Abstract
Wilson Disease (WD) is an autosomal recessive disorder caused by mutation in the ATP7B gene that affects copper transport in the body. ATP7B mutation damages copper transporter function, ultimately resulting in excessive copper accumulation and subsequent toxicity in both the liver and brain. Mechanisms of copper toxicity, however, are not well defined. The Atp7b−/− mouse model is well-characterized and presents a hepatic phenotype consistent with WD. In this study, we found that the untreated Atp7b−/− mice accumulate approximately 2-fold excess hepatic zinc compared to the wild type. We used targeted transcriptomics and proteomics to analyze the molecular events associated with zinc and copper accumulation in the Atp7b−/− mouse liver. Altered gene expression of Zip5 and ZnT1 zinc transporters indicated a transcriptional homeostatic response, while increased copper/zinc ratios in associated with high levels of metallothioneins 1 and 2, indicated altered Zn availability in cells. These data suggest that copper toxicity in Wilson Disease includes effects on zinc-dependent proteins. Transcriptional network analysis of RNA-seq data reveals an interconnected network of transcriptional activators with over-representation of zinc-dependent and zinc-responsive transcription factors. In the context of previous research, these observations support the hypothesis that mechanisms of copper toxicity include disruption of intracellular zinc distribution in liver cells. The translational significance of this work lies in oral zinc supplementation in treatment for WD, which is thought to mediate protective effects through the induction of metallothionein synthesis in the intestine. This work indicates broader impacts of altered zinc-copper balance in WD, including global transcriptional responses and altered zinc balance in the liver.
Keywords: Wilson Disease, gene expression, transcription network, zinc, copper, homeostasis, metalloprotein
Graphical Abstract
INTRODUCTION
Wilson disease (WD) is an autosomal recessive disorder that affects copper (Cu) distribution in the body, resulting in copper accumulation in the liver and brain and impacts on these organs. The disease is fatal in the absence of treatment. Treatment of WD employs Cu chelating agents that increase urinary excretion as well as high doses of oral zinc (Zn) to interfere with Cu absorption, while liver transplant is a last resort therapy.1 WD is caused by a mutation in the ATP7B gene, which encodes a Cu-transporting P-type ATPase 2 that supplies Cu to secreted cuproproteins. The protein maintains whole-body Cu levels through expression in hepatocytes and transport of excess Cu into vesicles for biliary export. 3 ATP7B mutation in WD damages Cu transporter function and ultimately results in excessive Cu accumulation with subsequent toxicity to both the liver and brain.4 Diagnosis and treatment of WD are challenging due to diverse clinical manifestations such as hepatitis, cirrhosis of the liver, and liver failure, as well as neurological symptoms including tremors, dystonia, and psychological conditions.4–6 Despite years of study, the mechanisms by which excess Cu induces the diverse set of pathologies are not clear and mechanisms of Cu toxicity in WD are not yet entirely defined. Similarly, treatments are life-long and improvements in health of WD patients are often not seen for months. As a consequence, compliance is frequently a problem.7 Therefore, a refinement of treatment options is much needed.
The Atp7b−/− mouse is an important model to define molecular effects of Cu accumulation in WD. The animal exhibits many similarities to WD patients, while also developing a consistent pathophysiology that facilitates study.8–9 Atp7b−/− mouse pups are born with liver Cu deficiency but quickly develop high hepatic Cu, with peak levels observed at approximately six weeks of age. Histopathological liver tissue damage is not usually observed until the mice are between 12 and 20 weeks of age,10–11 allowing for investigation of the molecular and metabolic effects of high Cu prior to the onset of significant pathology and potential secondary effects.
Important findings in the Atp7b−/− mouse include disruption of lipid metabolism and promotion of cell cycle progression,12 regulation of urinary Cu excretion13 and cardiolipin fragmentation with mitochondrial damage.10 Transcriptomic analysis of the spontaneous Atp7b mutant Long-Evans Cinnamon (LEC) rat and the Atp7b−/− mouse indicated changes in gene expression linked to lipid metabolism and regulated by ligand-activated nuclear receptors (NR), which require structural Zn cofactors.14 Further, our proteomics studies were the first to directly identify effects of Cu accumulation on NRs FXR/NR1H4 and GR/NR3C1 and interacting proteins in the Atp7b−/− mouse.15 More recently, Wooton-Kee and co-workers investigated the functional consequences of Cu accumulation in the Atp7b−/− mouse and reported decreased DNA binding of promoter elements by FXR/NR1H4 and other NRs, along with decreased mRNA expression from NR target genes.16 Hamilton and co-workers found that agonist-mediated activation of the LXR/RXR NR pathway mitigated liver disease pathology in the Atp7b−/− mouse.17
Other affected Zn-dependent proteins in Wilson Disease (or WD) mouse models were carbonic anhydrase III, in which protein levels were decreased in the ‘toxic milk’ (tx) mouse,18 and apparent attenuation of Zn-dependent sorbitol dehydrogenase function (SDH).15 These data suggest changes in cellular Zn-dependent processes as a consequence of Cu accumulation in WD. Therefore, this study tested the hypothesis that Zn homeostasis is disrupted by Cu accumulation in a WD model.
Our study presents a new concept in the copper toxicity of Wilson Disease. We found that six-weeks-old Atp7b−/− mice accumulate 2-fold excess hepatic Zn compared to WT animals, high Cu/Zn ratios in metallothionein (MT)-containing soluble liver fractions from the Atp7b−/− animals compared to wild type and sufficient increase in MT expression to bind the increased Cu and Zn. This change corresponded to impacts on gene expression in the Zn distribution system. Since MT is typically bound to Zn and provides Zn to other proteins, but also binds Cu with high affinity, we suggest a mechanism for Cu-Zn competition in protein metallation in WD-induced Cu toxicity. We further confirmed a transcriptional response that includes Zn-containing transcription factor global regulators.
EXPERIMENTAL
Animal
The generation of Atp7b−/− mice has been described in prior work.16 Liver tissue was isolated from animals housed at the University of Alaska Anchorage Vivarium. Wild type, heterozygous and Atp7b−/− Mice were maintained on strain C57BL x 129S6/SvEv. Samples used in RNAseq were a gift from Prof. Svetlana Lutsenko (Johns Hopkins Medical Institute). Liver Zn levels from animals housed at either facility were not different. Animal care and use was approved by respective Institutional Animal Care and Use Committees performed in accordance with US Public Health Service Policy as documented by The Guide for the Care and Use of Laboratory Animals 8th Edition.19 Housing was in shoebox cages and animals were fed Mazuri Rodent diet (PMI Nutrition, Inc., Richmond, Indiana), containing 16 ppm Cu, 100 ppm Zn, and 235 ppm Fe and water ad libitum, with a 12-hour light/dark cycle. Animals were sacrificed at six weeks of age; mice of both sexes were used for metal measurements and transcriptomic studies. Animals were euthanized at six weeks of age by CO2 asphyxiation followed by cervical dislocation. Animals were immediately exsanguinated and perfused with 0.9% saline. Hepatic RNA was isolated as described in Tallino et. al.20
Elemental analysis.
Approximately 30 mg of hepatic tissue was dehydrated for 24 hours at 37°C, after which tissue samples and serum were analyzed for transition metal content using inductively-coupled plasma mass spectrometry (ICP-MS) by the Elemental Analysis Core at the Oregon Health & Science University in Portland, OR.
