Abstract
Given that diseases associated with anti-SSA/Ro autoantibodies such as systemic lupus erythematosus and Sjögren’s syndrome are linked with an upregulation of IFN and type I IFN-stimulated genes, including Sialic Acid-Binding Ig-Like Lectin 1 (Siglec-1), a receptor on monocytes/macrophages, recent attention has focused on a potential role for IFN and IFN-stimulated genes in the pathogenesis of congenital heart block (CHB). Accordingly, three approaches were leveraged to address the association of IFN, IFN-stimulated genes, and the phenotype of macrophages in affected fetal cardiac tissue: a) cultured healthy human macrophages transfected with hY3, an anti-SSA/Ro-associated single-stranded RNA, b) RNA isolated from freshly sorted human leukocytes/macrophages after Langendorff perfusion of three fetal hearts dying with CHB and three healthy gestational age-matched hearts, and c) autopsy tissue from three additional human CHB hearts and one healthy heart. TLR ligation of macrophages with hY3 led to the upregulation of a panel of IFN transcripts, including SIGLEC1, a result corroborated using qPCR. Utilizing independent and agnostic bioinformatics approaches, CD45+CD11c+ and CD45+CD11c- human leukocytes flow sorted from the CHB hearts highly expressed type I IFN response genes inclusive of SIGLEC1. Furthermore, Siglec-1 expression was identified in the septal region of several affected fetal hearts. These data now provide a link between IFN, IFN-stimulated genes, and the inflammatory and possibly fibrosing components of CHB, positioning Siglec-1-positive macrophages as integral to the process.
Keywords: human, autoimmunity, monocytes/macrophages, autoantibodies, inflammation
1. Introduction
Neonatal lupus is often referred to as a model of passively acquired autoimmunity since disease in the developing offspring associates with circulating maternal autoantibodies to SSA/Ro and SSB/La transported via trophoblastic neonatal Fc receptors (1). These antibodies are thought to mediate cardiac manifestations, most notably advanced atrioventricular (AV) conduction abnormalities and less commonly cardiomyopathy (2). Immunohistochemistry of hearts from fetuses dying with the condition have consistently demonstrated fibrosis and calcification in the region of the AV node in association with macrophages and multinucleated giant cells (3). In vitro studies support that anti-SSA/Ro-associated ssRNA binds to macrophage TLRs, resulting in secretion of proinflammatory and profibrosing mediators which transdifferentiate fibroblasts into myofibroblasts consistent with a scarring phenotype (4–7).
While studies have implicated macrophage secretion of TNFα as a contributory mediator of injury (7, 8), to date little attention has been given to type I IFNs. Of relevance, mothers of affected children can be clinically asymptomatic or have defined diseases such as systemic lupus erythematosus or Sjögren’s syndrome, both of which are frequently characterized by high IFNα signatures (9, 10). Logically aligned, it was recently reported that plasma IFNα levels during pregnancy were higher in nine anti-SSA/Ro-positive females whose fetuses had congenital heart block (CHB) compared to similarly antibody-positive females carrying healthy fetuses, albeit neonatal bloods were not evaluated (11). Furthermore, substantiating downstream consequences of this cytokine profile, maternal cell expression of surface protein Sialic Acid–Binding Ig-Like Lectin 1 (Siglec-1) (sialoadhesin, CD169) was identified as a risk factor for the development of CHB. Siglec-1, which is monocyte/macrophage restricted, is a receptor that can bind to granulocytes, erythrocytes, B cells, and to CD43 on T cells and is upregulated by type I IFN (12, 13). The presence of Siglec-1–positive monocytes in patients with lupus nephritis and systemic sclerosis potentially implicates Siglec-1 in organ fibrosis (14).
