Abstract
The ESCRTs catalyze reverse-topology scission from the inner face of membrane necks in HIV budding, multivesicular endosome biogenesis, cytokinesis, and other pathways. We encapsulated ESCRT-III subunits Snf7, Vps24, and Vps2, and the AAA+ ATPase Vps4 such that membrane nanotubes reflecting the correct topology of scission could be pulled from giant vesicles. Upon ATP release by photo-uncaging, this system was capable of generating forces within the nanotubes in a manner dependent upon Vps4 catalytic activity and Vps4 coupling to the ESCRT-III proteins. Imaging of scission revealed Snf7 and Vps4 puncta within nanotubes whose presence followed ATP release, correlated with force generation and nanotube constriction, and preceded scission. These observations directly verify long-standing predictions that ATP-hydrolyzing assemblies of ESCRT-III and Vps4 sever membranes.
One Sentence Summary:
ESCRT-III and Vps4 were reconstituted from within the interior of nanotubes pulled from giant vesicles, revealing that this machinery harnesses ATP-dependent force production for membrane scission.
Main Text:
Cellular membranes are constantly remodeled in the course of vesicular trafficking, cell division, the egress of HIV, and many other processes. Membranes can bud and be severed either towards or away from the cytosol. The latter is referred to as “reverse topology” scission and is catalyzed by the ESCRT machinery, a set of ~18 proteins in yeast and ~28 in mammals (1–4). The core machinery of membrane scission by the ESCRTs consists of the ESCRT-III protein family. The most important components for membrane scission are Snf7, Vps24, and Vps2 (5, 6). When recruited to membranes, ESCRT-III proteins assemble into flat spiral discs (7–9), helical tubes (7, 10, 11), or conical funnels (11–13). ESCRT filaments have a preferred curvature (8, 9, 14). When they are bent to curvatures of higher or lower values, ESCRT filaments act as springs that restore their own shape to the preferred value (9, 15, 16). This spring-like behavior has led to the prediction that ESCRTs could exert measurable forces upon membranes, which we set out to test.
The AAA+ ATPase Vps4 (17) is intimately associated with the ESCRT machinery and is essential for the membrane scission cycle. Vps4 is recruited to scission sites by Vps2 (18, 19). Vps2 is thought to have a capping role whereby it inhibits Snf7 polymerization (6). By recycling Vps2 (20), Vps4 promotes Snf7 polymerization. Thus, Vps4 is critical for the recycling of ESCRT-III and the replenishment of the soluble cytoplasmic pool. Early attempts at in vitro reconstitution of ESCRT-mediated budding and scission using giant unilamellar vesicles (GUVs) suggested that the process was independent of Vps4 and ATP (21, 22), except for the final post-scission recycling step. Cell imaging studies (23–28), however, showed that Vps4 localization peaked prior to scission in HIV-1 budding and cytokinesis, consistent with its direct role in scission upon ATP hydrolysis. A second goal of this study was to determine if Vps4 and ATP hydrolysis were directly involved in membrane scission, as opposed to mere recycling.
We encapsulated the minimal ESCRT-III-Vps4 module containing yeast Snf7, Vps24, Vps2, and Vps4 (referred to hereafter as the “module;” Fig. S1), in POPC:POPS:Biotinyl-PE (80:20:0.1) GUVs at near-physiological ionic strength (~150 mM NaCl; Fig. 1D–G). We used optical tweezers to pull nanotubes extending between the surface of a GUV held by suction on an aspiration pipette and the surface of a streptavidin-coated polystyrene bead held by an optical trap (Fig. 1A–C). To fuel the AAA+ ATPase Vps4, we also encapsulated the caged ATP analog NPE-ATP. An optical fiber was used to UV-illuminate one GUV at a time so that experiments could be carried out sequentially on individual GUVs in the same microfluidic observation chamber. In control experiments, where all components were included except for ATP, UV illumination led to no change in the force exerted on the bead (Fig. 1H, 2B). In similar control experiments omitting only Vps4, UV illumination results in a slight drop in the pulling force (Fig. 1I, 2C), attributed to the generation of two product molecules upon NPE-ATP uncaging. Thus, in the absence of ESCRT activity the membrane nanotube is stable. When ATP was uncaged in the presence of the complete ESCRT module, a large rise in retraction force was indeed observed (Fig. 1J, 2I, Movie S1). Over ~2–10 min (Fig 1J), the force exceeded the trap maximum of ~65 pN and pulled the bead out of the laser trap (Movie S1). This showed that in the presence of ATP, the ESCRT module can exert forces on membranes.