Quantitative Real Time PCR (qRT-PCR)
Total RNA was extracted from livers using PureZOL RNA Isolation Reagent (Biorad), followed by a DNase cleanup procedure (Optizyme Recombinant DNase, Thermo Scientific). Total RNA concentrations were determined using a NanoDrop ND-1000 Spectrometer. Total RNA was then used to synthesize cDNA using reverse transcription of 2 µg RNA using RevertAid Reverse Transcriptase (Thermo Scientific), random hexamer primers (TaKaRa), and dNTPs (Promega). The PCR was prepared in a reaction mixture with final volume of 20 µl containing 10 µL Advanced SYBR green PCR super mix (BioRad), 8 µL RNase-free H2O, 1 µL of cDNA, 250 nM forward primer, and 250 nM reverse primer. Forward and reverse primers (Eurofins Genomics) for the Zn transporters: ZIP1, ZIP4, ZIP5, ZIP7, ZIP8, ZIP9, ZIP10, ZIP13, ZIP14, ZnT1, ZnT4, ZnT5, ZnT6, ZnT7, and ZnT10. Primer sequences specific to Mus musculus ZIP and ZnT transporters were reported in Sun et al. 21 and Lichten et al.22 RT-PCR analysis was performed on a BioRad CFX96 Real Time System with the following cycling conditions: 95° C for 3:00 minutes, 95° C for 0:30 minutes, 55° C for 0:30 minutes, 72° C for 1:00 minute, repeat steps 2 through 4 × 34 times, 72° C for 5:00 minutes, followed by an infinite hold at 12° C. The data were normalized to beta-actin mRNA using forward and reverse primers (forward: GCG CAA GTA CTC TGT GTG GA; reverse: AGC TCA GTA ACA GTC CGC CT). The amount of target mRNA for each transporter in Atp7b−/− mice relative to the mRNA amount of wild type mice was quantified using the PCR efficiency corrected Pfaffl method.23 PCR efficiencies were determined via baseline corrected and linear regression analysis using LinRegPCR (www.hartfaalcentrum.nl).24 A t-statistic was calculated for individual Atp7b−/−fold changes. Fold change was considered significant at Bonferonni-adjusted P value of < 0.01. Antibodies for Zn transporter detection were: rabbit anti-Zip5 (Novus NBP1–69246) and rabbit anti-Znt1 (Lifespan Bio LS-C161213–400).
Metallothionein fraction analysis
Approximately 200 mg of liver tissue from each of three wild type and three Atp7b−/− mice was homogenized with a Dounce homogenizer with tight-fitting glass pestle in 50 mM ammonium acetate pH 7.0. The homogenate was clarified by centrifugation at 500g for 10 minutes at 4 degrees Celsius. Supernatant was transferred to a new microcentrifuge tube and centrifuged at 3,000g for for 10 minutes at 4° C. This second supernatant was then transferred once again and centrifuged at 22,000g for 30 minutes at 4° C. 50 micoliters of the final supernatant was fractionated by size exclusion chromatography using a Tosoh BioAssist G3SWxl column and GE AKTA Explorer system with the homogenization buffer as mobile phase. Molecular weight standards (Biorad, Hercules, CA) thyroglobulin (bovine, 670 kDa), γ-globulin (bovine, 158 kDa), ovalbumin (chicken, 44 kDa), myoglobin (horse, 17 kDa) and vitamin B12 (1.35 kDa) were used to identify low-molecular weight fractions. Five 0.5 mL fractions included molecular weights from 1.35 and 44 kDa and were analyzed for Cu and Zn content by ICP-MS at the Oregon Health & Science University Metal Ion Core.
LC-MS/MS was performed by the Oregon Health & Science University Proteomics Shared Resource. Size exclusion fractions were dried in a Speedvac and digested in ProteaseMAX (Promega, Madison, WI). Proteins were reduced by addition of dithiothreitol to 20 mM and incubated at 50°C, then alkylated by addition of 50 mM iodoacetamide. Reduced and alkylated protein was digested with trypsin overnight at 37°C. Digestion was stopped by addition of neat 88% formic acid. Digests were then solid phase extracted (Sep Pak Light Cartridges, Waters Corp) and peptides were separated by reverse phase chromatography and 60 min of tandem mass spectrometry data was collected for each of the 35 fractions using an LTQ Velos linear ion trap (Thermo Scientific, San Jose, CA). Spectra were analyzed as described in Wilmarth et al. to generate peptide spectral counts.25
Corrected peptide counts were used for comparison of abundance in fractions from wild type and Atp7b−/− animals. One wild type sample had very low specific peptides identified and was not used for further analysis. After filtering contaminant and redundant proteins, we identified a total of 315 proteins. Total non-contaminant counts were normalized and analyzed by Student’s T-test with a false discovery rate at 5% to identify the most highly different proteins in the target fraction.
RNA sequencing and network analysis.
RNA sequencing was performed at NCGR using the GAIIx platform. Average read quality was 38. RNA-seq data from three wild type (WT) and three Atp7b−/− mice were analyzed. The data consisted in two pair-end reads runs, one for a WT sample and one for a KO sample; and 4 single-end reads runs, 2 for WT samples and 2 for KO (supplementary table 2). Read length was 54 bp in all cases.
Reads from each sample were mapped to Mus musculus reference genome assembly GRCm38 (released January 2012) using STAR version 2.4.26 STAR was provided with both fasta and GTF files for the reference genome downloaded from Ensembl and default parameters were used for the alignment. Genes and exons read counts were calculated using featureCounts from the R package Rsubread version 1.16.27 Read counts and mapping to reference are summarized in Table S1. Gene differential expression of KO vs WT samples and differential exon usage was calculated using the R package edgeR version 3.8.28 We used an FDR-corrected p-value cutoff of 0.05 in all cases. Gene ontology enrichment was performed by GOrilla.29 Data availability: RNA sequencing data is available in Gene Expression Omnibus (GEO), ID code GSE95260, https://www.ncbi.nlm.nih.gov/geo/: GSE95260.
Transcription factor clusters were assigned according to common enriched DNA binding site motif using iRegulon (organism: Mus musculus, default parameters).30 Transcriptional regulatory networks were generated by Cytoscape. Differentially expressed genes used in Sco1 and Zfp36l2 networks construction were obtained from GEO (Id: code GSE58997 and GSE15146 respectively).31–32
RESULTS
THE ATP7B−/− MOUSE LIVER ACCUMULATES EXCESS ZINC
In order to test the hypothesis that transition metal homeostasis is disrupted in the WD model, we analyzed the Cu, Zn and Fe in the livers of six-week-old Atp7b−/− mice. Cu in the Atp7b−/− mouse liver is approximately 28-fold above wild type levels (Table 1), confirming earlier data 33. Atp7b−/− mouse livers accumulate Zn to approximately 2-fold excess compared to wild type animals (P<0.0001, Student’s T-test). Fe levels were also increased in the Atp7b−/− mice (P <0.05) but were more variable than Zn levels. Zn levels in the wild type liver are 4–5-fold in excess of Cu, while Cu is 3-fold in excess of Zn in the Atp7b−/− mouse (Fig. 1A). Six-week-old animals were selected as a target age, as prior studies reported that liver Cu levels are highest at six weeks of age, while pathology is not apparent and secondary effects are not expected.11
Table 1.
Elemental content in dried liver tissue (n=5) and serum (n=4–5) from six-week-old Wilson Disease (Atp7b−/− ) and wild type (Atp7b+/+ ) mice.
| Liver | Serum | |||
|---|---|---|---|---|
| Atp7b−/− | Atp7b+/+ | Atp7b−/− | Atp7b+/+ | |
| Cu [µg/g] | 318.64 ± 28.81* | 11.32 ± 2.92 | 301.00 ± 86.53 | 475.6 ± 59.41 |
| Fe [µg/g] | 207.84 ± 70.04* | 117.02 ± 24.90 | 7725.3 ± 3422.2 | 9723.0 ± 4230.5 |
| Zn [µg/g] | 95.06 ± 7.27 * | 46.18 ± 2.70 | 817.75 ± 171.67 | 685.6 ± 209.25 |
Figure 1.
