Abstract
Objective:
Mechanisms of early and late improvements in cardiovascular risk after bariatric surgery and applicability to larger, at-risk populations remain unclear. We aimed to identify proteins altered after bariatric surgery and their relations to metabolic syndrome and diabetes.
Approach and Results:
We identified 19 proteins altered in 32 non-fasting plasma samples from a study of patients undergoing bariatric surgery who were evaluated pre-operatively (“Visit 1”) versus both early (“Visit 2”; ≈3 months) and late (“Visit 3”; ≈12 months) post-operative follow-up using pre-defined protein panels (Olink). Using in silico methods and publicly available gene expression repositories, we found that genes encoding 8/19 proteins had highest expression in liver relative to other assayed tissues, with the top biological and disease processes including major obesity-related vascular diseases. Of 19 candidate proteins in the surgical cohort, 6 were previously measured in over 3,000 Framingham Heart Study participants (IGFBP-1, IGFBP-2, P-selectin, CD163, LDL-receptor, and PAI-1). A higher concentration of insulin-like growth factor binding protein-2 (IGFBP-2) at baseline was associated with a lower risk of incident metabolic syndrome (OR per log-normal unit 0.45, 95% CI 0.32-0.64, P=7.7×10−6) and diabetes (OR=0.63, 95% CI 0.49-0.79, P=0.0001) after multivariable adjustment.
Conclusions:
Using a directed protein quantification platform (Olink), we identified known and novel proteins altered after surgical weight loss, including IGFBP-2. Future efforts in well-defined obesity intervention settings may further define and validate novel targets for the prevention of vascular disease in obesity.
Keywords: proteomics, metabolic syndrome, weight loss, vascular risk, Metabolism, biomarkers, proteomics
INTRODUCTION
Bariatric surgery in obese individuals has a demonstrable impact on cardiometabolic risk, with both early and late benefits on insulin resistance and cardiovascular disease risk. Accordingly, examination of circulating biomolecules (e.g., metabolites, circulating RNAs, and proteins) after bariatric weight loss surgery has yielded important insights into molecular pathways affected by changes in weight and accompanying improvement in insulin resistance. Indeed, clinical investigations using “multi-omic” platforms during medical weight loss have suggested the fruitfulness of this paradigm for identifying metabolic, genomic, and epigenetic alterations that may prove to be critical in the pathogenesis of cardiometabolic disease. Nevertheless, while these efforts have been important in identifying reversible molecular signatures of obesity, whether or not such biomarkers are applicable to larger populations at risk for metabolic syndrome and diabetes remains unclear.
In this investigation, we examined selected proteins using serial non-fasting blood draws from a sample of individuals from a large study of bariatric surgery pre- and post-operatively to identify specific proteins altered early (at approximately 3 months) and late (at approximately 12 months) post-operatively, relative to the pre-operative state. To extend these results to a large at-risk population, we studied several proteins identified in the bariatric surgical study in a large number of participants from the Framingham Heart Study (FHS) to assess their association with incident metabolic syndrome and diabetes. Finally, using in silico approaches at the mRNA level, we identified potential tissues of origin for expression of genes encoding these proteins and suggest potential pathways targeted by them. Our ultimate aim was to use an integrated approach originating in an extreme phenotype (bariatric surgical weight loss) to identify potential biomarkers relevant to the development of insulin resistance, a major contributor to obesity-related cardiometabolic disease.
RESEARCH DESIGN AND METHODS
Data from the Framingham Heart Study is publicly available (dbGaP; https://www.ncbi.nlm.nih.gov/gap); data from the bariatric surgical study will not be made publically available.
Patient cohorts
Bariatric surgery:
The analytic subsample for the present study was taken from a completed prospective cohort study aimed at understanding the effects of bariatric surgery on platelet function (U54-HL12311; PI: Freedman). The parent study enrolled 121 individuals scheduled for clinically indicated bariatric surgery (either sleeve gastrectomy or gastric bypass surgery). Of note, one individual enrolled in the study and had pre-operative measures, but did not proceed to surgery or study follow-up. Study participants were examined pre-operatively and at visits that were planned at approximately 3 and 12 months post-operatively, with 98 participants with follow-up and blood samples at all three time points. Non-fasting peripheral venous blood was drawn into citrate tubes, with plasma isolated as described1. Samples were immediately stored at our high-throughput biomarker facility (not thawed until immediately prior to assay). Study investigators performed a medical history (medications, demographics, metabolic risk factors) and a physical examination. Hypertension was defined as the use of anti-hypertension medication or a systolic blood pressure greater than 140 mmHg or a diastolic blood pressure greater than 90 mmHg. A diagnosis of diabetes was defined as type 1 on insulin or type 2 on any medications or a hemoglobin A1c greater than 6.5% at the baseline visit.
The sampling strategy for the discovery (bariatric surgery) cohort was complex (Supplementary Figure I). This project was part of pilot efforts in our laboratory to configure and optimize the Olink protein quantification platform. As such, only a specific set of proteins was quantified (see Protein quantification, below), and protein quantification occurred during two separate sessions (“runs”), leading to potential biases from batch effects. Accordingly, we only included individuals who had blood at all three time points (Visit 1, 2, and 3), with proteins quantified on the same run; this would allow us to avoid increased “noise” by comparing proteins in the same participant across runs (e.g., Visit 1 from Run 1 being compared to Visits 2 and 3 at Run 2). It is critical to note that samples chosen for each run were done so in a non-random fashion, attempting to enrich for relief in insulin resistance over time. Briefly, samples included in the current analysis from Run 1 were comprised to individuals selected based on age less than or equal to 50 years and resolution of diabetes at post-operative visit (N=4; selected based on absolute reduction in hemoglobin A1c from Visit 1 to Visit 3) versus no diabetes (N=6). Serial samples on Run 2 included individuals with diabetes at baseline (one with pre-diabetes) and a selection of individuals with greater magnitude of change in hemoglobin A1c over time post-operatively. Further details of selection strategy are in Supplementary Figure I. This approach yielded 10 subjects in the first run and 22 subjects in the second run. For the subjects included in our analytic cohort, the average time (with standard deviation) between baseline study visit 1 and visit 2 (approximately “3 month visit”) was 139±43 days; the average time between baseline study visit 1 and visit 3 (approximately “12 month visit”) was 417±49 days. All subjects provided informed consent in accordance with an approved Institutional Review Board protocol.