Fig. 1. ESCRT-III exerts an ATP dependent force on membrane tubes.
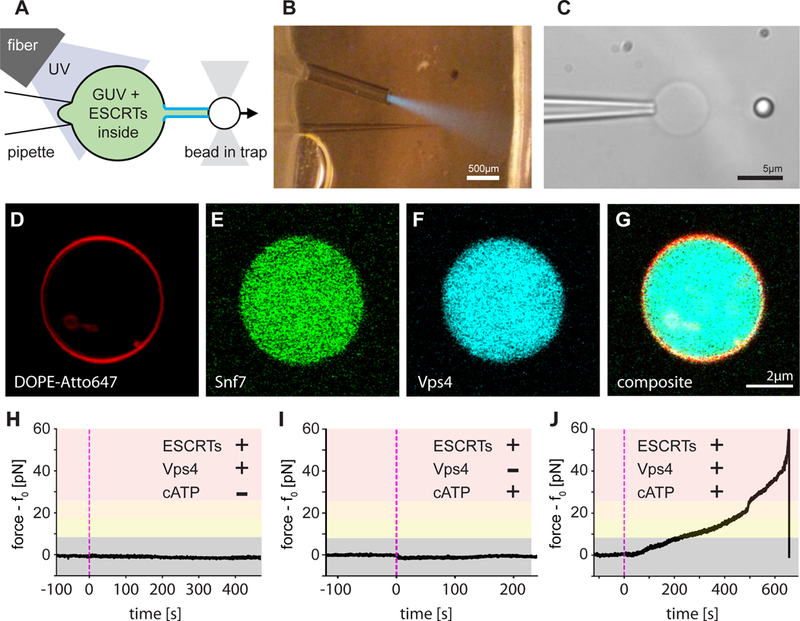
(A) Schematic of the experiment: a membrane tube (middle) is pulled out of a micropipette-aspirated GUV with a functionalized bead held in an optical trap, creating a reverse-curvature topology. Components of the ESCRT machinery and caged ATP are encapsulated in the lumen of the GUV. An optical fiber delivers UV light to uncage the ATP inside the vesicle and start the reaction. (B) Aspiration pipette, optical fiber, and UV light cone (blue) inside the microfluidic chamber for our experiments. (C) GUV (center) aspirated by the micropipette (left) and a tube-pulling bead (right). (D to G) Labeling different components of the ESCRT module (membrane label in (D), Snf7 in (E), Vps4 in (F), and merged in (G), 2 μM for all components) revealed a uniform distribution of proteins in the lumen of GUVs). (H to J) Force profile over time detected by optical tweezers on a membrane tube pulled from a GUV that encapsulates components of the ESCRT module. (H) Control experiment on a full ESCRT module but omitting ATP. No change in force measurable. (I) Control experiment on a full ESCRT module but omitting Vps4. Apart from a minute dip in the force profile, which is due to small changes in osmolarity upon ATP uncaging, no effects were detected. (J) ATP uncaging (black arrow head) in the presence of a full ESCRT module leads to a rise in force exerted on the tube, which is connected to the bead held by the optical trap. A large rise in force can overcome the trapping strength and pull the bead out of the trap.
Fig. 2. Molecular determinants of force generation.
