Abstract
The epidermal growth factor receptor (EGFR) is a promising target for cancer therapy. The presence of certain somatic mutations in the tyrosine kinase (TK) domain of the EGFR gene is associated with clinical response to TK inhibitors (TKI) in patients with lung adenocarcinoma. In this study we evaluated the status of somatic mutations in the entire TK domain of the EGFR gene by direct sequencing using early passage peritoneal mesothelioma cells, established cell lines as well as 33 peritoneal mesothelioma tumor samples. No novel mutations were found in the cell lines. Sequence analysis of the EGFR TK domain revealed the presence of a silent polymorphism (c.2607G→A, Q787Q) at exon 20 of both peritoneal mesothelioma cell lines as well as tumor specimens. The frequency of genotypes AA and GA was 42.8 and 57.2% in the cell lines and 33.3 and 57.6% in tumor specimens, respectively. The TKI erlotinib showed an IC50 in the range of 10–50 μM in five out of the seven cell lines with a GA genotype while all five cell lines with the AA genotype had an IC50 >50 μM. Of the 33 peritoneal mesothelioma tumor samples analyzed none had an EGFR TKI sensitizing mutation and only one specimen showed an earlier reported somatic mutation at codon 850 in exon 21 of the EGFR gene. Our data show that patients with peritoneal mesothelioma do not harbor somatic mutations in the EGFR TK domain that would make them sensitive to EGFR TKI.
Keywords: EGFR, peritoneal mesothelioma, somatic mutation, tyrosine kinase inhibitor, erlotinib
Introduction
Malignant mesothelioma is a tumor of the lining of the pleural and peritoneal cavities that is associated with prior asbestos exposure (1). However, 15–20% of cases with mesothelioma have no history of exposure to asbestos. An estimated 3,000 new cases per year are diagnosed in the United States out of which diffuse peritoneal mesothelioma accounts for approximately 10–30% of the cases (2). In patients with pleural mesothelioma aggressive surgical resection is an option in some patients with early stage disease. In a large randomized phase III clinical trial of patients with unresectable disease, systemic chemotherapy with pemetrexed and cisplatin resulted in improved overall survival as compared to cisplatin alone and is the current standard of care. However, the median survival of these patients was only 12.1 months (3). In contrast to patients with pleural mesothelioma, the prognosis of patients with peritoneal mesothelioma is significantly better, with a median overall survival greater than 5 years in those patients who are candidates for cytoreductive surgery and intraoperative chemotherapy (4,5).
Epidermal growth factor receptor (EGFR) is a member of the ErbB family of receptor tyrosine kinases (TK) and it plays a major role in proliferation, invasion, metastasis and survival of cancer cells (6). EGFR is expressed in a variety of human epithelial tumors including lung, head and neck, colorectal and breast cancers (7). EGFR is also overexpressed in 40–60% of the malignant mesothelioma cases (8,9). However, EGFR protein expression alone does not correlate with the clinical response to anti-EGFR therapy and thus does not show the dependence of the tumor on the EGFR pathway (8). Some patients with lung cancer have mutations in the EGFR that make them very sensitive to treatment with EGFR TK inhibitors (TKI), such as erlotinib and gefitinib (10,11). The presence of activating mutations (especially deletion in exon 19 and missense mutation in exon 21) in the TK domain enhances the kinase activity and maintains the EGFR in a constitutively active form with tumor growth dependent on EGFR signaling (12). Success in treatment with TKI in lung cancer patients has prompted the identification of these activating mutations in other tumors, including malignant mesothelioma. However, mutation in the TK domain of the EGFR gene are not present in malignant pleural mesothelioma (8,13,14). Furthermore, phase II clinical trials of TKI gefitinib (15) and erlotinib (16) in malignant pleural mesothelioma patients did not demonstrate any activity.