Framingham Heart Study:
This aspect of the investigation utilized data from the proteomics project of the “SABRe CVD initiative,” which included 2784 Offspring cohort participants from the Framingham Heart Study (FHS) who appeared for the 7th and 8th examinations and 3392 participants from the FHS Third Generation cohort who appeared for the 1st and 2nd on-site examinations2. Briefly, the FHS is a community-based, prospective cohort study of individuals in Framingham, MA, with examinations every 4-8 years, with detailed data and sample collection methods as well as assessment of cardiovascular diseases (CVD) risk factors 3, 4. We used the average of two measurements of resting, seated blood pressure as the blood pressure values in our analysis. Current smokers smoked at least one cigarette per day in the last year. Physical activity was described previously 5, and we generated a dichotomous variable for sedentary lifestyle (defined as a “physical activity score” of 30 or less). Finally, we abstracted self-reported educational attainment into a categorical variable of “no college degree,” “had college or similar degree,” and “completion of college or higher degree.” We excluded participants with missing data for age, sex, smoking, alcohol consumption, physical activity, education, fasting glucose, insulin measurements, and those with fasting status less than 8 hours. We defined “diabetes” as fasting blood glucose ≥126 mg/dl, use of medication for diabetes, or reported history of diabetes. We defined “metabolic syndrome” as the presence of at least 3 of 5 metabolic risk factors: (abdominal obesity: waist circumference ≥88cm for women and ≥102 cm for men; high triglyceride levels: ≥150 mg/dl; low HDL cholesterol: <50 mg/dl for women or <40 mg/dl for men; elevated blood pressure (BP): systolic BP ≥130 mmHg, diastolic BP ≥85 mmHg, or current anti-hypertensive medication use; high fasting blood glucose ≥100 mg/dl or current use of anti-dysglycemia medications). The final cohort for analysis consisted of 4877 participants for new-onset diabetes and 3971 for new-onset metabolic syndrome. Informed consent was provided by all study participants. The Boston University Medical Center Institutional Review Board approved the study protocol.
Protein quantification
Bariatric surgery:
We used the Olink platform for this study, which employs a quantitative polymerase chain reaction (PCR) technology called “Proximity Extension Assay” (PEA) for protein quantification (Olink, Uppsala, Sweden). For this study, we tested the “CVD II,” “CVD III”, and “Cardiometabolic” panels, but did not include results from the “CVD II” panel due to poor assay performance (specifically, poorly performing control values); the proteins in the included panels are shown in Supplementary Table I. A detailed protocol for Olink-based protein quantification (taken directly from the manufacturer and adapted slightly) is provided in the Online Supplement. The primary output from the Olink method was Cq values (quantification cycle number) corresponding to amplicons specific for each protein. These raw Cq values were imported to the Olink NPX Manager Software v1.0.0.2 and Normalized Protein Expression (NPX) values were calculated by normalizing the Cq values to inter-plate controls (included in triplicate in each run).
FHS:
Plasma concentrations of the candidate proteins were measured as part of the FHS SABRe CVD Initiative2. Concentrations for 71 plasma proteins were quantified using a modified approach of enzyme-linked immunosorbent assay on a Luminex xMAP platform (proteins and corresponding coefficients of variation provided in Supplementary Table II). Step-by-step procedures and protocol for reporting for the Luminex assays have been published 6. The practices to ensure adequate quality control included: (1) using triplicate approaches for defining acceptable ranges and distribution curve of protein concentrations; (2) preparing controls for low and high concentrations with multiple measurements by 5 scientists on different dates; (3) duplicate plasma sample measurements. The FHS assays demonstrated acceptable coefficients of variation (2.0-9.5%) across 71 proteins2. Proteins were log-transformed for analysis. The list of proteins used in this analysis of FHS participants was limited to a subset of proteins that was significantly altered over time after surgery in the bariatric surgical cohort. Results of the assays for the proteins targeted in this study are provided in Supplementary Table II.
RNA-seq analysis for tissues of origin
Whole transcriptome RNA sequencing data from the EMBL-EBI’s RNASeq Atlas (Array Express ID: E-MTAB-2836) that consists of 32 tissues from 122 individuals was used to look for the corresponding gene expression profiles of these dysregulated proteins in normal tissues. Briefly, the fastqs downloaded from EMBL were aligned to the human genome (hg19, ENSEMBL version 75) using STAR aligner7. Only reads that are uniquely mapped were considered for read annotation to gene features. Reads were annotated with the ENSEMBL 75 gene transfer format (gtf) file using Salmon8. Salmon provides both read counts and transcripts per million reads (TPMs) for each gene feature in the ENSEMBL gtf. Normalization of read counts was conducted via a median ratio method in DESeq29. Gender specific tissues such as endometrium, fallopian tube, placenta, ovary, testis and prostate gland were omitted. Fold change for each gene was computed by dividing expression in the tissue with the highest expression by the expression in the second highest expressing tissue across all tissues considered.
Pathway analysis
The list of 19 proteins with differences in the Visit 1 to Visit 2 and Visit 1 to Visit 3 time-point pre- versus post-bariatric surgery was processed for enrichment analysis. First, the list was expanded based on “predicted functional partners” provided by the STRING database10 to add 10 proteins with similar functional characteristics in addition to interactions among the combined set. In Cytoscape11, a coherent network of 26 proteins was identified (16 of the 19 original proteins plus 10 added by STRING). This set of 26 proteins served as the input to test for functional enrichment across a number of biological ontology and pathway resources using the online tool, Enrichr12. Tabular results were downloaded for Gene Ontology: Biological Process, Jensen Disease Ontology13 and WikiPathways14. Representative terms of interest were selected from these results and plotted as a bar graph ranked by a score calculated from both p-value and Z score.