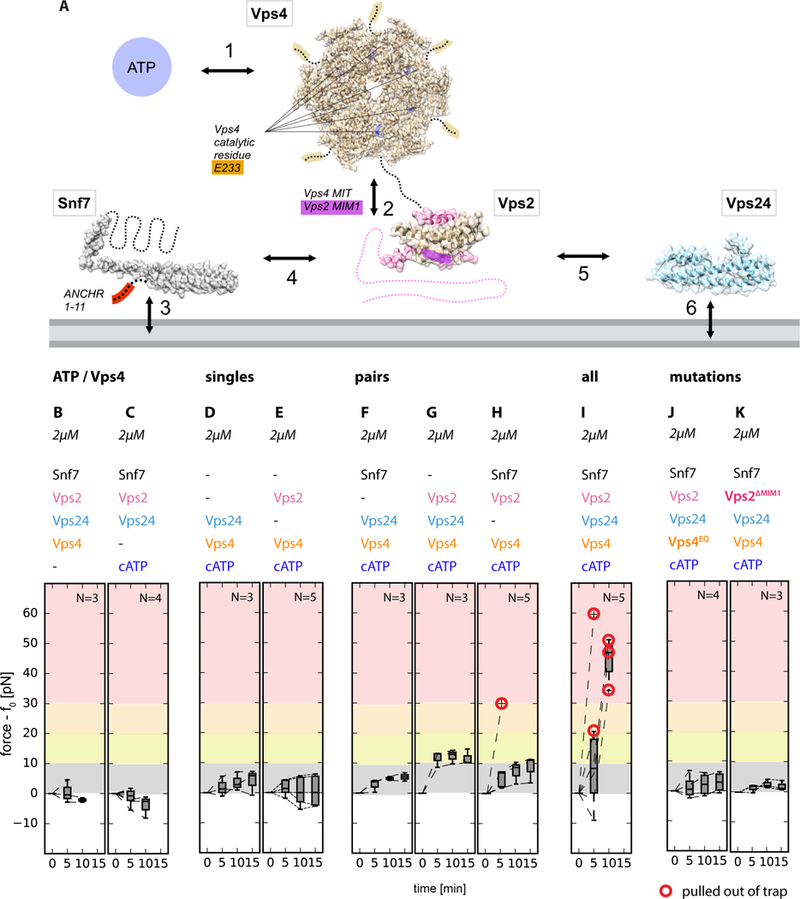
(A) Interaction network of the ESCRT module. ESCRT proteins (filled-space structures and dashed lines) interact with the membrane (grey, bottom) as well as with each other. Key components (ATP), catalytic sites, and interacting motifs are highlighted in colors. (B to K) Individual components of the module contribute differently to the force exerted on membrane tubes: ATP/Vps4 are essential for force generation (B, C), so is ATP hydrolysis (J, catalytically dead Vps4EQ mutant) and critical protein domains (K, Vps2 ΔMIM1). Only the full module (I), or pairings of Vps2+Vps24 and Snf7+Vps2 (G, H), lead to significant force generation. Our data are consistent with the known interactions underpinning the activity of the ESCRT machinery.
We sought to determine which components of the ESCRT module were required for force generation (Fig. 2A). With Vps2 or Vps24 as the only ESCRT-III subunits, essentially no force was generated (Fig. 2D, E). In the presence of Snf7, omission of Vps2 or Vps24 led to little or no force generation (Fig. 2F, H), consistent with the role of Vps2 in coupling of ATP hydrolysis by Vps4 to ESCRT-III remodeling, and a role of Vps24 in co-polymerizing with Vps2. When both Vps2 and Vps24 were present, but missing Snf7, a force rise of up to 12 pN was produced, consistent with the ability of Vps24 and Vps2 to co-polymerize (10, 20) (Fig. 2G). The inactivated mutant E233Q of Vps4 (17) failed to generate force (Fig. 2J). Deletion of the Vps4-coupling MIM1 motif of Vps2 (Fig. 2K) (18, 19) essential for biological function abrogated force production. Thus, the ability of the ESCRT module to exert forces on nanotubes, in an ATP-dependent manner (Fig. 2B, C), correlates closely with the presence of all the components that are crucial for ESCRT-mediated membrane scission and their individual integrity (Fig. 2I).
We integrated a confocal microscope with optical tweezing capability to image membrane nanotubes pulled from GUVs containing fluorophore-labeled ESCRTs (Fig. 3A–E, Fig. S2). By pulling on bare membranes, we obtained the bending modulus κ and standardized the calculation of the membrane nanotube radius (Fig. S3). To maximize the signal in these experiments, Snf7 was labeled with the photo-stable dye, Lumidyne-550, and imaged with a resonant scanner and a GaAsP detector. We quantitated Snf7, Vps4, and membrane intensity using Gaussian fitting to the diffraction limited tube profile (Fig. S4).
Fig. 3. Confocal imaging of ATP-dependent membrane tube scission by ESCRTs.