However, Foster et al described the presence of somatic EGFR TK mutations in 9 of 29 (31%) patients with peritoneal mesothelioma (17). They screened the whole TK domain (exons 18–24) using denaturing high performance liquid chromatography and confirmed the presence of mutation by sequencing. They found the presence of 7 novel mutations and one already known sensitizing mutation (L858R). All of these novel mutations were shown to activate the TK activity in a transfected cell line and were sensitive to the EGFR inhibitor erlotinib in vitro (18). Given the potential clinical implications of these findings, we conducted this study to evaluate EGFR mutations/polymorphisms in the TK domain of early passage peritoneal mesothelioma cell cultures, in established malignant peritoneal mesothelioma cell lines and in peritoneal mesothelioma tumor samples by completing direct sequencing. We also wished to correlate the effect of EGFR TKI erlotinib on cell proliferation and its correlation with any potential mutation/polymorphism encountered.
Materials and methods
Reagents and cell culture.
Erlotinib was procured from Active Biochem (Maplewood, NJ). Cell culture related reagents were purchased from Invitrogen/Life Technologies, Inc. (Rockville, MD). FBS was purchased from Lonza Walkersville, Inc. (Walkersville, MD). Cells were cultured in RPMI-1640 supplemented with 10% FBS, 2 mM glutamine and 10 μg/ml penicillin/streptomycin at 37 °C in 5% CO2 humidified air.
Patient samples.
Archival tumor samples were obtained from patients with peritoneal mesothelioma treated on clinical protocols approved by the National Cancer Institute (NCI) Institutional Review Board. Early passage peritoneal mesothelioma cell cultures were established from the ascites of patients with peritoneal mesothelioma treated on NCI IRB approved protocols. In addition, established peritoneal mesothelioma cell lines were also evaluated for EGFR TK mutations.
Pathological examination of tumor samples.
The patients whose tumor samples were used to determine somatic EGFR mutations were obtained from the archival files of the NCI. The diagnosis of peritoneal mesothelioma was confirmed by the pathologist (M.R.). Unstained slides were cut from the tumor blocks that showed mesothelioma. These unstained slides were then evaluated by the pathologist to identify the tumor area for DNA extraction.
Polymerase chain reaction and direct sequencing of EGFR.
Genomic DNA was isolated from cell lines, as well as from tumors embedded in paraffin blocks using ultra clean tissue and cells with the DNA isolation kit from Mo Bio Laboratories (Carlsbad, CA) and the QIAamp DNA FFPE tissue kit from Qiagen (Valencia, CA), respectively. Briefly, DNA was amplified in a total volume of 25 μl, containing 1.5 mM MgCl2, 200 μM of each dNTP (dATP, dCTP, dGTP and dTTP), 10 pM of each primer, 0.3 units Taq DNA polymerase, and 100 ng of DNA in 1X Taq polymerase buffer. The primers used to amplify exons 18–24 of the EGFR gene are listed in Table I. DNA was first denatured at 95 °C for 5 min, followed by 35 cycles of PCR with denaturation at 94 °C for 45 sec, primer annealing for 45 sec at 56 °C or 58 °C and primer extension for 45 sec at 72 °C. At the end of the last cycle, the mixture was incubated at 72 °C for 5 min. The sequencing reaction was performed in a final volume of 20 μl containing 20 pmol of one primer (forward or reverse), 3 μl of BigDye (version 1.1) and 2 μl of purified PCR product. Twenty-five cycles were performed: denaturation at 96 °C for 10 sec, primer annealing at 55 °C for 10 sec and extension at 60 °C for 4 min. Direct sequencing of amplified PCR products was performed on an ABI Prism sequencing apparatus (ABI Prism 310 Genetic Analyzer, Applied Biosystems, Foster City, CA).
Table I.