Statistical analysis
The statistical methods for this analysis consisted of several distinct steps: (1) analysis of differential expression of circulating proteins from bariatric surgical cohort (including correction for batch effects); (2) validation of selected overlapping, candidate proteins with new-onset diabetes in the FHS cohort; (3) analyses of potential tissue of origin, using RNA-seq, and potential pathways implicated by these proteins. Steps 1 and 2 are described below. Step 3 is described above.
Differential expression of proteins at different time-points after bariatric surgery
Selection of proteins and handling below-detection limit values:
Plasma proteins from the Cardiometabolic panel (N=92 total proteins included as per manufacturer) and the Cardiovascular III panel (N=91 total proteins included as per manufacturer) were analyzed in this study. Raw Cq values were imported to the Olink NPX Manager Software v1.0.0.2 and Normalized Protein Expression (NPX) values are calculated by normalizing the Cq values to inter-plate controls (included in triplicate in each run) using a proprietary method. We filtered the initial number of proteins by 43 (those with expression below limit of assay in >50% of all samples included), leading to 139 total proteins included in our final analysis (86 from Cardiometabolic panel; 53 from Cardiovascular III panel).
Batch effect correction and differential expression:
The protein measurements were performed at two different time points (“runs”) for both panels. Before correcting for potential batch effects, the proteins MEPE and NT-proBNP were removed from the analysis from the Cardiovascular III panel (position of MEPE on the plate in the two runs was switched; the assay performance of the NT-proBNP was improved by the vendor in Run 2). Characteristics of the 181 proteins (92 from Cardiometabolic, 91 from Cardiovascular Disease III, then excluding MEPE and NT-proBNP) is presented in Supplementary Table III along with the number of samples with expression below detection limit, mean expression value and inter-quartile range for Visit 1, Visit 2 and Visit 3 samples. As mentioned previously, 43 total proteins were filtered due to below detection limit expression in >50% of included samples (4 from Cardiovascular Disease III panel and 39 from the Cardiometabolic panel). Of the remainder, proteins with below detection limit expression data were included in analysis with a value of “0”. While we recognize For all proteins included, principal components analysis (PCA) on log2-transformed protein levels for the Cardiovascular III panel revealed that there was a significant batch effect between Run 1 and Run 2, while there was no discernible batch effect for the Cardiometabolic panel (Supplementary Figure II). The batch effect correction between the two runs was performed for both panels using ComBat from the SVA package in R that uses an empirical Bayesian framework15. In the end, the combined data from both runs and panels yielded 139 proteins that were used for differential expression analysis using LIMMA package in R 16 (Supplementary Table III), a standard approach in gene expression from microarray experiments. We identified proteins differentially expressed between Visit 1 (pre-operative) and Visit 2 (early post-operative) and between Visit 1 and Visit 3 (late post-operative) using a paired analysis with a Bonferroni correction to address type 1 error and multiplicity.
Validation of selected candidate proteins with new-onset metabolic syndrome or diabetes in the FHS
Clinical and demographic characteristics of the FHS study sample were calculated as mean (with standard deviation) or median (with 25% and 75% interquartile values) for continuous variables and frequency for categorical variables. Of the 139 proteins included in differential expression analyses (above), 25 were measured in the FHS; of these, we found an overlap of 6 proteins (IGFBP-1, IGFBP-2, P-selectin, CD163, LDL-receptor, and PAI-1) with those differentially expressed between Visit 1 and 2 and between Visit 1 and 3 in the bariatric surgery study after Bonferroni correction for multiple testing. These 6 proteins were included in subsequent analyses in the FHS. We applied logistic regression models to measure the association of each candidate protein with new-onset metabolic syndrome or diabetes. Models were adjusted for baseline age, sex, education, current smoking status (as defined above), alcohol consumption, body mass index, fasting insulin, and physical activity. For new-onset diabetes, we also adjusted for baseline fasting glucose. For new-onset metabolic syndrome, we also adjusted for waist circumference, systolic and diastolic blood pressures, HDL cholesterol, triglyceride, and fasting glucose level at baseline. A two-sided P<0.05 was statistical significant and analyses were performed in SAS 9.4 (SAS Institute, Cary, NC) or R (http://www.r-project.org).
RESULTS
Clinical characteristics of the bariatric surgery cohort
Baseline characteristics of the 32 participants from the bariatric surgery study included in our analysis, relative to the other non-included participants, are shown in Table 1. The study sample consisted primarily of middle-aged Caucasian individuals (median age 45 years; 47% female), with a median body mass index (BMI) 45.0 kg/m2 (interquartile range 40.4-50.2 kg/m2). Over 50% of participants included in this study had diabetes or other obesity-related comorbidities (e.g., hypertension). Consistent with the non-random selection of samples for this sub-study (see Methods, above, under Patient cohorts), relative to subjects not included in this sub-study, we found that subjects included were less likely to be female and had a more even distribution of type of bariatric surgery. The prevalence of diabetes was higher in the included subjects, as was the hemoglobin A1c (both P<0.0001), whereas the BMI was similar between the two groups. In the 32 subjects included in this analysis, BMI had fallen by a median 9.5 kg/m2 by Visit 2 (interquartile range of weight loss 11.4 to 7.7 kg/m2); by Visit 3, the median BMI reduction was 13.0 kg/m2 (interquartile range of weight loss 15.8 to 9.7 kg/m2). (Similar ranges of reduction in BMI where available were observed in the non-included group: median reduction by Visit 2 was 8.7 kg/m2 [N=81; P=0.10 vs. included group]; median reduction by Visit 3 was 13.4 kg/m2 [N=68, P=0.84 vs. included group].)