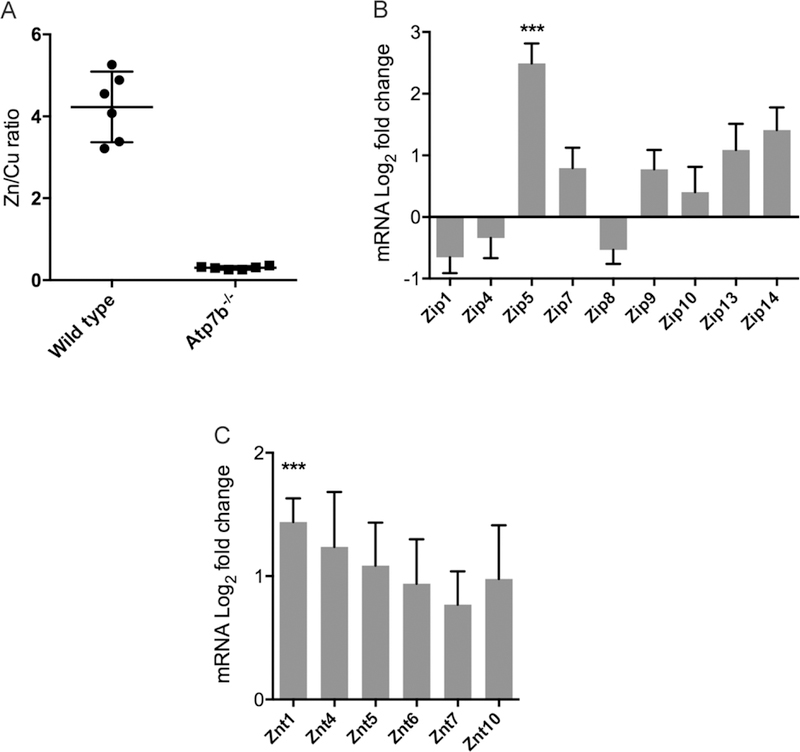
Zn/Cu ratio and transporter expression in Atp7b−/− compared to wild type liver. A. Zn/Cu ratio in lier for wild type and Atp7b−/− mice (n=6), mean and 95% CI indicated. B. Log2 fold change of ZIP (SLC39 family) transporters (n=6) by quantitative RT-PCR. C. Log2 fold change of ZnT (SLC30 family) transporters. Mean and SEM are shown for gene expression; “***” indicates P<0.01.
The specific increase in hepatic Zn led us to test the hypothesis that expression of specific Zn transporters will change in Atp7b−/− compared to wild type livers. We analyzed the gene expression levels of putative and verified Zn importers (SLC39 family, a.k.a. Zip or Zrt-Irt-like protein) and exporters (SLC30 family, a.k.a. ZnT, Zn transporter) that are known to be expressed in mouse liver.21 We found up-regulation of Zip5 (mean fold-change: 6.55, P<0.01), a transcriptional homeostatic response consistent with altered Zn distribution (Fig. 1B). The ZnT1 Zn exporter was moderately increased in expression (mean fold-change: 3.15, P<0.01, Fig. 1C), suggesting a response to enhance Zn supply in the early secretory pathway, where ZnT1 and ZnT4 cooperate with MT to deliver Zn for enzyme metalation.34 Both Zip5 and ZnT1 have been confirmed to transport Zn.22, 35 These observations indicate that a transcriptional homeostatic response to acquire Zn for cytosolic and/or secretory pathway processes is activated in Atp7b−/− animals in the early stage of disease. We analyzed the levels of the Zip5 and Znt1 protein by Western blot and immunodetection and found no significant changes in protein levels (Supp. Data S1).
We retained a small number of Atp7b−/− and WT/het mice to >20 weeks of age to analyze Zn and Cu levels at later stages of disease. As expected, Cu levels decreased with age. In these animals, we find that Zn and Cu both remain elevated compared to wild type, but also become more variable compared to six-week-old animals (F-Test for unequal variances (F > Fcritical). In some Atp7b−/− animals, Zn levels were near the six-week Atp7b−/− level, while some were near wild type levels. Zn transporter expression was also more variable with moderate decreases in Zip9 (−1.8 fold, P<0.05), ZnT4 (−1.3-fold, P<0.05), ZnT6 (−1.67 fold, P<0.05) and ZnT10 (−2.7-fold, P<0.05). These data indicate that Zn balance remains disrupted in later stages of disease, while heterogeneity of the responses increases.
METALLOTHIONEINS 1 AND 2 ARE ASSOCIATED WITH ALTERED Cu/Zn RATIO
MTs perform functions that include intracellular Zn homeostasis and distribution as well as protection against the accumulation of excessive amounts of both essential and non-essential metals.36 MTs accomplish these functions through the binding of metals within the two domains of the protein to create metal-cysteine thiolate clusters. Zn2+ is a direct activator of the MTF-1 transcription factor and induces MT expression.37 Excess metals such as Zn and Cd, as well as oxidative stress promote MT expression.38 However, since Cu efficiently competes with Zn for binding to MT, and MT is important to modulate Zn availability,36 it is possible that very high levels of Cu may disrupt Zn distribution by associating with MT. Previous profiling studies in the Atp7b−/− mouse revealed increased levels of Mt1, Mt2 and Mtf-1 transcripts and increased Mt1 and Mt2 proteins.12 To test the hypothesis that excess Cu in the Atp7b−/− liver is associated with Mt, and thus has the potential to affect Zn distribution, we analyzed Cu and Zn content in size-exclusion fractions of soluble liver extracts encompassing 1,350 to 44,000 Daltons. This method allowed us to analyze Cu and Zn specifically associated with proteins and confirm Mt presence in high metal fractions. We compared Cu and Zn levels in these fractions and used quantitative label-free LC-MS/MS 15, 25 to identify the proteins in the highest Cu fraction, which was also the highest Zn fraction (all proteins identified are detailed in Supp. Data S2). Significant differences were observed in the abundance of 18 specific proteins (Table 2). Importantly, Mt1 and Mt2 were identified at low levels in the fraction from wild type liver extracts but were abundant in liver extracts from Atp7b−/− animals. Assuming there are a maximum of 7 Zn ions utilized per single WT metallothionein protein 39 Mt levels would need to increase three-fold for the sum of increased cellular Zn concentration to be bound to Mts, unavailable for use as free zinc by various proteins/enzymes. Mt in Atp7b−/− would include both Cu and Zn. We found 15-fold increases in Mt1 and an approximate 40-fold increase in Mt2 in the Atp7b−/− mice (Table 2), sufficient to include both the increase in Zn and Cu. This observation suggests that Mt1 and Mt2 indeed bind excess Cu in addition to Zn.
Table 2.