Primers and PCR conditions for 7 exons of the EGFR TK domain.
| Exons | Primer sequence 5’→3’ | Annealing temperature (°C) | PCR product size (bp) |
|---|---|---|---|
| Exon 18 | |||
| Forward | GCT GAG GTG ACC CTT GTC TC | 58 | 250 |
| Reverse | CCA CAG CTT GCA AGG ACT CTG G | ||
| Exon 19 | |||
| Forward | ACA ATT GCC AGT TAA CGT CTT CC | 58 | 199 |
| Reverse | CCA AAA GGT GGG CCT GAG GTT CA | ||
| Exon 20 | |||
| Forward | CCA CCA TGC GAA GCC ACA CT | 58 | 261 |
| Reverse | CCT CCC CTC CCC GTA TCT CCC T | ||
| Exon 21 | |||
| Forward | TGA TCT GTC CCT CAC AGC AG | 58 | 226 |
| Reverse | CCA ATG CTG GCT GAC CTA AAG C | ||
| Exon 22 | |||
| Forward | TCC AGA GTG AGT TAA CTT TTT CCA | 56 | 277 |
| Reverse | TTG CAT GTC AGA GG A TAT AAT GTA A | ||
| Exon 23 | |||
| Forward | CTA AAG CAA GGG ATT GTG ATT G | 58 | 259 |
| Reverse | CCA GGC TCA GCT AGG CAG TGT GGA | ||
| Exon 24 | |||
| Forward | GCA ATG CCA TCT TTA TCA TTT C | 56 | 281 |
| Reverse | GCT GGC ATG TGA CAG AAC AC |
Growth inhibition of early passage mesothelioma cell cultures and established cell lines.
Cells were seeded in 96-well plates at 1×104 cells/well in 200 μl of medium. After seeding for 24 h, cells were treated with various concentrations (0.1–100 μM) of erlotinib for 72 h. The inhibition of cell growth was assessed using the WST assay from Dojindo Molecular Technologies (Rockville, MD). Relative growth inhibition rates for the vehicle control (DMSO) were calculated and IC50 values were determined as the drug concentrations associated with 50% survival of cells.
Results
Absence of EGFR mutations in early passage and in established cell lines.
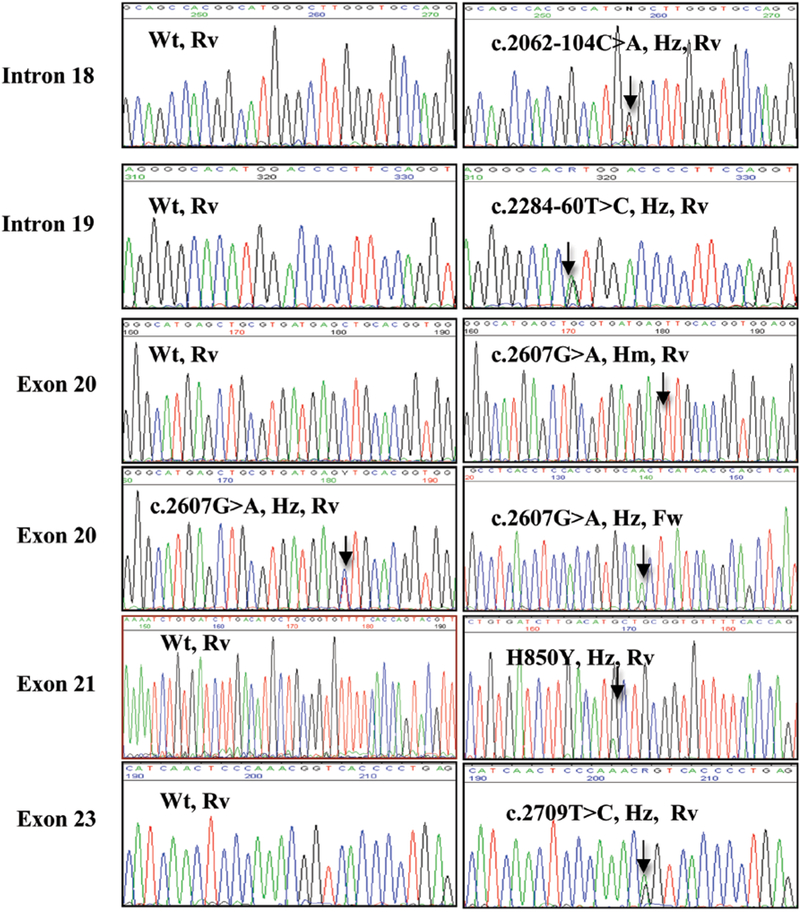
There were no mutations present in the TK domain of peritoneal mesothelioma cell lines. However, we found the commonly known single nucleotide polymorphism (SNP) c.2607G→A (rs1050171, Q787Q glutamine, CAG to CAA) in exon 20 of all cell lines either in the heterozygous (GA, 57.2%) or the homozygous form (AA, 42.8%) (Table II and Fig. 1). None of the cell lines had the GG genotype. The other SNPs that were found in these cells included earlier reported c.2709T→C (rs1140475, T903T, threonine, ACT to ACC) in exon 23, c.2284–60T→C (rs10241451) and c.2062104C→A (rs17337100) in the intronic region of the EGFR gene (Fig. 1). The c.2062–104C→A and c.2709T→C were present in 13.3% and the c.2284–60T→C in 46.6% of the peritoneal mesothelioma cell lines, respectively, in the heterozygous form. To our knowledge this is the first report identifying the presence of these SNPs in mesothelioma since earlier studies only looked for the presence of specific mutations which are commonly found in lung cancer.