Accordingly, this study sought to address the hypothesis that TLR-ligated macrophages, in association with type I IFN, play an important role as effector cells in the inflammatory and fibrotic sequelae of CHB. Several approaches were taken. An in vitro approach employed human macrophages transfected with hY3, a single-stranded RNA serving as a proxy for SSA/Ro-associated immune complexes, or treated with IFN, and the resulting differentially expressed transcripts were assigned to informatics categories based on their function. A complementary in vivo approach interrogated the transcriptomes of leukocytes flow-sorted from hearts of three fetuses dying with CHB and three healthy electively terminated, age-matched hearts. In addition archived immunohistochemical fetal autopsy slides from affected and unaffected hearts were interrogated to provide tissue-based evidence of IFN signaling in the disease pathogenesis.
2. Materials and methods
2.1. Specimens
Hearts from healthy and CHB-affected fetuses were obtained following elective termination and after written informed consent from the NYU Institutional Review Board as part of the Research Registry for Neonatal Lupus. Fetal hearts between the ages of 20–25 weeks of gestation were used in one of two evaluations: immunohistochemistry or flow sorting and RNA isolation. For immunohistochemistry, archived cardiac tissue sections from three fetuses dying with CHB previously described (3, 15) and one electively terminated heathy fetus were interrogated. For the studies in which RNA was extracted from fresh cells isolated following the Langendorff preparation and then flow sorted, three CHB hearts (described in detail below) and three healthy hearts of gestational ages 18, 22, and 23 weeks were utilized. CHB-1, previously described (16), was obtained from a 19-gestational-week fetus of a 35-year-old anti-SSA/Ro-positive clinically asymptomatic African American female, G3P1, with two previous children, one having died with CHB (16). CHB-2 was obtained from a 23-gestational-week fetus of a 26-year-old clinically asymptomatic anti-SSA/Ro-positive Asian female, G1P0. This fetus was found to be bradycardic at 20 weeks during routine monitoring and on echocardiogram confirmed to have complete AV block with endocardial fibroelastosis and severely diminished left ventricular function. Despite maternal treatment with dexamethasone (8 mg daily reduced to 4 mg), hydroxychloroquine (400 mg) added after CHB diagnosis, IVIG (600 mg), and albuterol (2 mg, four times daily), the fetus continued to deteriorate and was electively terminated at 23 weeks. CHB-3 was obtained from a 21-gestational-week fetus of a 35-year-old Asian anti-SSA/Ro-positive female with Sjögren’s Syndrome, G1P0. This fetus was found to be bradycardic at 20 weeks after a normal echo at 18 weeks. The echocardiogram at 20 weeks showed 2:1 AV block; shortly thereafter the echocardiogram revealed complete AV block with a heart rate of 75 and no signs of a cardiomyopathy. The mother was on no prior medications, and she declined dexamethasone and IVIG and elected to terminate the pregnancy.
2.2. Isolation, preparation, and treatment of cardiac leukocytes
The Langendorff preparation was used to generate cell suspensions from the CHB and healthy hearts as described previously (4). In brief, the hearts were cannulated through the aorta, and the coronary arteries were continuously perfused with proteolytic enzymes to yield a single cell suspension of primary human fetal cardiac cells for the culturing experiments or flow cytometry. For the three healthy hearts and one CHB heart (CHB-1), flow sorting was employed to yield a DAPI negative, CD45 positive, podoplanin negative, and CD31 negative single cell suspension. For CHB-2 and CHB-3, DAPI negative CD45 positive, podoplanin negative and CD31 negative cells were yielded and then further subsetted into CD11c positive or negative populations.