Table 1.
Baseline characteristics of the bariatric surgical participants included in this study, relative to those not included. Data on the 120 individuals who underwent surgery are reported here, with 32 individuals included in the protein quantification analysis. Values are reported as median (interquartile range) or number (percentage). P values refer to Chi square comparisons (categorical) or Wilcoxon comparisons (continuous). Of note, covariates were available for all participants in the “Included” group, but missing for several in the non-included group (e.g., hypertension, N=4; hyperlipidemia or hypercholesterolemia, N=1; hemoglobin A1c, N=1).
| Characteristic | Included (N=32) | Not included (N=88) | P |
|---|---|---|---|
| Age | 45 (38-51) | 42 (30-51) | 0.16 |
| Female sex | 15 (47%) | 71 (81%) | 0.0003 |
| Race Caucasian Hispanic African-American |
30 (94%) 2 (6%) 0 (0%) |
69 (78%) 7 (8%) 12 (14%) |
0.08 |
| Type of surgery Sleeve gastrectomy Gastric bypass |
18 (56%) 14 (44%) |
72 (81%) 16 (18%) |
0.004 |
| Diabetes | 17 (53%) | 15 (17%) | <0.0001 |
| Hypertension | 20 (63%) | 41 (49%) | 0.19 |
| Hypercholesterolemia or hyperlipidemia | 15 (47%) | 30 (34%) | 0.22 |
| Body mass index | 45.0 (40.4-50.2) | 43.9 (40.6-49.9) | 0.78 |
| Systolic BP | 130 (118-138) | 126 (120-136) | 0.72 |
| Diastolic BP | 79 (73-83) | 80 (73-86) | 0.46 |
| Hemoglobin A1c | 6.6 (5.9-8.4) | 5.7 (5.4-6.1) | <0.0001 |
Plasma proteins dysregulated early and late post-bariatric surgery, tissues of origin, and relevant biological pathways
We found 27 proteins to be differentially expressed between Visit 1 and Visit 2 and 24 proteins between Visit 1 and Visit 3 (at a type 1 Bonferroni-adjusted P<0.05; Supplementary Table III). Figure 1A shows a Volcano plot demonstrating relative fold-change between Visit 1 and Visit 2 relative to significance level. Of note, 19 of 27 proteins differentially expressed between Visit 1 and Visit 2 were also differentially expressed at Visit 3 (Figure 1B), with a consistent directionality over time (Figure 1C). A box plot of expression of these proteins is shown in Supplementary Figure III.
Figure 1.
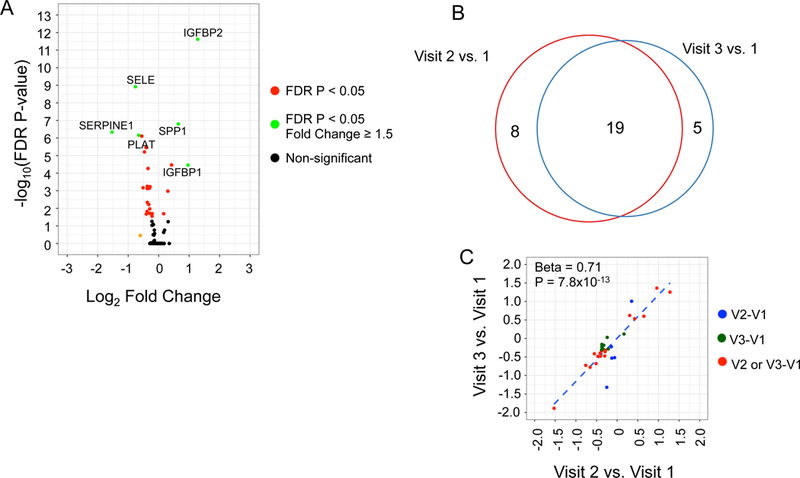
Differentially expressed plasma proteins after bariatric surgery. (A) Volcano plot shows the log-fold change and Bonferroni adjusted P-value between Visit 1 and Visit 2. The proteins highlighted in green have >1.5-fold change with statistical significance. (B) Venn diagram displaying the number of differentially expressed proteins between Visit 1 and the two post-operative visits after bariatric surgery. (C) Comparison of fold change (log2-scale) between proteins differentially expressed between Visit 1 and the two post-operative time points. The points shaded in red are differentially expressed between Visit 1 and both post-operative time points; the points in blue and green are differentially expressed for Visit 1 versus Visit 2 or Visit 3, respectively.
To address the functional significance of these results, we sought next to understand the tissue of origin for these 19 proteins, and the potential functional significance of these proteins to human disease processes via pathway analysis. First, to address the tissues of origin for these 19 differentially abundant proteins post-bariatric surgery, we analyzed whole transcriptome RNA-seq data from the EMBL-EBI’s RNASeq Atlas, consisting of 32 tissues from 122 individuals 17-19. Figure 2A shows the fold change for these 19 genes between the highest expressing tissue and the next highest expressing tissue in the human RNASeq Atlas. Out of the genes corresponding to the 19 proteins, 8 genes had the highest expression in liver relative to other non-gender specific tissues assayed in the RNAseq Atlas. The list of top two tissues expressing these 19 proteins and their corresponding fold changes are presented in Supplementary Table IV.
Figure 2.