Table II.
Presence of EGFR mutation/polymorphisms in malignant peritoneal mesothelioma cells and sensitivity to erlotinib.
| Peritoneal mesothelioma cells | c.2062–104C→A (Intronic change) | c.2284–60T→C (Intronic change) | Q787Q (c.2607 G→A) (Exon 20) | T903T (c.2709 T→C) (Exon 23) | Sensitivity of peritoneal mesothelioma cells to erlotinib (IC50) |
|---|---|---|---|---|---|
| Early passage cells (n=8) | |||||
| NCI-M-03 | Wt | Wt | Hz | Wt | ND |
| NCI-M-05 | Wt | Wt | Hz | Hz | >50 μM |
| NCI-M-07 | Hz | Wt | Hm | Wt | >100 μM |
| NCI-M-09 | Wt | Wt | Hz | Hz | 10 μM |
| NCI-M-10 | Wt | Hz | Hz | Wt | 20 μM |
| NCI-M-11 | Hz | Hz | Hm | Wt | ND |
| NCI-M-14 | Wt | Wt | Hz | Wt | >50 μM |
| NCI-M-17 | Wt | Hz | Hm | Wt | >50 μM |
| Established cell lines (n=6) | |||||
| ORT | Wt | Wt | Hz | Wt | 10 μM |
| ROB | Wt | Wt | Hz | Wt | 10 μM |
| YOU | Wt | Hz | Hz | Wt | 40 μM |
| PET | Wt | Hz | Hm | Wt | >50 μM |
| HAY | Wt | Hz | Hm | Wt | >100 μM |
| HEC | Wt | Hz | Hm | Wt | >50 μM |
ND, not done; Wt, wild-type; Hz, heterozygous; Hm, homozygous.
Figure 1.
Identification of mutation/polymorphisms in malignant mesothelioma. Respective chromatogram of each mutation/polymorphism has been shown. Change of position is indicated by an arrow. Wt, wild-type; Hz, heterozygous; Hm, homozygous; Fw, forward primer; Rv, reverse primer
Although the significance of these SNPs is unknown, we wanted to examine whether these SNPs might play some role in the sensitivity of these cell lines to the TKI erlotinib. We found that peritoneal mesothelioma cell lines having homozygous AA genotype for c.2607G→A were completely resistant (IC50 >50 μM) to erlotinib while 5 out of the 7 cell lines with heterozygous GA genotype showed moderate sensitivity (IC50 <50 μM) to treatment with erlotinib although this is not in the clinically relevant dose range (Table II). No correlation could be seen with the presence of other SNPs and the sensitivity of these cell lines towards erlotinib. There was also no correlation between the expression of EGFR protein in these cell lines and their sensitivity to erlotinib (data not shown). Our results show that none of the cell lines is highly sensitive to erlotinib indicating that absence of sensitizing mutations in the EGFR gene contributes towards resistance to TKI therapy.