2.3. Treatment of macrophages
Human macrophages were derived from healthy control monocytes in accordance with a previously published protocol. Macrophages were transfected with or without 2.5 ug/mL hY3 as previously described (7). hY3 plasmids kindly provided by Dr. Sandra Wolin (Yale University, New Haven, CT) were digested with HindIII restriction enzyme for linearization and subjected to transcription with the TranscriptAid transcription kit (Fermentas Life Sciences, Burlington, Ontario, Canada). Transcripts were purified by phenol/chloroform extraction, resuspended in water at 2.5 ug/ml, and evaluated for quality by RNA Agilent Technologies (Genomics Facility, New York University Medical Center). 2.5 μg ssRNA were mixed with 15 μl of the DOTAP reagent following the specific manufacturer’s instructions using a commercial kit (DOTAP Liposomal Transfection Reagent, Roche, Germany) to a final volume of 75 μl reaction buffer, and incubated at 22°C for 15 minutes. The mixture was then added to IFN-γ–primed macrophages. In separate experiments the macrophages were treated with recombinant human IFNα (30 ng/mL, Life Technologies, PHC 4014). After 18 hours, RNA was isolated from the macrophages as described previously (6). Note that the microarray experiment (GEO Accession Number GSE121704 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121704]) was previously used to determine the association of levels of plaquenil-sensitive transcripts and treatment, and the methods used have been described in detail (17).
2.4. Immunohistochemistry
Septal tissue sections inclusive of the AV node from both CHB and control hearts were prepared as described (3). A standard protocol was selected to evaluate Siglec-1 staining as detected by rabbit anti-human antibody HPA053457 (purchased from Sigma). The secondary antibody was anti-rabbit IgG (H+L) peroxidase conjugate purchased from Invitrogen (31470).
2.5. RNA Sequencing
For each in vivo and in vitro condition, leukocytes were lysed in QIAzol solution (Qiagen) for RNA extraction using the Direct-zol RNA Miniprep kit (ZymoResearch) in accordance with the manufacturer’s instructions. Leukocyte RNA was then quantified using a Nanodrop ND-2000 spectrophotometer. Downstream assays were performed in the NYU Genome Technology Center. To assess protein-coding expression profiles, RNA sequencing (RNA-seq) libraries were made using the SMARTer Ultra Low RNA kit by Illumina employing 10 ng of total RNA to prepare cDNA libraries. Each condition was barcoded. Libraries were run on Illumina 2500 and 4000 sequencing systems using a singe-read 50 bp protocol. Sequencing reads were then demultiplexed and converted to FASTQ format using Illumina FASTQ software.
Single-read raw sequencing data were received in FASTQ format. Quality control metrics were measured on the FASTQ files using the FASTQC 0.11.4 tool. Read mapping was then performed using Tophat 2.0.9 against the hg19 human reference genome. The resulting binary alignment map files were processed using the HTSeq 0.6.1 python framework and respective hg19 gene transfer format annotation, obtained from the UCSC database. The Bioconductor package DESeq2 (3.2) was then used to identify differentially expressed genes. Data are available at GEO Accession Number GSE121699 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121699).
Genes were defined as upregulated if their expression increased by at least 2 fold (log2 transformed ratio > 1) and downregulated if decreased by at least 0.5 fold (log2 transformed ratio ≤ −1) in transcripts generated from hY3-transfected macrophages relative to untreated macrophages. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used in conjunction with a statistical routine (18) to assign differentially regulated genes to functional clusters. The statistical analysis examined associations between modular gene clusters and change in expression, which was developed by Dr. Chaussabel and coworkers using mathematics and computer science to model pairwise relations between objects, a process that was used to formulate the interferon module (19–21). A module representing housekeeping genes was selected as a control based on prior literature (22).
2.6. qPCR
Total RNAs were used to prepare cDNA libraries. For real-time quantitative PCR (qPCR) analysis of SIGLEC1, primers were CCTCGGGGAGGAACATCCTT (forward) and AGGCGTACCCCATCCTTGA (reverse). Levels of expression were normalized by parallel amplification and quantification of the GAPDH mRNA level with primers ACCACAGTCCATGCCATCAC (forward) and TCCACCACCCTGTTGCTGTA (reverse). Brilliant SYBR Green RT-PCR (Invitrogen) was used as qPCR reaction mix.
2.7. Statistics
The Mann-Whitney test was used to compare the relative gene expression of SIGLEC1 in the leukocytes of the different groups. Values of P ≤ 0.05 were considered significant.