Proteins identified with differential abundance in low molecular weight size-exclusion fraction with high Cu/Zn ratio in Atp7b−/− mouse liver homogenate. Metallotioneins 1 and 2 are shaded. Mean spectral counts from LC-MS/MS are reported.
| Uniprot Accession |
Description | P value | Wild type |
Atp7b−/− | Mean Difference |
t ratio |
|---|---|---|---|---|---|---|
| P01942 | Hemoglobin subunit alpha | <0.001 | 105.19 | 336.663 | −231.473 | 37.9065 |
| P02088 | Hemoglobin subunit beta-1 | <0.001 | 82.085 | 258.08 | −175.995 | 28.8212 |
| P16015 | Carbonic anhydrase 3 | <0.001 | 147.41 | 73.8267 | 73.5833 | 12.0501 |
| P04247 | Myoglobin | <0.001 | 60.06 | 0.976667 | 59.0833 | 9.67559 |
| P02089 | Hemoglobin subunit beta-2 | <0.001 | 8.05 | 66.8067 | −58.7567 | 9.62209 |
| Q63836 | Selenium-binding protein 2 | <0.001 | 52.44 | 3.15 | 49.29 | 8.07181 |
| P02802 | Metallothionein-1 | <0.001 | 2.42 | 37.2167 | −34.7967 | 5.69836 |
| P17742 | Peptidyl-prolyl cis-trans isomerase A |
<0.001 | 91.595 | 57.12 | 34.475 | 5.64568 |
| P19157 | Glutathione S-transferase P 1 | <0.001 | 66.92 | 33 | 33.92 | 5.5548 |
| P05202 | Aspartate aminotransferase, mitochondrial |
<0.001 | 96.305 | 63.1333 | 33.1717 | 5.43225 |
| Q64374 | Regucalcin | <0.001 | 114.41 | 83.4767 | 30.9333 | 5.0657 |
| P08228 | Superoxide dismutase [Cu- Zn] |
<0.001 | 60.445 | 30.73 | 29.715 | 4.86618 |
| P12710 | Fatty acid-binding protein, liver |
<0.001 | 121.82 | 92.6267 | 29.1933 | 4.78075 |
| P30115 | Glutathione S-transferase A3 | <0.001 | 72.515 | 46.37 | 26.145 | 4.28155 |
| P52196 | Thiosulfate sulfurtransferase | <0.001 | 15.335 | 38.32 | −22.985 | 3.76406 |
| P02798 | Metallothionein-2 | 0.0017 | 0.485 | 19.6 | −19.115 | 3.13031 |
| P00920 | Carbonic anhydrase 2 | 0.0023 | 12.775 | 31.43 | −18.655 | 3.05497 |
| P10649 | Glutathione S-transferase Mu | 0.00284 | 162.35 | 144.08 | 18.27 | 2.99193 |
This proteomics analysis identified additional proteins with differential abundance that provide insight into the metabolic pathology of WD. Specifically, we observed decreased abundance of carbonic anhydrase III (CAIII) in Atp7b−/− animals. CAIII decrease is consistent with the tx mouse, and suggests that Zn availability for formation of the stable holoenzyme is limited,18 while in vitro studies support CAIII Zn acquisition from MT, with kinetics dependent on the number of MT-bound Zn ions.40 In this study, wild type liver contains Zn in approximately 4-fold excess of Cu; however, Atp7b−/− livers contain over 3-fold Cu in excess of Zn. The MT fraction indicates even greater Cu excess over Zn (Fig. 2). Thus, despite increased Zn and MT, the availability of Zn to proteins may be limited due to excess Cu compared to Zn associated with MT.
Figure 2.
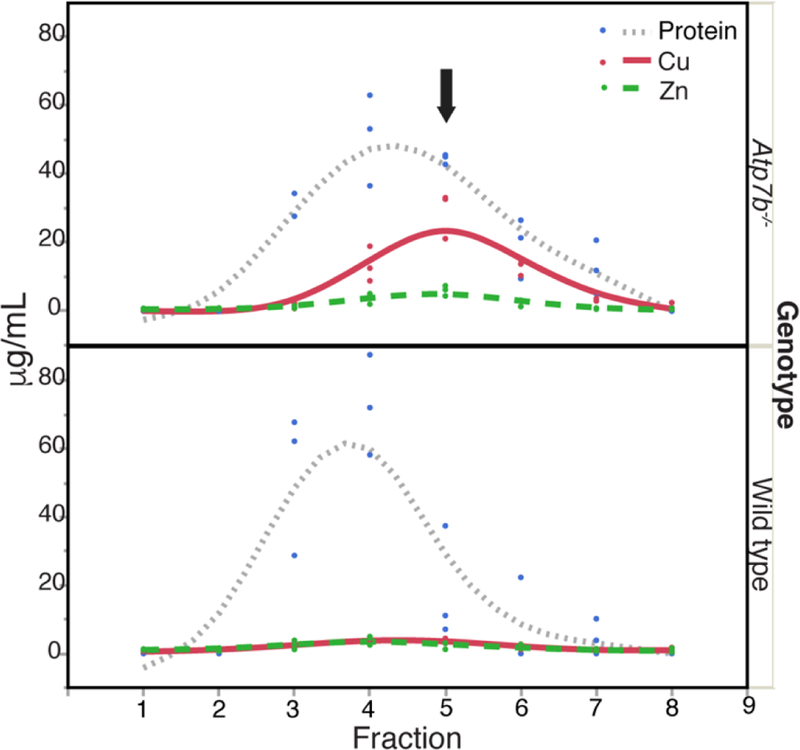
Size-exclusion fractionation to analyze metals and relative MT content. Cu, Zn, and protein concentrations for size exclusion fractions encompassing low molecular weight soluble proteins extracted from Atp7b−/− and wild type liver. Fractions include sizes from approximately 44 to 1.3 kDa as estimated from standards. Black arrow indicates the fraction with highest Cu/Zn concentrations in Atp7b−/− samples.
TRANSCRIPTIONAL NETWORK ANALYSIS REVEALS IMPACTS ON Zn-CENTERED CLUSTERS IN THE ATP7B−/−MOUSE LIVER
In order to provide biologically important data and mitigate the limitations of focus on individual genes,41 we used a systems approach to RNAseq data analysis, in which overall enrichment of certain gene families were examined and gene expression networks were generated. RNAseq analysis revealed a total of 296 differentially expressed transcripts in the Atp7b−/− mouse compared to age-matched wild type animals (Supp. Data S3, S4). Comparison with published microarray analysis of the Atp7b−/− mouse at the same age was used for validation of the dataset (Supp. Data S5). Additional RNAseq analysis is provided in Supplementary RNA-seq analysis (Supp. Data S5, S6).
We integrated the Atp7b−/− RNAseq results into a global model of gene regulation in mice 30 to generate the first transcriptional regulatory network in a WD model experiment (Figure 3, Supp. Data S5). This approach was employed to test the hypothesis that Cu accumulation in the Atp7b−/− mouse liver is associated with a global impact on Zn dependent systems. Nine of the 15 transcription factor clusters in the network include Zn-containing transcription factors. The most highly connected clusters (M1, M2, M3) include global regulators with targets in cell cycle (M1, ex. E2F transcription factors) and regulators of metabolism (M2, including the nuclear receptor GR/Nr3c1, and Nf1a/b), which are consistent with the initial transcriptional studies of this model.12 Cluster M21 includes the transcription factor Mtf-1, which controls the expression of metallothioneins (Mt1, Mt2) in the liver as well as the Zn exporter ZnT1.37, 42–43 Mtf-1 includes labile Zn fingers integral to its transactivation activity,which mechanistically links altered Zn levels to downstream transcription events.37 MTF-1 also has Cu-sensing activity through displacement of Zn by Cu in MT.44
Figure 3.