Absence of somatic EGFR mutations in peritoneal mesothelioma tumor samples.
Out of the 33 cases analyzed, 23 were male and 10 were female ranging from 19–75 years of age (Table III). All patients had epithelial histology with mostly high-grade tumors. Out of these 33 patients, 25 underwent cytoreductive surgery with intraperitoneal hyperthermic chemotherapy while 8 patients had exploratory laparotomy.
Table III.
Presence of EGFR mutation/polymorphisms in malignant peritoneal mesothelioma tumor samples
| Patient | Gender | Age (years) | c.2062–104C→A (Intronic change) | c.2284–60T→C (Intronic change) | Q787Q (c.2607 G→A) (Exon 20) | H850Y(c.2548 C→T) (Exon 21) | T903T (c.2709 T→C) (Exon 23) |
|---|---|---|---|---|---|---|---|
| 1 | M | 65 | Wt | Wt | Hm | Wt | Wt |
| 2 | F | 71 | Wt | Hz | Hz | Wt | Wt |
| 3 | F | 51 | Wt | Hz | Hm | Wt | Wt |
| 4 | M | 41 | Wt | Wt | Hz | Wt | Hz |
| 5 | M | 58 | Wt | Wt | Hz | H850Y | Hz |
| 6 | M | 53 | Hz | Wt | Hz | Wt | Wt |
| 7 | F | 19 | Wt | Hz | Hz | Wt | Hz |
| 8 | M | 35 | Wt | Wt | Hz | Wt | Wt |
| 9 | M | 78 | Wt | Hz | Hm | Wt | Wt |
| 10 | M | 31 | Wt | Wt | Hz | Wt | Wt |
| 11 | M | 51 | Wt | Wt | Hz | Wt | Wt |
| 12 | M | 50 | Wt | Wt | Hz | Wt | Hz |
| 13 | M | 57 | Wt | Wt | Wt | Wt | Wt |
| 14 | F | 54 | Wt | Wt | Hm | Wt | Hm |
| 15 | M | 75 | Wt | Wt | Wt | Wt | Wt |
| 16 | M | 42 | Wt | Hz | Hm | Wt | Wt |
| 17 | F | 63 | Wt | Wt | Hm | Wt | Hz |
| 18 | F | 65 | Wt | Wt | Wt | Wt | Hz |
| 19 | M | 60 | Wt | Wt | Hz | Wt | Wt |
| 20 | F | 72 | Wt | Wt | Hz | Wt | Hz |
| 21 | M | 69 | Wt | Wt | Hm | Wt | Wt |
| 22 | M | 29 | Wt | Hz | Hm | Wt | Wt |
| 23 | M | 54 | Wt | Wt | Hm | Wt | Wt |
| 24 | M | 54 | Wt | Wt | Hm | Wt | Wt |
| 25 | M | 38 | Wt | Wt | Hz | Wt | Wt |
| 26 | M | 57 | Wt | Wt | Hz | Wt | Wt |
| 27 | F | 54 | Wt | Wt | Hm | Wt | Wt |
| 28 | M | 63 | Wt | Wt | Hz | Wt | Wt |
| 29 | M | 68 | Wt | Wt | Hm | Wt | Wt |
| 30 | F | 57 | Wt | Wt | Hm | Wt | Wt |
| 31 | M | 56 | Wt | Wt | Hz | Wt | Hz |
| 32 | F | 75 | Wt | Wt | Hz | Wt | Wt |
| 33 | M | 59 | Wt | Wt | Hz | Wt | Hz |
M, male; F, female; Wt, wild-type; Hz, heterozygous; Hm, homozygous.
