMed] [Google Scholar]
- Berger AH, and Pandolfi PP (2011). Haplo-insufficiency: a driving force in cancer. J. Pathol 223, 137–146. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP et al. (2012). Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau D, and Longy M (2000). Mutations of the human PTEN gene. Hum. Mutat 16, 109–122. [DOI] [PubMed] [Google Scholar]
- Bradford PT, Goldstein AM, Tamura D et al. (2011). Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J. Med. Genet 48, 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal RD, Antonescu CR, Wolchok JD et al. (2011). KIT as a therapeutic target in metastatic melanoma. JAMA 305, 2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WH Jr, Elder DE, Guerry D, Epstein MN, Greene MH, and Van HM (1984). A study of tumor progression: the precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol 15, 1147–1165. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T et al. (2005). Distinct sets of genetic alterations in melanoma. N.Engl. J. Med 353, 2135–2147. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, and Bastian BC (2006). Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol 24, 4340–4346. [DOI] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, You MJ, DePinho RA, McMahon M, and Bosenberg M (2009). Braf (V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet 41, 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C et al. (2002). Mutations of the BRAF gene in human cancer. Nature 417, 949–954. [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean L, and Sarasin A (2005). The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat. Res 571, 43–56. [DOI] [PubMed] [Google Scholar]
- D’Errico M, Calcagnile A, Canzona F et al. (2000). UV mutation signature in tumor suppressor genes involved in skin carcinogen-esis in xeroderma pigmentosum patients. Oncogene 19, 463–467. [DOI] [PubMed] [Google Scholar]
- DiGiovanna JJ, and Kraemer KH (2012). Shining a light on xeroderma pigmentosum. J. Invest. Dermatol 132, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, and Kunkel TA (2006). Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl Acad. Sci. USA 103, 18083–18088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner JJ, Parker SC, Prickett TD et al. (2013). Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc. Natl Acad. Sci. USA 110, 13481–13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). (2013). SEER Cancer Statistics Review, 1975–2010 (Bethesda, MD: National Cancer Institute; ), http://seer.cancer.gov/csr/1975_2010/ (based on November 2012 SEER data submission, posted to the SEER web site, April 2013). [Google Scholar]
- Kamiya H, Iwai S, and Kasai H (1998). The (6–4) photoproduct of thymine-thymine induces targeted substitution mutations in mammalian cells. Nucleic Acids Res. 26, 2611–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowniczek M, Spittle CS, Morrison T, Wu H, and Henske EP (2008). mTOR is activated in the majority of malignant melanomas. J. Invest. Dermatol 128, 980–987. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Andrews AD, and Lambert WC (1994). The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch. Dermatol 130, 1018–1021. [PubMed] [Google Scholar]
- LeClerc JE, Borden A, and Lawrence CW (1991). The thymine-thymine pyrimidine-pyrimidone (6–4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3’ thymine-to-cytosine transitions in Escherichia coli. Proc. Natl Acad. Sci. USA 88, 9685–9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Takeuchi F, Wang JA, Fan Q, Komurasaki T, Billings EM, Pacheco-Rodriguez G, Moss J, and Darling TN (2008). Mesenchymal-epithelial interactions involving epiregulin in tuberous sclerosis complex hamartomas. Proc. Natl Acad. Sci. USA 105, 3539–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kelly JW, Trivett M et al. (2007). Distinct clinical and pathological features are associated with the BRAF (T1799A (V600E)) mutation in primary melanoma. J. Invest Dermatol 127, 900–905. [DOI] [PubMed] [Google Scholar]
- Marsh DV, Deuker MM, Bosenberg MW, Phillips W, and McMahon M (2013). Differential AKT dependency displayed by mouse models of BRAFV600E-initiated melanoma. J. Clin. Invest 123, 5104–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, and Mihm MC Jr (2006). Melanoma. N.Engl. J. Med 355, 51–65. [DOI] [PubMed] [Google Scholar]
- Nikolaou VA, Stratigos AJ, Flaherty KT, and Tsao H (2012). Melanoma: new insights and new therapies. J. Invest Dermatol 132, 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Populo H, Soares P, Faustino A, Rocha AS, Silva P, Azevedo F, and Lopes JM (2011). mTOR pathway activation in cutaneous melanoma is associated with poorer prognosis characteristics. Pigment Cell Melanoma Res. 24, 254–257. [DOI] [PubMed] [Google Scholar]
- Populo H, Soares P, and Lopes JM (2012). Insights into melanoma: targeting the mTOR pathway for therapeutics. Expert Opin. Ther. Targets 16, 689–705. [DOI] [PubMed] [Google Scholar]
- Poynter JN, Elder JT, Fullen DR, Nair RP, Soengas MS, Johnson TM, Redman B, Thomas NE, and Gruber SB (2006). BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 16, 267–273. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, and Sunyaev S (2002). Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Escudero I, Oliver MD, Andres-Pons A, Molina M, Cid VJ, and Pulido R (2011). A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum. Mol. Genet 20, 4132–4142. [DOI] [PubMed] [Google Scholar]
- Sanger Institute (2012). Catalogue of Somatic Mutations in Cancer. http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- Stern JB, Peck GL, Haupt HM, Hollingsworth HC, and Beckerman T (1993). Malignant melanoma in xeroderma pigmentosum: search for a precursor lesion. J. Am. Acad. Dermatol 28, 591–594. [DOI] [PubMed] [Google Scholar]
- Tsao H, Zhang X, Fowlkes K, and Haluska FG (2000). Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res. 60, 1800–1804. [PubMed] [Google Scholar]
- Tsao H, Mihm MC Jr, and Sheehan C (2003). PTEN expression in normal skin, acquired melanocytic nevi, and cutaneous melanoma. J. Am. Acad. Dermatol 49, 865–872. [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, and Haluska FG (2004). Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J. Invest Dermatol. 122, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewoestyne M, Van NF, Van HD, and Deforce D (2012). Evaluation of three DNA extraction protocols for forensic STR typing after laser capture microdissection. Forensic Sci. Int. Genet 6, 258–262. [DOI] [PubMed] [Google Scholar]
- Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, and Maher VM (2007). Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 67, 3018–3026. [DOI] [PubMed] [Google Scholar]
- Wang Y, DiGiovanna JJ, Stern JB, Hornyak TJ, Raffeld M, Khan SG, Oh KS, Hollander MC, Dennis PA, and Kraemer KH (2009). Evidence of ultraviolet type mutations in xeroderma pigmentosum melanomas. Proc. Natl Acad. Sci. USA 106, 6279–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lee JE, Gershenwald JE et al. (2003). Repair of UV light-induced DNA damage and risk of cutaneous malignant melanoma. J. Natl Cancer Inst. 95, 308–315. [DOI] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC et al. (2011). Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet 43, 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman DC, Pavan WJ, and Bastian BC (2011). The melanomas: a synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment Cell Melanoma Res. 24, 879–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Ponten F, Moberg C, Soderkvist P, Uhlen M, Ponten J, Sitbon G, and Lundeberg J (1999). A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am. J. Pathol 155, 1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebary A, Omholt K, Vassilaki I, Hoiom V, Linden D, Viberg L, Kanter-Lewensohn L, Johansson CH, and Hansson J (2013). KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J. Dermatol. Sci 72, 284–289. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Leffell DJ, Kunala S et al. (1993). Mutation hotspots due to sunlight in the p53 gene of nonmelanoma skin cancers. Proc. Natl Acad. Sci. USA 90, 4216–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.