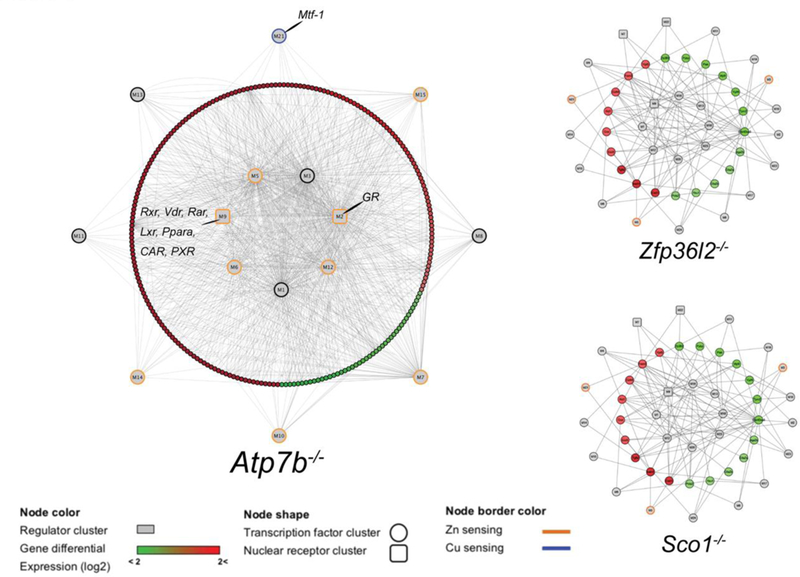
Transcriptional regulatory networks generated from iRegulon clusters (Mus musculus) and Cytoscape for differential expressed genes in Atp7b−/−, Sco1−/− and Zfp36l2−/− mice. MTF-1 (M21) and nuclear receptor (M2, M9) clusters are indicated in the Atp7b−/− network. Zn sensing clusters are defined by inclusion of Zn-containing transcription factors.
We identified two major clusters that include nuclear receptors. Cluster M2 makes 138 connections and includes glucocorticoid receptor (GR/NR3C1), which that was identified as downregulated at the protein level in our previous study,15 as well as Zn-containing Zbtb14 and Trp53 transcription factors. The M9 cluster includes 34 connections and contains transcription factor binding sites for the nuclear receptors Vdr, Rxrβ, Rxrα, Nr1i3, Nr1i2, Rarα, Pparα, Rarβ, Nr1h4, Rarγ, and Nr2f1. Our previous work identified downregulation of Nr1h4 (Farnesoid-x-receptor/FXR) at the protein level,15 consistent with these results. These NRs are involved in the regulation of a wide range of functions including cellular differentiation, development, and carbohydrate, lipid, and protein metabolism. The network also contains a total of seven clusters that include non-nuclear receptor Zn finger transcription factors (Fig. 3), with representatives of the Zbtb and Zfp families, that also includes the most highly connected regulators in the network.
In order to analyze the specificity of the network to and potential activation by other disruptions of Cu or Zn machinery, we compared these results with other genetically engineered mouse models with metalloprotein defects. We used the same strategy to construct two additional networks based on published transcriptomes, integrating the global transcriptional response of mouse livers deficient in specific metalloproteins Sco1 (mitochondrial Cu chaperone) or Zfp36l2 (CCCH tandem Zn finger RNA-binding protein) (Figure 3). Networks assembled from these two mutants are more limited compared to the Atp7b−/− network, with fewer nodes and lower transcriptional activator connectivity when compared to the Atp7b−/− mouse. The number of central targeted transcription factors in the Atp7b−/− network is smaller than in the other two models, while the Atp7b−/− central factors are connected to a very high number of downstream targets represented in the differentially expressed genes. This characteristic indicates a global response in Atp7b−/− with fewer targeted transcriptional activators, supporting the observation that the absence of Atp7b−/− impacts the regulation of a high number of genes.
DISCUSSION
Zn Homeostatic response to Cu accumulation Atp7b−/−mouse
Our study demonstrated that the Atp7b−/− mouse has significantly higher hepatic Zn compared to wild-type mice (Table 1) in addition to expected elevated Cu. Mildly increased Zn was previously reported in the tx mouse;45 though no further analysis was mentioned. The significant increase in Zip5 expression, as well as only minimal changes in expression of the Zn-exporting ZnT transporters, suggest a gene expression response to import Zn. Zip5, which is located on the basolateral membrane of hepatocytes,22, 46 functions directly in importing Zn from the extracellular space into the cytoplasm of cells. The lack of increased Zip5 protein is consistent with transcriptional regulation mediated in the 3’-UTR of the message.47
Metallothionein Ion Content Disruption and Zn homeostasis
It has been challenging to experimentally link competition between Cu and Zn in tissues, though thermodynamic effects favor Cu binding to available transition metal sites.48 In healthy cells, Cu is bound to specific proteins while excess is exported, preventing adventitious interaction.49 The WD condition in hepatocytes results in excess Cu, providing potential to displace other metals from proteins.
Interestingly, prior work by Schilsky et al.50 showed that exposing HepG2 cells to Zn prior to incubation in media supplemented with copper chloride resulted in decreased copper toxicity compared to cells that had no initial zinc treatment. It was determined that this protective effect was not due to reduced copper uptake, but likely due to MT induction and Cu sequestration in the cultured hepatocytes as a result of Zn exposure. This mechanism is thought to be important in oral Zn supplementation as a current treatment for Wilson disease. It has been established in both the tx mouse and the Atp7b−/− mouse that liver Mt1 and Mt2 expression increases,12, 51 likely due to induction by Cu accumulation. Excess Cu bound to MT might promote presentation of Cu instead of Zn to critical transcription factors or enzymes, due to Cu’s ability to displace bound Zn from Zn-MT (1.93 μM Cu EC50 vs. 8.06 μM Zn EC50 for bound Zn displacement).52
Our data suggests that increases in the ratio of Cu+ to Zn2+ in MT might have several consequences as illustrated in Fig. 4: 1) available Zn will bind to labile Zn sites in MTF-1,38 inducing transcription of additional MT (I and II) mRNA, as observed in our transcriptomic data and increasing MT levels in this animal in previous work.12 2) Increased MT abundance will coordinate increasing amounts of both Cu(I) and Zn(II), most likely resulting in mixed Cu/Zn-MT in the alpha and beta domains of MT, such as (Cu-3;Zn-2)alpha(Cu-4;Zn-1)beta-MT from (Zn-4)alpha(Cu-4;Zn-1)beta-MT.53 3) High levels of Cu in MT relative to Zn in MT disrupt normal Zn distribution, resulting in Zn-limitation for Zn-specific proteins, particularly those that receive Zn from Zn-MT (ADH, SDH).36, 54 With the above stoichiometry and our observed high Cu/Zn ratios, increased Mt1 and Mt2 expression could result in an increase Zn bound to MT, even though MT will contain more Cu than Zn. Given the estimates of nearly 3000 Zn proteins in the human proteome,55 disruption of Zn distribution has potential to impact diverse cellular functions.
Figure 4.
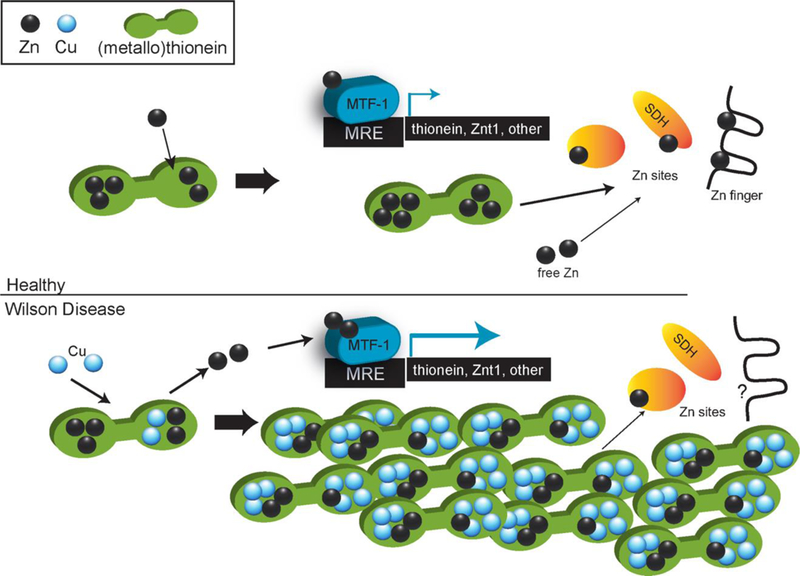
Proposed schematic of copper interference in Zn distribution. Top: schematic of metallothionein-centered Zn distribution in healthy cells. Bottom: schematic of metallothionein-centered Zn distribution in Wilson Disease. Despite an increase in total Zn, proteins may encounter more Cu than Zn in MT.