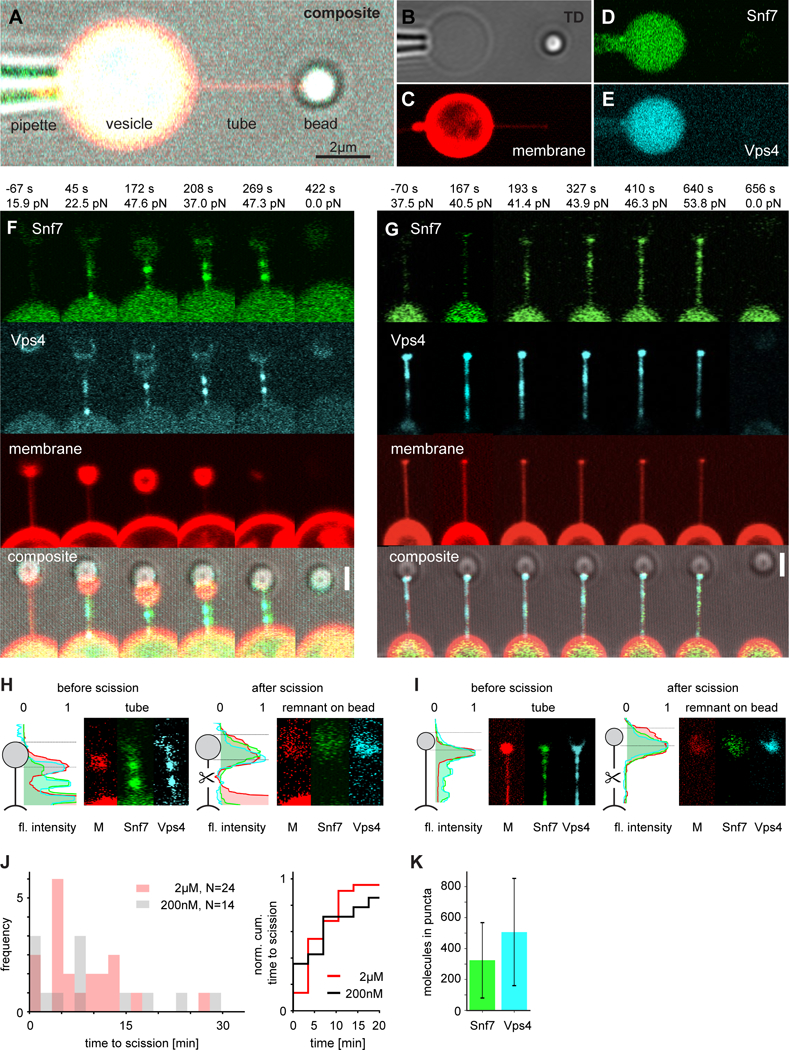
(A to E) Micrograph of the experiment setup: the GUV (center, yellow) encapsulating the ESCRT module and caged ATP is aspirated by a micropipette (left). A membrane nanotube was pulled from the GUV using a bead (right) in an optical trap. Membrane, Snf7, and Vps4 are respectively labeled with different fluorophores (composite (A) individual channels shown in (B to E), scale bar: 2μm). (F and G) Progression of two representative membrane scission events (2 of 17). UV illumination at t = 0 s. Snf7 (green) and Vps4 (cyan) puncta become visible at the vesicle-tube-junction and migrate into the tube. Scission happens at ~410 s and ~650 s, respectively. Scale bar 1.5μm. (H and I) Tube remnants on the bead identify scission in the middle of the tube. Brightness adjusted compared to F and G. (J) Quantification of tube scission. (K) Protein copy number in puncta.