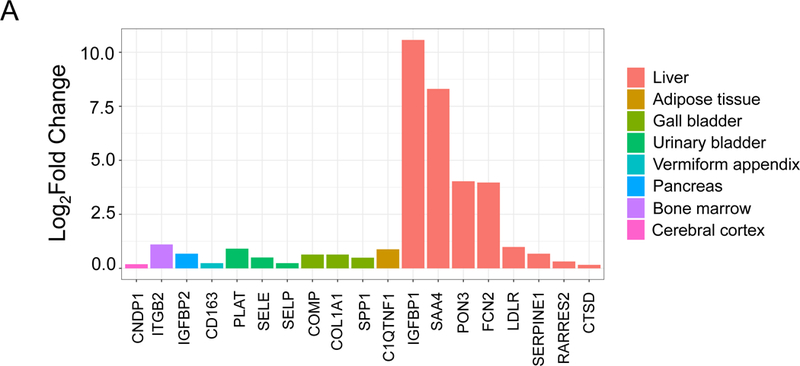
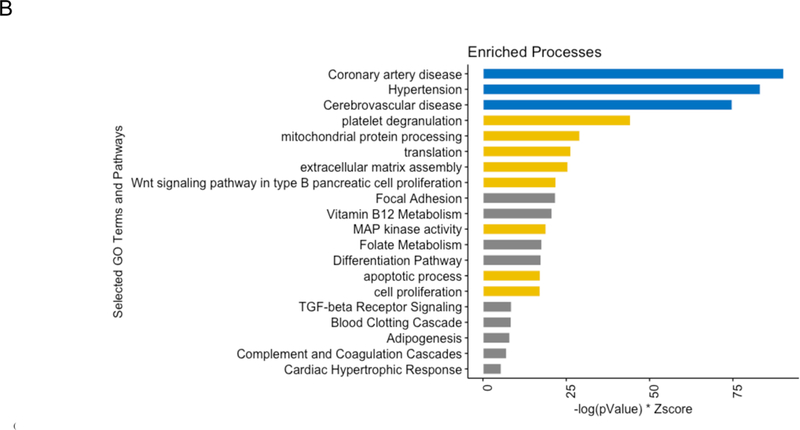
Functional analysis of 19 proteins differentially expressed in plasma after bariatric surgery. (A) Gene expression in tissue of 19 proteins dysregulated between Visit 1 versus both post-operative time points. The height of each bar represents the fold change between the tissue that has the highest expression of the gene coding the respective protein and the tissue that has the next highest expression of the gene. (B) Bar graph of significantly enriched processes. The results of an enrichment analysis performed on the set 26 proteins expanded from 19 proteins with differences in terms of pre- versus post-bariatric surgery are shown here, ranked by a score of -log(pValue) * Zscore. Three different sources of biological process terms were included: Jensen Disease Ontology (blue), Gene Ontology (gold), and WikiPathways (gray).
Given that pathway analytics from 19 proteins may be limited, we expanded the list of candidate targets via the framework discussed in Methods (above). Supplementary Table V lists an expanded list of 26 targets. From these targets, Figure 2B demonstrates the top biological and disease processes specified by these proteins, highlighting major obesity-related cardiometabolic disease illnesses (e.g., cerebrovascular disease, coronary artery disease and hypertension).
Association of candidate proteins with incident diabetes and metabolic syndrome in the FHS
Characteristics of FHS participants stratified by new-onset diabetes or metabolic syndrome are shown in Supplementary Table VI. Among the 4877 diabetes-free participants at study baseline, 192 (3.9%) developed new-onset diabetes at an average 6.3 years of follow-up. In contrast to 4685 diabetes-free participants, those who developed diabetes during follow-up had significantly lower baseline concentrations of IGFBP2, IGFBP1, and PAI-1 (adjusted for age and sex). In multivariable logistic regression for incident diabetes, only IGFBP-2 was associated negatively with risk of incident diabetes (Table 2). Of note, IGFBP-2 increased after bariatric surgery (Supplemental Table II).
Table 2.
Association of 6 candidate proteins with new-onset diabetes and new-onset metabolic syndrome in FHS. Each odds ratio is expressed per log-normalized unit of the protein biomarker. CI indicates confidence interval.
| Protein | New-onset diabetes | New-onset metabolic syndrome | ||||||
|---|---|---|---|---|---|---|---|---|
| Odds ratio | 95% CI | P | Odds ratio | 95% CI | P | |||
| CD163 | 0.84 | 0.61 | 1.15 | 0.28 | 0.83 | 0.67 | 1.03 | 0.08 |
| GMP-140 | 0.91 | 0.54 | 1.56 | 0.74 | 0.99 | 0.70 | 1.40 | 0.95 |
| IGFBP-1 | 1.05 | 0.85 | 1.31 | 0.63 | 0.87 | 0.75 | 1.01 | 0.06 |
| IGFBP-2 | 0.45 | 0.32 | 0.64 | 7.7×10−6 | 0.63 | 0.49 | 0.79 | 0.0001 |
| LDL-R | 0.86 | 0.72 | 1.03 | 0.10 | 0.88 | 0.78 | 1.00 | 0.04 |
| PAI-1 | 0.98 | 0.66 | 1.46 | 0.93 | 0.91 | 0.70 | 1.18 | 0.48 |
After we excluded participants with prevalent metabolic syndrome at baseline, we observed an 11% (428 in 4391) incidence of new-onset metabolic syndrome during an average follow-up of 6.3 years. Relative to those participants without metabolic syndrome, individuals who developed metabolic syndrome during follow-up were older and had more significant cardiometabolic risk factors at baseline (e.g., higher BMI, glucose, blood pressure), as well as different baseline concentrations of several candidate proteins (higher GMP-140, lower IGFBP1, IGFBP2; Supplementary Table VI). We observed that a higher concentration of IGFBP-2 at baseline was associated with a lower risk of incident metabolic syndrome after multivariable adjustment (Table 2).