DNA was extracted from these 33 formalin-fixed paraffinembedded tumor samples and the quality of the DNA was checked using PCR amplification of β-actin as a quality control gene (data not shown). Of the 33 specimens we found only one case with a single mutation in exon 21 at codon 850 (H850Y, CAT to TAT, c.2548C→T) of the EGFR gene (Table III and Fig. 1). No other previously reported or novel EGFR mutations were found. However, we found the same pattern of SNPs that were present in peritoneal mesothelioma cell lines. The SNP c.2607G→A was present in 91% of malignant mesothelioma cases. Eleven of the 33 samples had the AA genotype (33.3%) and 19 had the AG genotype (57.6%) while 3 patients (9.1%) had the prototype GG genotype (Table III). The TC genotype of c.2709 T→C was present in 9/33 cases (27.3%) and the CC genotype in 1/33 cases (3.03%). The intronic polymorphisms c.2062–104C→A and c.2284–60T→C were present in 1/33 (3.03%) and 6/33 (19%) of cases, respectively, in the heterozygous form (Table III).
Discussion
EGFR somatic mutations are present in some patients with lung cancer and are associated with sensitivity to EGFR TKI. However, no such mutations are found in patients with pleural mesothelioma and clinical trials of EGFR TKI have no activity in this disease. However, a recent report suggested that EGFR somatic mutations are frequent in peritoneal mesothelioma patients (17). Since these findings could have clinical implications we conducted a comprehensive analysis of EGFR TK mutation analysis by direct sequencing of the entire TK domain of the EGFR gene using early passage peritoneal mesothelioma cell cultures, established peritoneal mesothelioma cell lines and tumor samples from patients with peritoneal mesothelioma.
In both the mesothelioma cells and tumor specimens we observed several SNPs. The most commonly found polymorphism in peritoneal mesothelioma cases was c.2607G→A, present in exon 20 of the EGFR gene. This polymorphism has been reported earlier in a wide variety of tumors including gastric (19), lung (20), cervical (21), pancreatic (22) and head and neck (23). Zhang et al have shown the association of the AA genotype of c.2607G→A with an increased risk of lung cancer (24), although they did not show any correlation between this SNP and somatic EGFR mutations found in those samples. In head and neck cancer cell lines the GA genotype showed higher sensitivity to gefitinib than the GG genotype (25). A study by Sasaki et al has shown a weak association between the SNP (GA/AA genotype) and a worse outcome with gefitinib treatment in Japanese lung cancer patients (26). All of these findings indicate that this SNP may affect either the stability of the EGFR protein, the rate of translation, or possibly gene transcription (24,26). We also found a moderate response to erlotinib in vitro in some of the peritoneal mesothelioma cell lines with the GA genotype while cells with the AA genotype were completely resistant to erlotinib.
Direct sequencing of the entire TK domain of the 33 tumor samples from patients with malignant peritoneal mesothelioma did not identify any novel mutations or presence of mutations that are associated with sensitivity to EGFR TKI. Only once case showed the presence of a mutation in exon 21 (H850Y) of the EGFR gene. Mutation at codon 850 has been reported previously in other cancers although the change in amino acid was different from what we found in our study. Jia and Chen reported the presence of H850D mutation in one patient with adenosquamous carcinoma of the lung (20). The mutation coexisted with the silent SNP c.2607G→A. Yang et al showed the presence of the H850R mutation together with the EGFR sensitizing mutation L858R in two patients with lung cancer (27).
The results of our study are in contrast to the report by Foster et al who described the presence of somatic EGFR mutations in 9 out of 29 cases with peritoneal mesothelioma (17,18). The reason for these desperate results is not clear. However, our results are in agreement with the study by Perrone et al who found no EGFR mutations in 20 patients with malignant peritoneal mesothelioma (28). Perrone et al performed the direct sequencing only for exons 18–21 where the activating mutations of the EGFR gene are present, whereas we performed the DNA sequencing of the entire TK domain (exons 18–24) to exclude the presence of any novel mutation.