We monitored the response of a total of 46 nanotubes pulled from ESCRT module-filled GUVs following UV illumination. The nanotubes manifested a force increase and accumulation of Snf7 and Vps4 (Fig. 3F–I, Movie S2). Of the 46 trials, 38 (83%) led to scission (Fig. 3J) as judged by simultaneous disappearance of the tube, sudden decrease in the force to zero, and appearance of membrane-, Snf7-, and Vps4-containing remnants on the trapped bead (Fig. 3H, I). Tube lifetimes were widely distributed (Fig. 3J), with a mean lifetime of 425 s before scission. The distributions of lifetimes were similar at the superphysiological concentration of 2 μM, which was used to facilitate visualization, and the near-physiological value of 200 nM (Fig. 3J). One to two diffraction-limited puncta of Snf7 intensity appeared at > 7 SD in 11 out of 17 events analyzed. Typically the puncta nucleated at the tube-vesicle junction (Fig. 3F, G, Movie S2). Subsequently, the puncta sometimes moved or disappeared within the tubes. Snf7 puncta were essentially always co-localized with Vps4 (Fig. 3F, G). Working at 2 μM, puncta contained in the range of 100–600 copies of Snf7 (Fig. 3K), which could exceed the minimum needed for scission, yet is also roughly consistent with estimates in yeast cells (29).
In order to understand the relationship between the observed force (Fig. 4A, B) and bulk and microscopic properties of the system, we quantitated the nanotube radius over time. Nanotubes used in this study were typically of radius r ~ 20 nm prior to ATP release (Fig. 4C, D). The tubes began to narrow almost immediately following ATP release. Narrowing was associated with an increase in the amount of Snf7 and Vps4 seen in the tubes (Fig. 4E, F), whilst essentially no change was seen in the intensity of the GUV membrane or the amount of Snf7 and Vps4 associated with the GUV (Fig. 4G, H). This behavior was consistent over 17 traces (Fig. 4 K–N). Final values of r reached 5–10 nm by the time of scission. The uncertainty in the final values of r is substantial because the membrane fluorescence signal is weak when r < 10 nm. In the case of constant membrane tension σ and bending modulus κ, the initial and final force and radius are related to each other by and Fig. 4A–D shows the values of f (Fig. 4A, B) and r (Fig. 4C, D) computed from the experimental values of r and f, respectively, with no adjustable parameters. The correlation between the measured and computed values is generally excellent (Fig. 4O). Consistent with an apparently constant κ, there is no accumulation of Snf7 on the GUV membrane over the course of the scission events (Fig. 4I). Consistent with a constant σ, the length of the membrane tongue in the pipet was typically constant (Fig. 4J). The formation of ESCRT puncta imposes nanotube radii r smaller than the equilibrium radius r0, which is associated with an increased pulling force f > f0. These findings are consistent with force generation by punctate microscopic assemblies of ESCRTs within the nanotubes, leading to the constriction of membrane tubes followed by scission.
Fig. 4. ESCRT membrane scission is mediated by tube constriction.
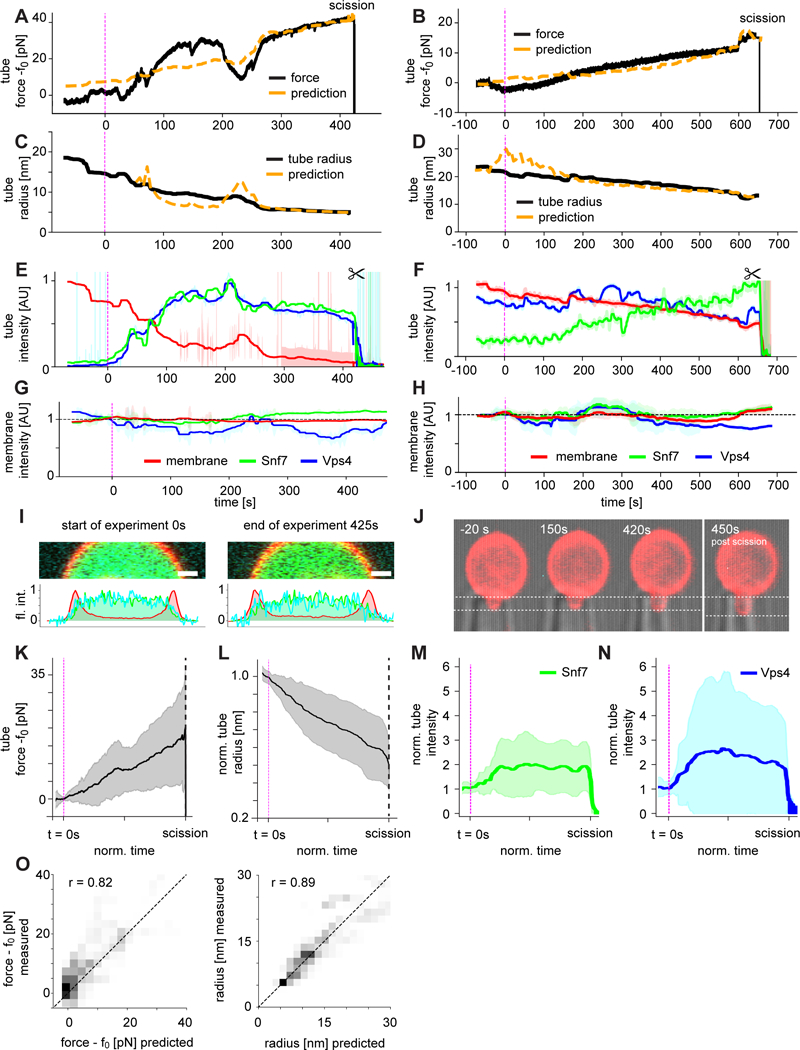