DISCUSSION
We studied a targeted set of cardiometabolic disease-associated proteins in individuals before and after bariatric surgery, finding a set of proteins altered following early and late post-operative weight loss. Using RNA-seq tissue transcriptome data across multiple tissues in healthy individuals, we found that a significant proportion of genes encoding proteins differentially expressed post-bariatric surgery were expressed in the liver and adipose tissue, two major metabolic tissues known to change with weight loss. Pathway analyses suggested important roles of these genes/proteins in obesity-related cardiovascular disease, including vascular disease and hypertension. Finally, in a large longitudinal study of individuals in the community, we found that IGFBP-2 (altered after bariatric surgery) was inversely associated with incident metabolic syndrome and diabetes, after adjusting for established cardiometabolic disease risk factors. Collectively, these findings suggest the utility of discovery efforts rooted in extreme metabolic phenotypes (obesity and weight loss surgery) to implicate known and novel proteins for further mechanistic and prognostic study in obesity.
With the advent of rapid, quantitative profiling techniques, there has been a shift from studies characterizing a handful of circulating biomarkers to metabolome- 20-22, microbiome-23, 24, proteome- 25-27, and transcriptome-wide 28 approaches, including “integrating” all of these technologies 29. To our knowledge, large studies of individuals undergoing medical or surgical weight loss with monitoring of the plasma proteome have not been published, limiting direct comparison with the results presented here. In a study of serial proteomic alterations seven times during a modest (12%) dietary weight reduction followed weight maintenance in 43 obese subjects, Geyer and colleagues observed decreased plasma abundance of pro-inflammatory and pro-insulin resistance proteins with weight loss (e.g., C-reactive protein) 27. In a separate study, Piening and co-workers assessed genomic variation, metabolome, transcriptome, and proteome during dietary weight gain, maintenance, and loss in the same individuals over a 30-60 day period29. This “integrated”29 approach used a similar platform for proteome assessment (Olink) as performed here, with a wider coverage. Interestingly, despite subjects entering at a BMI 23-25 kg/m2 with a 3 kg average excursion during weight loss and maintenance, this study identified 27 proteins (at a false discovery rate of 20%) that were associated with BMI change over time, including MPO and P-selectin, both of which were identified in our study. There were few similarities in the proteins uncovered in this study and our study, likely owing to the differences in magnitude and duration of weight loss (with surgical vs. medical approaches).
Importantly, several proteins in our panel have been previously implicated in obesity and metabolic pathophenotypes in a directionally consistent fashion to what we observed, though large studies reaffirming their relationship with metabolic outcomes remain limited. While we interrogated a subset of proteins pre-selected for their involvement in cardiovascular disease (proprietary panels from Olink), we identified proteins that were known to have dynamic expression with weight change, as well as those not previously known to be responsive to intervention. One of the top targets identified in our study was IGFBP-2. IGFBP-2 expression in the liver is regulated by leptin30, a key hormone implicated in obesity and weight change. In turn, increased IGFBP-2 after leptin administration in rodent models of insulin resistance results in decreased hepatic glucose production31, linking weight change, leptin, IGFBP-2, and dysglycemia, a major component of metabolic syndrome. IGFBP-2 expression in mouse models decreases obesity and hypertension and improves insulin sensitivity32. In humans, IGFBP-2 may impact visceral fat metabolism (adipogenesis) 33. Consistent with our results in bariatric surgery and a more heterogeneous community study (FHS), lower IGFBP-2 levels have been associated with metabolic syndrome 34 and its phenotypes across the age spectrum35 and may increase early after bariatric surgery (24 hours), with a sustained response to 1 year36.
Similarly, CD163 had been associated with visceral fat content and distribution37 and responds to dietary weight reduction38, with recent studies suggesting that inflammatory cell expression of CD163 may be an even more sensitive marker for insulin resistance39. Finally, the selectins40, 41 and PAI-142, 43 have well-established associations with obesity, insulin resistance, and responsiveness to interventions targeting metabolic disease44, 45. While they provide important measure of external validity to the results here, these proteins only encompass a fraction of the overall discovered set in our bariatric cohort (Supplementary Table III). Future investigations in animal systems that target those proteins heretofore not extensively implicated in metabolic disease or obesity previously will provide an important avenue of data for further investigation in both animal and human obesity states.
Our pilot study has several limitations that impact generalizability. Limitations include (1) the fact that we used a pre-selected manufacturer’s protein panel with limited coverage of the proteome that only included those proteins known to be involved in cardiovascular and cardiometabolic disease and (2) the non-random selection strategy of non-fasting samples for protein quantification (Supplementary Figure I). The protein discovery in the bariatric surgery cohort was biased, in that we selected certain panels from the manufacturer that pre-determined the set of proteins quantified; as such, we were predestined to identify relevant pathways from vascular disease in our pathway analytics. A more unbiased discovery (e.g., mass spectrometry) might have not only uncovered more novel targets, but also might have provided coverage of post-translational protein modifications (potentially relevant in post-operative state). Sample selection in the bariatric study was non-random (as described above) and was a small discovery population, which may limit generalizability of the 19 or more targets that were uncovered. In addition, samples were run in two batches, with a clear batch effect for which we statistically corrected (alongside a proprietary normalization strategy), though residual batch and normalization effects may remain. Finally, we did not consider “split samples” across runs (e.g., pre-operative visit proteins quantified on Run 1, while post-operative on Run 2) to avoid additional “noise” from normalization. A more ideal study design would have employed quantification of all bariatric surgery participants with serial samples at the same time (run) to maximize power and internal validity. Certainly, sample selection in part enriched for insulin resistance, which limits the generalizability of all proteins to the overall benefits of bariatric surgery (e.g., on outcome). Future larger studies with a greater range of outcome are necessary to address this important point, as well as detection of lower abundance proteins that may be biologically relevant (but not included due to protein selection scheme here). Finally, given that we observed changes in weight at our early time-point and did not specifically investigate the effects of type of bariatric surgery (banding vs. bypass), we cannot pinpoint whether the changes observed are due to type of surgery or weight loss. Studies targeting an earlier time-point post-operatively would address this important physiologic concern. Finally, additional studies that evaluate serial tissue expression across multiple tissue types in humans may help address the tissue of origin for the differentially expressed proteins observed in this study. With regard to the FHS component, FHS independently selected their proteins for quantification, and we did not have the opportunity to validate all of the proteins identified in our bariatric surgery discovery. Despite these limitations, we did demonstrate (1) several directionally consistent relationships in a handful of proteins with incident metabolic diseases in FHS and (2) noted several identified proteins may have prior biological validity in insulin resistance, suggesting generalizability.