Zn-dependent systems
Our previous metabolite analysis indicates that some Zn-dependent enzymes (i.e. sorbitol dehydrogenase) are attenuated: we previously reported a 2-fold increase in sorbitol and xylitol levels in the livers of Atp7b−/− mice, along with a decrease in glyceraldehyde-3-phosphate.15 Glyceraldehyde-3-phosphate is synthesized from glucose via sorbitol and fructose,56 indicating an attenuated Zn-dependent metabolic process. Hamiltion and co-workers also found metabolic changes in the Atp7b−/− mouse model that were consistent with the disruption of Zn-dependent enzymes downstream of glyceraldehyde-3-phosphate, evidenced by decreased glycerol-3-phosphate and glycerol-2-phosphate levels due to an inability to synthesize the molecules.17
There are other similar parallels between Zn deficiency and Wilson disease that have been reported. Per Huster et al.,12 genes associated with cholesterol biosynthesis are significantly down-regulated in the Atp7b−/− mouse model. Of note, the cholesterol synthesis rate limiting enzyme 3-hydroxy-3-methylglutaryl-coenzyme-A reductase had decreased expression. This finding was also observed in Zn deficient lung tissue of rats.57 The enzyme 3-hydroxy-3-methylglutaryl-coenzyme-A reductase is regulated by SREBP-2, which is itself activated by a Zn metalloprotease, whereby the authors postulated that a decrease in enzyme expression could be due to the decrease in available cellular Zn.58 It is possible that a similar Zn deficient-like response, in which Zn is less available for major Zn dependent proteins, occurs in the Atp7b−/− mouse, despite the elevated levels of Zn. This may explain the continual importation of Zn and upregulation of mRNA for Zn transporters. It is not yet known whether the same Zn transporter expression response is observed in WD patients, especially at pre-symptomatic disease stages since most of these patients are not yet diagnosed. However, a recent report indicates decreased expression of ZIP5 and ZIP8 in WD patient liver biopsies analyzed by microarray, suggestive of a different transcriptional response at later stages in the disease.59
Systems-level analysis connects Zn to gene expression
The Atp7b−/− liver transcriptional network indicates global targeting of Zn-containing transcriptional activators in response to Cu excess. Research indicating reversible exchange of Zn between MT and the estrogen receptor DNA binding domain indicates that MT can be a source of Zn for transcription factors other than MTF-1.60 NR involvement identified in WD is consistent with this observation: our previous work found that NRs and NR-interacting proteins are less abundant in Atp7b−/− hepatic nuclei compared to wild type.15 Synthesis of earlier transcriptomic surveys in the LEC rat and Atp7b−/− mouse indicated changes in NR-regulated transcripts;14 while others have functionally implicated NRs FXR, RXR, HNF4α, and LRH-1 in mouse models and WD patients.16–17 These observations led to the hypothesis that NRs are specifically and selectively impacted by excess Cu. However, our results demonstrate that several types of Zn-containing transcription factors (NR, Mtf-1, Zbtb, Zfp) are impacted by excess Cu, which is not surprising if Zn distribution is altered. The NR-containing nodes are consistent with NR targeting observed in the Atp7b−/− and LEC models, as well as WD patients, and provide specific explanation of disrupted lipid metabolism in the model as well as WD patients. Further, Zn supplementation in a non-Wilson disease rat primary hepatocyte model has been shown to promote expression of lipid metabolism genes that are attenuated in the Atp7b−/− mouse.61 Similarly, Ralle and coworkers 33 reported that down-regulation of cholesterol synthesis genes persists in the Atp7b−/− mouse as the disease progressed. Analysis of these genes for specific transcription factor elements 30 revealed persistence of GR/Nr3c1 targets, indicated in cluster M2 of the Atp7b−/− network (Fig. 3), as well as additional NR-binding elements and other Zn-dependent transcription factors (including Gli, Gata, Yy1 and Zic families). These transcriptional responses along with above-mentioned metabolite observations are supportive of a Zn-centered mechanism to explain how excess Cu promotes WD pathology. The working model suggests that increased Cu leads to increased MT expression, which binds large amounts of Cu along with available Zn, leading to a ‘sensed’ Zn deficiency.
Extending this model to specific cellular processes related to pathology, it is possible that the hepatocyte damage is due attenuation of specific Zn-dependent functions, which include but are not limited to enzymatic function and Zn-dependent transcriptional activator function. The former is indicated by our previous work indicating an increase in hepatic polyols sorbitol and xylitol and decrease in glyceraldehyde-3-phosphate. 15 Sorbitol dehydrogenase, in the sorbitol pathway of lipid synthesis,62 uses a Zn cofactor that can be acquired from MT. Attenuation of Zn-dependent transcriptional activators and potential involvement in pathology is illustrated in the nuclear receptor class of transcription factors. Published studies with liver-specific nuclear receptor knockouts are limited; however, a number of studies indicate that functional nuclear receptors are important in prevention and mitigation of liver damage, 63–64 while FXR is established with a role in bile acid and lipid homeostasis. 65 Treatment with an FXR agonist attenuated liver pathology in the Atp7b−/− mouse model, while FXR also suppresses NFκB in a hepatic inflammatory response. 66 Thus, suppressed FXR activity can be specifically linked to observed pathology. Taken together, these mechanisms indicate that altered Zn distribution may promote pathology, even if Cu is effectively sequestered in MT.
CONCLUSION
Our findings demonstrate that excess Cu in the Atp7b−/− mouse disrupts Zn balance and impacts the proportion of Cu vs. Zn associated with MT. Transcript network analysis indicates impacts on global regulators that, many of which require Zn cofactors and are important in lipid metabolism and cell cycle gene expression. This new insight into the mechanism of Cu toxicity in WD has potential to provide for more effective monitoring of therapy and pathology, as well as lead to new insights into WD treatment such as the recent proposal of zinc as a maintenance therapy and alternative to high-cost chelators for stable WD patients;67 however, the importance of Zn balance in WD pathology has yet to be fully defined.
Supplementary Material
SIGNIFICANCE TO METALLOMICS.
Zinc balance and zinc-dependent systems are disrupted in the Atp7b−/− mouse model of Wilson Disease. This work reveals a biological interaction between copper and zinc in a Wilson Disease model that provides insight into the underlying mechanism of copper toxicity in the liver. A schematic is proposed where total zinc is increased and bound to metallothionein, but zinc distribution is limited due to an excess of copper.
ACKNOWLEDGMENTS
We are grateful to Svetlana Lutsenko for sharing the Atp7b−/− mouse strain. The National Center for Genome Resources (NCGR), including Faye D. Schilkey, was critical for assistance with RNA-seq study design as well as data and technical assistance from Ingrid Lindquist and Ritu Barthi. Larry David, Phillip Wilmarth and Ashok Reddy of the Oregon Health & Science University Proteomics Shared Resource were essential for mass spectrometry experiments.