(A and B) Force profiles of the same events shown in Fig. 3 were normalized to their respective baseline tube holding force f0. ATP released by UV uncaging leads to a rise in force and eventually scission (indicated by the sharp drop of force at the end of the trace, 2 of 17). (C and D) Simultaneous observation of the tube radius revealed a continuous radius decrease. Force and radius plots are overlaid with predictions from the Helfrich model for a tube of unchanged bending rigidity under constant tension (dashed orange lines, see Methods). (E and F) Normalized and bleaching-corrected fluorescence intensity profiles of the scission events for the tube in the three fluorescence channels (Snf7, green; Vps4, cyan; membrane, red; ± SD shown as shaded area). After ATP uncaging, the tube membrane fluorescence intensity decreased while the tube Snf7 and Vps4 intensities increased. (G and H) The GUV membrane fluorescence intensity profile over the time course of the scission event was essentially constant and showed no correlation with the force (Snf7, green; Vps4, cyan; membrane, red; ± SD shown as shaded area). (I) Fluorescence intensity profiles of the GUV membrane at the start and at the end of the scission trace. No increase in protein fluorescence on the membrane was detected. (J) GUV aspiration projection at the beginning, middle and end of the scission trace, showing essentially no change. (K to N) Averages (N = 17 ± SD) of force, radius and tube fluorescence show consistent force rise, radius decrease and fluorescence increase in the tube. (O) Correlation (r) between experiment and prediction for radius and force, N=17.
It has been inferred that the core ESCRT-III proteins Snf7, Vps24, and Vps2, together with Vps4, comprise the minimal ATP-dependent scission machinery (1, 4, 30, 31). Here, we directly confirmed this idea by visualizing scission in a minimal system that replicates a wide range of biologically validated structure-function relationships.
The most striking finding from the reconstituted system is that the core ESCRT-III proteins and Vps4 together exert an ATP-dependent axial force on the nanotube before severing. It had previously been proposed (8) and then demonstrated (9) that Snf7 filaments have a preferred curvature and may exert forces when bent above or below their preferred value. It had also been hypothesized that breakage or remodeling of ESCRT filaments by Vps4 could contribute to force generation (4, 15, 20, 32). Our observations now provide experimental confirmation that ESCRTs indeed can generate force from within a narrow membrane tube, and show that this force contributes to membrane constriction and is correlated with reverse-topology membrane scission.
ESCRTs (7, 10, 11) and the “normal topology” scission factor dynamin (33) have been visualized as cylindrical membrane coats, and Snf7 has also been seen in the form of large spirals of hundreds to thousands of copies (9). In the case of dynamin, extended coating of the tube is not needed, and one or a few rings appear capable of mediating scission (34). Our measurements of scission by diffraction-limited puncta of Snf7 are consistent with imaging in cells (29) and with scission by rings or cones of molecular dimensions. The mechanical nature of constriction and force generation remains to be elucidated through structural approaches.
Supplementary Material
Acknowledgments:
We thank J.-Y. Lee, H. Aaron, S. Ruzin and D. Schichnes for assistance with imaging, M. Vahey, D. Fletcher, and P. Lishko for advice on the aspiration pipette set-up, A. Lee for assistance with the optical trap force calibration and C. Glick for advice with the microfluidics.