In conclusion, using a directed protein quantification platform in a group of individuals before and after bariatric surgery, we identified known and novel proteins altered after surgical weight loss, with implicated roles in cardiovascular and metabolic diseases. One of these proteins—IGFBP-2—was inversely associated with incident metabolic syndrome and diabetes after adjustment for clinical risk factors in a large population study. These efforts lay the groundwork for large-scale molecular profiling efforts in well-defined obesity interventions and large epidemiologic cohorts to define novel targets for investigation, manipulation, and clinical surveillance.
Supplementary Material
Highlights.
Using a targeted platform to quajntify proteins involved in cardiovascular and metabolic disease, we found a select group of proteins that are altered before and after bariatric surgery.
Insulin-like growth factors binding protein-2 (one of the proteins found in the bariatric surgery study) was associated with metabolic diseases in the Framingham Heart Study.
Identifying markers of metabolic function during weight loss may be a fruitful way to develop important insights into the biology of insulin resistance in humans.
ACKNOWLEDGEMENTS
none
Sources of Funding: This work was supported by U54HL112311 and U01HL126495 (to Dr. Freedman), and from NHLBI, Framingham Heart Study (National Heart, Lung, and Blood Institute/NIH contract No HHSN268201500001I). The Framingham Heart Study is funded by National Institutes of Health contract N01-HC-25195. This project was funded in part by the Division of Intramural Research, National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, MD. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
ABBREVIATIONS
- RNA
ribonucleic acid
- FHS
Framingham Heart Study
- PCR
polymerase chain reaction
- NT-proBNP
N-terminal pro-B-type natriuretic peptide
- IGFBP-2
Insulin-like growth factor binding protein-2
Footnotes
Disclosures: Dr. Shah receives grants from the National Institutes of Heath, and within the last 12 months has received funds from Amgen (scientific advisory board), Myokardia (consulting), and Best Doctors (consulting), none of whom had a role in the study. Dr. Shah is a co-inventor on a patent for ex-RNAs signatures of cardiac remodeling. Dr. Murthy has minor stock holdings in General Electric.
REFERENCES
- 1.Tanriverdi K, Kucukural A, Mikhalev E, Tanriverdi SE, Lee R, Ambros VR, Freedman JE. Comparison of RNA isolation and associated methods for extracellular RNA detection by high-throughput quantitative polymerase chain reaction. Analytical biochemistry. 2016;501:66–74. [DOI] [PubMed] [Google Scholar]
- 2.Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, Hwang S, Massaro J, Larson M, Levy D. Protein Biomarkers of Cardiovascular Disease and Mortality in the Community. Journal of the American Heart Association. 2018;e008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. [DOI] [PubMed] [Google Scholar]
- 4.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RB Sr., Fox CS, Larson MG, Murabito JM, O’Donnell CJ, Vasan RS, Wolf PA, Levy D. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. [DOI] [PubMed] [Google Scholar]
- 5.Agresti A, Kateri M. Ordinal probability effect measures for group comparisons in multinomial cumulative link models. Biometrics. 2017;73:214–219. [DOI] [PubMed] [Google Scholar]
- 6.dupont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66:175–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patro R, Duggal G, Kingsford C. Salmon: Accurate, Versatile and Ultrafast Quantification from RNA-seq Data using Lightweight-Alignment. bioRxiv 21592. [Google Scholar]
- 9.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research. 2017;45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pletscher-Frankild S, Palleja A, Tsafou K, Binder JX, Jensen LJ. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–9. [DOI] [PubMed] [Google Scholar]
- 14.Kutmon M, Riutta A, Nunes N, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic acids research. 2016;44:D488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djureinovic D, Hallstrom BM, Horie M, et al. Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight. 2016;1:e86837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habuka M, Fagerberg L, Hallstrom BM, Ponten F, Yamamoto T, Uhlen M. The Urinary Bladder Transcriptome and Proteome Defined by Transcriptomics and Antibody-Based Profiling. PLoS One. 2015;10:e0145301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 20.Narath SH, Mautner SI, Svehlikova E, Schultes B, Pieber TR, Sinner FM, Gander E, Libiseller G, Schimek MG, Sourij H, Magnes C. An Untargeted Metabolomics Approach to Characterize Short-Term and Long-Term Metabolic Changes after Bariatric Surgery. PLoS One. 2016;11:e0161425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberbach A, von Bergen M, Bluher S, Lehmann S, Till H. Combined serum proteomic and metabonomic profiling after laparoscopic sleeve gastrectomy in children and adolescents. Journal of laparoendoscopic & advanced surgical techniques Part A. 2012;22:184–8. [DOI] [PubMed] [Google Scholar]
- 22.Laferrere B, Reilly D, Arias S, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Science translational medicine. 2011;3:80re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Hong J, Xu X, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nature medicine. 2017;23:859–868. [DOI] [PubMed] [Google Scholar]