FUNDING
Research supported in this publication was supported by the National Institutes of Health [1R15DK114747-01 to JLB]; an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103395; NCGR and NMINBRE [NIGMS 8P20GM103451-12]; [GM090016 to MR]; Alaska Heart Institute [Fellowship to KM]; and Center for Genome Regulation, Basal grant of the Center for Mathematical Modeling UMI2807 UCHILE-CNRS [Fondecyt 11150679 to ML]. ICP-MS measurements were performed at the OHSU Elemental Analysis Core with partial support from the National Institutes of Health [S10RR025512 to MR]; Proteomics was partially supported by National Institutes of Health [P30EY010572, P30CA069533 and R01DC002368-15S1 to Larry David]. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH.
REFERENCES
- 1.Weiss KH; Stremmel W, Evolving perspectives in Wilson disease: diagnosis, treatment and monitoring. Curr Gastroenterol Rep 2012, 14 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- 2.Bull PC; Thomas GR; Rommens JM; Forbes JR; Cox DW, The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet 1993, 5 (4), 327–37. [DOI] [PubMed] [Google Scholar]
- 3.Harris ED, Cellular copper transport and metabolism. Annual review of nutrition 2000, 20, 291–310. [DOI] [PubMed] [Google Scholar]
- 4.Rosencrantz R; Schilsky M, Wilson disease: pathogenesis and clinical considerations in diagnosis and treatment. Semin Liver Dis 2011, 31 (3), 245–59. [DOI] [PubMed] [Google Scholar]
- 5.Strausak D; Mercer JF; Dieter HH; Stremmel W; Multhaup G, Copper in disorders with neurological symptoms: Alzheimer’s, Menkes, and Wilson diseases. Brain Res Bull 2001, 55 (2), 175–85. [DOI] [PubMed] [Google Scholar]
- 6.Lutsenko S, Modifying factors and phenotypic diversity in Wilson’s disease. Ann N Y Acad Sci 2014, 1315, 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poujois A; Woimant F, Wilson’s disease: A 2017 update. Clin Res Hepatol Gastroenterol 2018. [DOI] [PubMed]
- 8.Buiakova O; Xu J; Lutsenko S; Zeitlin S; Das K; Das S; Ross B; Mekios C; Scheinberg I; Gilliam T, Null mutation of the murine ATP7B (Wilson disease) gene results in intracellular copper accumulation and late-onset hepatic nodular transformation. Hum Mol Genet 1999, 8 (9), 1665–71. [DOI] [PubMed] [Google Scholar]
- 9.Lutsenko S, Atp7b−/−mice as a model for studies of Wilson’s disease. Biochem Soc Trans 2008, 36 (Pt 6), 1233–8. [DOI] [PubMed] [Google Scholar]
- 10.Huster D, Structural and metabolic changes in Atp7b−/−mouse liver and potential for new interventions in Wilson’s disease. Ann N Y Acad Sci 2014, 1315, 37–44. [DOI] [PubMed] [Google Scholar]
- 11.Huster D; Finegold M; Morgan C; Burkhead J; Nixon R; Vanderwerf S; Gilliam C; Lutsenko S, Consequences of copper accumulation in the livers of the Atp7b−/−(Wilson disease gene) knockout mice. Am J Pathol 2006, 168 (2), 423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huster D; Purnat T; Burkhead J; Ralle M; Fiehn O; Stuckert F; Olson N; Teupser D; Lutsenko S, High copper selectively alters lipid metabolism and cell cycle machinery in the mouse model of Wilson disease. J Biol Chem 2007, 282 (11), 8343–55. [DOI] [PubMed] [Google Scholar]
- 13.Gray LW; Peng F; Molloy SA; Pendyala VS; Muchenditsi A; Muzik O; Lee J; Kaplan JH; Lutsenko S, Urinary copper elevation in a mouse model of Wilson’s disease is a regulated process to specifically decrease the hepatic copper load. PLoS One 2012, 7 (6), e38327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkhead J; Gray L; Lutsenko S, Systems biology approach to Wilson’s disease. Biometals 2011. [DOI] [PMC free article] [PubMed]
- 15.Wilmarth P; Short K; Fiehn O; Lutsenko S; David L; Burkhead J, A systems approach implicates nuclear receptor targeting in the Atp7b(−/−) mouse model of Wilson’s disease. Metallomics 2012, 4 (7), 660–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wooton-Kee CR; Jain AK; Wagner M; Grusak MA; Finegold MJ; Lutsenko S; Moore DD, Elevated copper impairs hepatic nuclear receptor function in Wilson’s disease. J Clin Invest 2015, 125 (9), 3449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton JP; Koganti L; Muchenditsi A; Pendyala VS; Huso D; Hankin J; Murphy RC; Huster D; Merle U; Mangels C; Yang N; Potter JJ; Mezey E; Lutsenko S, Activation of liver X receptor/retinoid X receptor pathway ameliorates liver disease in Atp7B(−/−) (Wilson disease) mice. Hepatology 2016, 63 (6), 1828–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimes A; Paynter J; Walker ID; Bhave M; Mercer JF, Decreased carbonic anhydrase III levels in the liver of the mouse mutant ‘toxic milk’ (tx) due to copper accumulation. Biochem J 1997, 321 (Pt 2), 341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Committee, Guide for the Care and Use of Laboratory Animals: Eighth Edition. In Guide for the Care and Use of Laboratory Animals, 2011; p 118.