In conclusion, direct sequencing of the entire EGFR TK domain, using 14 mesothelioma cell lines and 33 patient samples, did not reveal any EGFR sensitizing mutations. Therefore, we conclude that EGFR TKI will not be useful for the treatment of patients with peritoneal mesothelioma.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations:
- EGFR
epidermal growth factor receptor
- TK
tyrosine kinase
- TKI
TK inhibitor
References
- 1.Robinson BW and Lake RA: Advances in malignant mesothelioma. N Engl J Med 353: 1591–1603, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Price B and Ware A: Time trend of mesothelioma incidence in the United States and projection of future cases: an update based on SEER data for 1973 through 2005. Crit Rev Toxicol 39: 576–588, 2009. [DOI] [PubMed] [Google Scholar]
- 3.Vogelzang NJ, Rusthoven JJ, Symanowski J, Denham C, Kaukel E, Ruffie P, Gatzemeier U, Boyer M, Emri S, Manegold C, et al. : Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol 21: 2636–2644, 2003. [DOI] [PubMed] [Google Scholar]
- 4.Yan TD, Deraco M, Baratti D, Kusamura S, Elias D, Glehen O, Gilly FN, Levine EA, Shen P, Mohamed F, et al. : Cytoreductive surgery and hypothermic intraperitoneal chemotherapy for malignant peritoneal mesothelioma: multi-institutional experience. J Clin Oncol 27: 6237–6242, 2009. [DOI] [PubMed] [Google Scholar]
- 5.Baratti D, Kusamura S and Deraco M: Diffuse malignant peritoneal mesothelioma: systematic review of clinical management and biological research. J Surg Oncol 103: 822–831, 2011. [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T and Yatabe Y: Epidermal growth receptor in relation to tumor development EGFR gene and cancer. FEBS J 277: 301–308, 2010. [DOI] [PubMed] [Google Scholar]
- 7.Arteaga CL: Epidermal growth factor receptor dependence in human tumors: more than just expression? Oncologist 7: 31–39, 2002. [DOI] [PubMed] [Google Scholar]
- 8.Destro A, Ceresoli GL, Falleni M, Zucali PA, Morenghi E, Bianchi P, Pellegrini C, Cordani N, Vaira V, Alloisio M, et al. : EGFR overexpression in malignant pleural mesothelioma. An immunohistochemical and molecular study with clinico-pathological correlations. Lung Cancer 51: 207–215, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Trupiano JK, Geisinger KR, Willingham MC, Manders P, Zbieranski N, Case D and Levine EA: Diffuse malignant mesothelioma of the peritoneum and pleura, analysis of markers. Mod Pathol 17: 476–481, 2004. [DOI] [PubMed] [Google Scholar]
- 10.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, et al. : National Cancer Institute of Canada Clinical Trials Group: Erlotinib in previously treated non-small cell lung cancer. N Engl J Med 353: 123–132, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. : Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139, 2004. [DOI] [PubMed] [Google Scholar]
- 12.Gazdar AF, Shigematsu H, Herz J and Minna JD: Mutations and addiction to EGFR: the Achilles ‘heal’ of lung cancers? Trends Mol Med 10: 481–486, 2004. [DOI] [PubMed] [Google Scholar]
- 13.Cortese JF, Gowda AL, Wali A, Eliason JF, Pass HI and Everson RB: Common EGFR mutations conferring sensitivity to gefitinib in lung adenocarcinoma are not prevalent in human malignant mesothelioma. Int J Cancer 118: 521–522, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Okuda K, Sasaki H, Kawano O, Yukiue H, Yokoyama T, Yano M and Fujii Y: Epidermal growth factor receptor gene mutation, amplification and protein expression in malignant pleural mesothelioma. J Cancer Res Clin Oncol 134: 1105–1111, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govindan