- 24.Liou AP, Paziuk M, Luevano JM Jr., Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Science translational medicine. 2013;5:178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell LE, Langlais PR, Day SE, Coletta RL, Benjamin TR, De Filippis EA, Madura JA 2nd, Mandarino LJ, Roust LR, Coletta DK. Identification of Novel Changes in Human Skeletal Muscle Proteome After Roux-en-Y Gastric Bypass Surgery. Diabetes. 2016;65:2724–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo P, Yu H, Zhao X, Bao Y, Hong CS, Zhang P, Tu Y, Yin P, Gao P, Wei L, Zhuang Z, Jia W, Xu G. Metabolomics Study of Roux-en-Y Gastric Bypass Surgery (RYGB) to Treat Type 2 Diabetes Patients Based on Ultraperformance Liquid Chromatography-Mass Spectrometry. Journal of proteome research. 2016;15:1288–99. [DOI] [PubMed] [Google Scholar]
- 27.Geyer PE, Wewer Albrechtsen NJ, Tyanova S, Grassl N, Iepsen EW, Lundgren J, Madsbad S, Holst JJ, Torekov SS, Mann M. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Molecular systems biology. 2016;12:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinhel MAS, Noronha NY, Nicoletti CF, de Oliveira BAP, Cortes-Oliveira C, Pinhanelli VC, Salgado Junior W, Machry AJ, da Silva Junior WA, Souza DRS, Marchini JS, Nonino CB. Changes in Global Transcriptional Profiling of Women Following Obesity Surgery Bypass. Obesity surgery. 2018;28:176–186. [DOI] [PubMed] [Google Scholar]
- 29.Piening BD, Zhou W, Contrepois K, et al. Integrative Personal Omics Profiles during Periods of Weight Gain and Loss. Cell systems. 2018;6:157–170 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma A, Bartell SM, Baile CA, Chen B, Podolsky RH, McIndoe RA and She JX. Hepatic gene expression profiling reveals key pathways involved in leptin-mediated weight loss in ob/ob mice. PLoS One. 2010;5:e12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedbacker K, Birsoy K, Wysocki RW, Asilmaz E, Ahima RS, Farooqi IS, Friedman JM. Antidiabetic effects of IGFBP2, a leptin-regulated gene. Cell metabolism. 2010;11:11–22. [DOI] [PubMed] [Google Scholar]
- 32.Wheatcroft SB, Kearney MT, Shah AM, Ezzat VA, Miell JR, Modo M, Williams SC, Cawthorn WP, Medina-Gomez G, Vidal-Puig A, Sethi JK, Crossey PA. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes. 2007;56:285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yau SW, Russo VC, Clarke IJ, Dunshea FR, Werther GA, Sabin MA. IGFBP-2 inhibits adipogenesis and lipogenesis in human visceral, but not subcutaneous, adipocytes. Int J Obes (Lond). 2015;39:770–81. [DOI] [PubMed] [Google Scholar]
- 34.Heald AH, Kaushal K, Siddals KW, Rudenski AS, Anderson SG, Gibson JM. Insulin-like growth factor binding protein-2 (IGFBP-2) is a marker for the metabolic syndrome. Experimental and clinical endocrinology & diabetes : official journal, German Society of Endocrinology [and] German Diabetes Association. 2006;114:371–6. [DOI] [PubMed] [Google Scholar]
- 35.Yau SW, Harcourt BE, Kao KT, Alexander EJ, Russo VC, Werther GA, Sabin MA. Serum IGFBP-2 levels are associated with reduced insulin sensitivity in obese children. Clinical obesity. 2018;8:184–190. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Martin J, Poirier P, Caron-Cantin SM, Hould FS, Marceau S, Marceau P, Picard F. Upregulation of plasma insulin-like growth factor binding protein 2 levels after biliopancreatic diversion in humans. Obesity (Silver Spring). 2012;20:1469–73. [DOI] [PubMed] [Google Scholar]
- 37.Sorensen LP, Parkner T, Sondergaard E, Bibby BM, Moller HJ, Nielsen S. Visceral obesity is associated with increased soluble CD163 concentration in men with type 2 diabetes mellitus. Endocrine connections. 2015;4:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fjeldborg K, Christiansen T, Bennetzen M, H JM, Pedersen SB, Richelsen B. The macrophage-specific serum marker, soluble CD163, is increased in obesity and reduced after dietary-induced weight loss. Obesity (Silver Spring). 2013;21:2437–43. [DOI] [PubMed] [Google Scholar]
- 39.Kawarabayashi R, Motoyama K, Nakamura M, Yamazaki Y, Morioka T, Mori K, Fukumoto S, Imanishi Y, Shioi A, Shoji T, Emoto M, Inaba M. The Association between Monocyte Surface CD163 and Insulin Resistance in Patients with Type 2 Diabetes. Journal of diabetes research. 2017;2017:6549242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto K, Sera Y, Abe Y, Tominaga T, Horikami K, Hirao K, Ueki Y, Miyake S. High serum concentrations of soluble E-selectin correlate with obesity but not fat distribution in patients with type 2 diabetes mellitus. Metabolism. 2002;51:932–4. [DOI] [PubMed] [Google Scholar]
- 41.Glowinska B, Urban M, Peczynska J, Florys B. Soluble adhesion molecules (sICAM-1, sVCAM-1) and selectins (sE selectin, sP selectin, sL selectin) levels in children and adolescents with obesity, hypertension, and diabetes. Metabolism. 2005;54:1020–6. [DOI] [PubMed] [Google Scholar]
- 42.Ma LJ, Mao SL, Taylor KL, Kanjanabuch T, Guan Y, Zhang Y, Brown NJ, Swift LL, McGuinness OP, Wasserman DH, Vaughan DE, Fogo AB. Prevention of obesity and insulin resistance in mice lacking plasminogen activator inhibitor 1. Diabetes. 2004;53:336–46. [DOI] [PubMed] [Google Scholar]
- 43.Schneider DJ, Sobel BE. PAI-1 and diabetes: a journey from the bench to the bedside. Diabetes Care. 2012;35:1961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanni MV, Stanley TL, Makimura H, Chen CY, Grinspoon SK. Effects of TNF-alpha antagonism on E-selectin in obese subjects with metabolic dysregulation. Clinical endocrinology. 2010;73:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skurk T, Hauner H. Obesity and impaired fibrinolysis: role of adipose production of plasminogen activator inhibitor-1. Int J Obes Relat Metab Disord. 2004;28:1357–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