- 20.Tallino S; Duffy M; Ralle M; Cortes MP; Latorre M; Burkhead JL, Nutrigenomics analysis reveals that copper deficiency and dietary sucrose up-regulate inflammation, fibrosis and lipogenic pathways in a mature rat model of nonalcoholic fatty liver disease. J Nutr Biochem 2015, 26 (10), 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q; Li Q; Zhong W; Zhang J; Sun X; Tan X; Yin X; Sun X; Zhang X; Zhou Z, Dysregulation of hepatic zinc transporters in a mouse model of alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol 2014, 307 (3), G313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichten LA; Ryu MS; Guo L; Embury J; Cousins RJ, MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS One 2011, 6 (6), e21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW, A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001, 29 (9), e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruijter JM; Ramakers C; Hoogaars WM; Karlen Y; Bakker O; van den Hoff MJ; Moorman AF, Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009, 37 (6), e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilmarth P; Riviere M; David L, Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J Ocul Biol Dis Infor 2009, 2 (4), 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A; Davis CA; Schlesinger F; Drenkow J; Zaleski C; Jha S; Batut P; Chaisson M; Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29 (1), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y; Smyth GK; Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30 (7), 923–30. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MD; McCarthy DJ; Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26 (1), 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eden E; Navon R; Steinfeld I; Lipson D; Yakhini Z, GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009, 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janky R; Verfaillie A; Imrichova H; Van de Sande B; Standaert L; Christiaens V; Hulselmans G; Herten K; Naval Sanchez M; Potier D; Svetlichnyy D; Kalender Atak Z; Fiers M; Marine JC; Aerts S, iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 2014, 10 (7), e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hlynialuk CJ; Ling B; Baker ZN; Cobine PA; Yu LD; Boulet A; Wai T; Hossain A; El Zawily AM; McFie PJ; Stone SJ; Diaz F; Moraes CT; Viswanathan D; Petris MJ; Leary SC, The Mitochondrial Metallochaperone SCO1 Is Required to Sustain Expression of the High-Affinity Copper Transporter CTR1 and Preserve Copper Homeostasis. Cell Rep 2015. [DOI] [PubMed]
- 32.Stumpo DJ; Broxmeyer HE; Ward T; Cooper S; Hangoc G; Chung YJ; Shelley WC; Richfield EK; Ray MK; Yoder MC; Aplan PD; Blackshear PJ, Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 2009, 114 (12), 2401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ralle M; Huster D; Vogt S; Schirrmeister W; Burkhead J; Capps T; Gray L; Lai B; Maryon E; Lutsenko S, Wilson disease at a single cell level: intracellular copper trafficking activates compartment-specific responses in hepatocytes. J Biol Chem 2010, 285 (40), 30875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimoto S; Itsumura N; Tsuji T; Anan Y; Tsuji N; Ogra Y; Kimura T; Miyamae Y; Masuda S; Nagao M; Kambe T, Cooperative functions of ZnT1, metallothionein and ZnT4 in the cytoplasm are required for full activation of TNAP in the early secretory pathway. PLoS One 2013, 8 (10), e77445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F; Kim BE; Petris MJ; Eide DJ, The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J Biol Chem 2004, 279 (49), 51433–41. [DOI] [PubMed] [Google Scholar]
- 36.Krezel A; Maret W, Thionein/metallothionein control Zn(II) availability and the activity of enzymes. J Biol Inorg Chem 2008, 13 (3), 401–9. [DOI] [PubMed] [Google Scholar]
- 37.Radtke F; Heuchel R; Georgiev O; Hergersberg M; Gariglio M; Dembic Z; Schaffner W, Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J 1993, 12 (4), 1355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B; Georgiev O; Hagmann M; Gunes C; Cramer M; Faller P; Vasak M; Schaffner W, Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol Cell Biol 2003, 23 (23), 8471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maret W, Redox biochemistry of mammalian metallothioneins. J Biol Inorg Chem 2011, 16 (7), 1079–86. [DOI] [PubMed] [Google Scholar]
- 40.Pinter TB; Stillman MJ, Kinetics of Zinc and Cadmium Exchanges between Metallothionein and Carbonic Anhydrase. Biochemistry 2015, 54 (40), 6284–93. [DOI] [PubMed] [Google Scholar]
- 41.Robert C; Watson M, Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol 2015, 16, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardyman JE; Tyson J; Jackson KA; Aldridge C; Cockell SJ; Wakeling LA; Valentine RA; Ford D, Zinc sensing by metal-responsive transcription factor 1 (MTF1) controls metallothionein and ZnT1 expression to buffer the sensitivity of the transcriptome response to zinc. Metallomics 2016, 8 (3), 337–43. [DOI] [PubMed] [Google Scholar]
- 43.Langmade SJ; Ravindra R; Daniels PJ; Andrews GK, The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J Biol Chem 2000, 275 (44), 34803–9. [DOI] [PubMed] [Google Scholar]
- 44.Selvaraj A; Balamurugan K; Yepiskoposyan H; Zhou H; Egli D; Georgiev O; Thiele DJ; Schaffner W, Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes Dev 2005, 19 (8), 891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howell J; Mercer J, The pathology and trace element status of the toxic milk mutant mouse. J Comp Pathol 1994, 110 (1), 37–47. [DOI] [PubMed] [Google Scholar]
- 46.Geiser J; De Lisle RC; Andrews GK, The zinc transporter Zip5 (Slc39a5) regulates intestinal zinc excretion and protects the pancreas against zinc toxicity. PLoS One 2013, 8 (11), e82149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weaver BP; Andrews GK, Regulation of zinc-responsive Slc39a5 (Zip5) translation is mediated by conserved elements in the 3’-untranslated region. Biometals 2012, 25 (2), 319–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster AW; Osman D; Robinson NJ, Metal preferences and metallation. J Biol Chem 2014, 289 (41), 28095–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison MD; Jones CE; Dameron CT, Copper chaperones: function, structure and copper-binding properties. J Biol Inorg Chem 1999, 4 (2), 145–53. [DOI] [PubMed] [Google Scholar]
- 50.Schilsky ML; Blank RR; Czaja MJ; Zern MA; Scheinberg IH; Stockert RJ; Sternlieb I, Hepatocellular copper toxicity and its attenuation by zinc. J Clin Invest 1989, 84 (5), 1562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng D; Ono S; Koropatnick J; Cherian M, Metallothionein and apoptosis in the toxic milk mutant mouse. Lab Invest 1998, 78 (2), 175–83. [PubMed] [Google Scholar]
- 52.Waalkes MP; Harvey MJ; Klaassen CD, Relative in vitro affinity of hepatic metallothionein for metals. Toxicol Lett 1984, 20 (1), 33–9. [DOI] [PubMed] [Google Scholar]
- 53.Bofill R; Capdevila M; Cols N; Atrian S; Gonzalez-Duarte P, Zinc(II) is required for the in vivo and in vitro folding of mouse copper metallothionein in two domains. J Biol Inorg Chem 2001, 6 (4), 405–17. [DOI] [PubMed] [Google Scholar]
- 54.Jiang LJ; Maret W; Vallee BL, The glutathione redox couple modulates zinc transfer from metallothionein to zinc-depleted sorbitol dehydrogenase. Proceedings of the National Academy of Sciences of the United States of America 1998, 95 (7), 3483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreini C; Banci L; Bertini I; Rosato A, Counting the zinc-proteins encoded in the human genome. J Proteome Res 2006, 5 (1), 196–201. [DOI] [PubMed] [Google Scholar]
- 56.Berry MN; Kun E; Werner HV, Regulatory role of reducing-equivalent transfer from substrate to oxygen in the hepatic metabolism of glycerol and sorbitol. Eur J Biochem 1973, 33 (3), 407–17. [DOI] [PubMed] [Google Scholar]
- 57.Gomez NN; Biaggio VS; Rozzen EJ; Alvarez SM; Gimenez MS, Zn-limited diet modifies the expression of the rate-regulatory enzymes involved in phosphatidylcholine and cholesterol synthesis. Br J Nutr 2006, 96 (6), 1038–46. [DOI] [PubMed] [Google Scholar]
- 58.Brown MS; Goldstein JL, A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proceedings of the National Academy of Sciences of the United States of America 1999, 96 (20), 11041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lutsenko S, Copper trafficking to the secretory pathway. Metallomics 2016, 8 (9), 840–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cano-Gauci DF; Sarkar B, Reversible zinc exchange between metallothionein and the estrogen receptor zinc finger. FEBS Lett 1996, 386 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- 61.Li X; Guan Y; Shi X; Ding H; Song Y; Li C; Liu R; Liu G, Effects of high zinc levels on the lipid synthesis in rat hepatocytes. Biol Trace Elem Res 2013, 154 (1), 97–102. [DOI] [PubMed] [Google Scholar]
- 62.Jeffery J; Jornvall H, Enzyme relationships in a sorbitol pathway that bypasses glycolysis and pentose phosphates in glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America 1983, 80 (4), 901–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L; Wang YD; Chen WD; Wang X; Lou G; Liu N; Lin M; Forman BM; Huang W, Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology 2012, 56 (6), 2336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayhurst GP; Lee YH; Lambert G; Ward JM; Gonzalez FJ, Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001, 21 (4), 1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sinal CJ; Tohkin M; Miyata M; Ward JM; Lambert G; Gonzalez FJ, Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 2000, 102 (6), 731–44. [DOI] [PubMed] [Google Scholar]
- 66.Wang YD; Chen WD; Wang M; Yu D; Forman BM; Huang W, Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 2008, 48 (5), 1632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schilsky ML; Roberts EA; Hahn S; Askari F, Costly choices for treating Wilson’s disease. Hepatology 2015, 61 (4), 1106–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