R, Kratzke RA, Herndon JE II, Niehans GA, Vollmer R, Watson D, Green MR and Kindler HL: Gefitinib in patients with malignant mesothelioma: a phase II study by the Cancer and Leukemia Group B. Clin Cancer Res 11: 2300–2304, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Garland LL, Rankin C, Gandara DR, Rivkin SE, Scott KM, Nagle RB, Klein-Szanto AJ, Testa JR, Altomare DA and Borden EC: Phase II study of erlotinib in patients with malignant pleural mesothelioma: a Southwest Oncology Group Study. J Clin Oncol 25: 2406–2413, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Foster JM, Gatalica Z, Lilleberg S, Haynatzki G and Loggie BW: Novel and existing mutations in the tyrosine kinase domain of the epidermal growth factor receptor are predictors of optimal respectability in a malignant peritoneal mesothelioma. Ann Surg Oncol 16: 152–158, 2009. [DOI] [PubMed] [Google Scholar]
- 18.Foster JM, Radhakrishna U, Govindarajan V, Carreau JH, Gatalica Z, Sharma P, Nath SK and Loggie BW: Clinical implications of novel activating EGFR mutations in malignant peritoneal mesothelioma. World J Surg Oncol 8: 88, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Liu L, Li M, Wang Z, Feng L, Zhang Q, Cheng S and Lu S: Epidermal growth factor receptor mutation in gastric cancer. Pathology 43: 234–238, 2011. [DOI] [PubMed] [Google Scholar]
- 20.Jia XL and Chen G: EGFR and KRAS mutations in Chinese patients with adenosquamous carcinoma of the lung. Lung Cancer 74: 396–400, 2011. [DOI] [PubMed] [Google Scholar]
- 21.Longatto-Filho A, Pinheiro C, Martinho O, Moreira MA, Ribeiro LF, Queiroz GS, Schmitt FC, Baltazar F and Reis RM: Molecular characterization of EGFR, PDGFRA and VEGFR2 in cervical adenosquamous carcinoma. BMC Cancer 9: 212, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzeng CW, Frolov A, Frolova N, Jhala NC, Howard JH, Buchsbaum DJ, Vickers SM, Heslin MJ and Arnoletti JP: Epidermal growth factor receptor (EGFR) is highly conserved in pancreatic cancer. Surgery 141: 464–469, 2007. [DOI] [PubMed] [Google Scholar]
- 23.Naji F, Attaleb M, Laantri N, Benchakroun N, El Gueddari B, Benider A, Azeddoug H, Ennaji MM, El Mzibri M and Khyatti M: Identification of G2607A mutation in EGFR gene with a significative rate in Moroccan patients with nasopharyngeal carcinoma. Cell Mol Biol (Noisy-le-grand) 56 (Suppl): OL1442–OL1426, 2010. [PubMed] [Google Scholar]
- 24.Zhang W, Stabile LP, Keohavong P, Romkes M, Grandis J, Traynor AM and Siegfried JM: Mutation and polymorphism in the EGFR-TK domain associated with lung cancer. J Thorac Oncol 1: 635–647, 2006. [PubMed] [Google Scholar]
- 25.Taguchi T, Tsukuda M, Imagawa-Ishiguro Y, Kato Y and Sano D: Involvement of EGFR in the response of squamous cell carcinoma of the head and neck cell lines to gefitinib. Oncol Rep 19: 65–71, 2008. [PubMed] [Google Scholar]
- 26.Sasaki H, Endo K, Takada M, Kawahara M, Tanaka H, Kitahara N, Matsumura A, Iuchi K, Kawaguchi T, Okuda K, et al. : EGFR polymorphism of the kinase domain in Japanese lung cancer. J Surg Res 148: 260–263, 2008. [DOI] [PubMed] [Google Scholar]
- 27.Yang CH, Yu CJ, Shih JY, Chang YC, Hu FC, Tsai MC, Chen KY, Lin ZZ, Huang CJ, Shun CT, et al. : Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol 26: 2745–2753, 2008. [DOI] [PubMed] [Google Scholar]
- 28.Perrone F, Jocollè G, Pennati M, Deraco M, Baratti D, Brich S, Orsenigo M, Tarantino E, De Marco C, Bertan C, et al. : Receptor tyrosine kinase and downstream signalling analysis in diffuse malignant peritoneal mesothelioma. Eur J Cancer 46: 2837–2848, 2010. [DOI] [PubMed] [Google Scholar]