Funding:
Research was supported by a Marie Skłodowska-Curie postdoctoral fellowship ‘smStruct’ (J. S.), an NSF predoctoral fellowship (M. R. P.), NIH grant R01AI112442 (J.H.H.), the Max Planck Society (A.H.B. and G.H.), and the German Research Foundation CRC 807 (A.H.B. and G.H.).
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: All data are available in the main text or the supplementary materials. Code is available at https://github.com/JohSchoeneberg/Confleezers.
Supplementary Materials:
Materials and Methods
Figures S1–S4
Movies S1–S2
References
- 1.McCullough J, Colf LA, Sundquist WI, Membrane Fission Reactions of the Mammalian ESCRT Pathway. Annu. Rev. Biochem 82, 663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olmos Y, Carlton JG, The ESCRT machinery: new roles at new holes. Curr. Opin. Cell Biol 38, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campsteijn C, Vietri M, Stenmark H, Novel ESCRT functions in cell biology: spiraling out of control? Curr. Opin. Cell Biol 41, 1 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Schöneberg J, Lee I-H, Iwasa JH, Hurley JH, Reverse-topology membrane scission by the ESCRT proteins. Nature Reviews Molecular Cell Biology 18, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD, ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3, 271 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Teis D, Saksena S, Emr SD, Ordered Assembly of the ESCRT-III Complex on Endosomes Is Required to Sequester Cargo during MVB Formation. Dev. Cell 15, 578 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Hanson PI, Roth R, Lin Y, Heuser JE, Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. The Journal of Cell Biology 180, 389 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Q-T et al. , Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. The Journal of Cell Biology 206, 763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiaruttini N et al. , Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 163, 866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lata S et al. , Helical Structures of ESCRT-III are Disassembled by VPS4. Science 321, 1354 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullough J et al. , Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobro MJ et al. , Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Molecular Biology of the Cell 24, 2319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cashikar AG et al. , Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife e02184, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenz M, Crow DJG, Joanny JF, Membrane Buckling Induced by Curved Filaments. Phys. Rev. Lett 103, (2009). [DOI] [PubMed] [Google Scholar]
- 15.Carlson L-A, Shen Q-T, Pavlin MR, Hurley JH, ESCRT Filaments as Spiral Springs. Dev. Cell 35, 397 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Chiaruttini N, Roux A, Dynamic and elastic shape transitions in curved ESCRT-III filaments. Curr. Opin. Cell Biol 47, 126 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Babst M, Wendland B, Estepa EJ, Emr SD, The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17, 2982 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuchell-Brereton M et al. , ESCRT-III recognition by VPS4 ATPases. Nature 449, 740 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Obita T et al. , Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Mierzwa BE et al. , Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol 19, 787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH, Membrane scission by the ESCRT-III complex. Nature 458, 172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wollert T, Hurley JH, Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumgartel VBV et al. , Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol 13, 469 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Jouvenet NJN, Zhadina M, Bieniasz PD, Simon SM, Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol 13, 394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elia N, Sougrat R, Spurlin T, Hurley JH, Lippincott-Schwartz J, Dynamics of ESCRT machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A 108, 4846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guizetti J et al. , Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Bleck M et al. , Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc. Natl. Acad. Sci. U. S. A 111, 12211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adell MAY et al. , Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. The Journal of Cell Biology 205, 33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adell MAY et al. , Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. Elife 6, e31652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson PI, Cashikar A, Multivesicular Body Morphogenesis. Annu. Rev. Cell. Dev. Biol 28, 337 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Henne WM, Stenmark H, Emr SD, Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harbor Perspectives on Biology 5, a016766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J, Computational Model of Cytokinetic Abscission Driven by ESCRT-III Polymerization and Remodeling. Biophys. J 102, 2309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonny B et al. , Membrane fission by dynamin: what we know and what we need to know. EMBO J 35, 2270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shnyrova AV et al. , Geometric Catalysis of Membrane Fission Driven by Flexible Dynamin Rings. Science 339, 1433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prevost C et al. , IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nature Communications 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorre B et al. , Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. U. S. A 106, 5622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derényi IJ, F.; Prost, J., Formation and interaction of membrane tubes. Phys. Rev. Lett 88, 238101 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