3. Results
3.1. Macrophage transfection with ssRNA
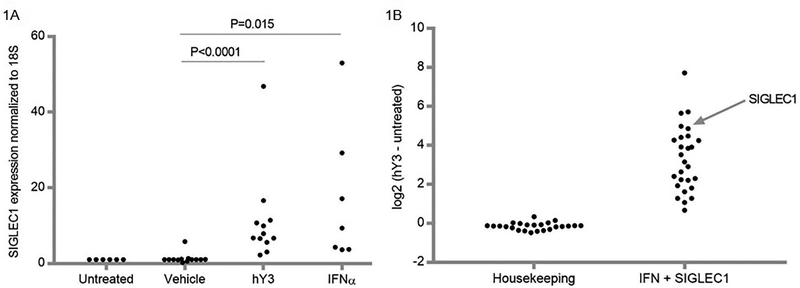
In vitro experiments were performed in accordance with our previous studies in which a model of anti-SSA/Ro-associated CHB injury exploits macrophage TLR 7/8 ligation with hY3, the ssRNA associated with Ro60, as a proxy for immune complex engagement. Compared to human PBMC-derived macrophages transfected with a lipid-based transfection reagent alone, hY3 transfection resulted in the enrichment of 26 type I IFN-related transcripts (listed in Table 1) plus SIGLEC1 versus 24 housekeeping genes (listed in Table 2; log2(transcript units of hY3-treated macrophages - transcript units of macrophages alone), N=4, P<0.001, Figure 1A). Comparing the mean expression +/−SEM, 44% of the enriched IFN-related transcripts were significantly increased in hY3-treated macrophages compared to those treated with lipid alone, while none of the housekeeping transcript increases were significant (Tables 1–2). To authenticate the finding related to SIGLEC1, qPCR was used to determine levels of this lectin receptor, which were significantly elevated in hY3-transfected macrophages versus macrophages given vehicle alone (N=13, P<0.0001, Figure 1B). Serving as a positive control, exposure of macrophages to IFNα resulted in increased expression of SIGLEC1 (N=6, P=0.015, Figure 1B).
Table 1.
hY3-treated macrophages demonstrate increased expression of IFN genes (20) compared to control macrophages in vitro
| Gene Name | Control | SD | hY3 Rx | SD | p-value |
|---|---|---|---|---|---|
| RTP4 | 7.46 | 0.11 | 10.97 | 0.85 | 2.5336E-05 |
| BATF2 | 6.87 | 0.07 | 11.11 | 1.05 | 5.4204E-05 |
| XAF1 | 9.18 | 0.25 | 12.08 | 0.69 | 0.0004 |
| OAS1 | 9.70 | 0.22 | 11.99 | 0.53 | 0.0004 |
| SPATS2L | 10.70 | 0.06 | 11.97 | 0.30 | 0.0005 |
| HERC5 | 8.22 | 0.29 | 12.48 | 1.04 | 0.0005 |
| OASL | 7.50 | 0.49 | 13.21 | 1.38 | 0.0006 |
| CXCL10 | 5.43 | 0.73 | 13.15 | 1.85 | 0.0010 |
| EPSTI1 | 8.43 | 0.43 | 12.33 | 0.93 | 0.0011 |
| IFIT3 | 10.08 | 0.28 | 13.22 | 0.77 | 0.0012 |
| LAMP3 | 6.42 | 0.41 | 10.82 | 0.96 | 0.0013 |
| DDX60 | 9.92 | 0.24 | 12.15 | 0.53 | 0.0013 |
| IFITM3 | 11.66 | 0.27 | 13.59 | 0.45 | 0.0019 |
| ISG15 | 9.99 | 0.38 | 13.90 | 0.97 | 0.0019 |
| IFI44 | 10.59 | 0.27 | 12.79 | 0.53 | 0.0020 |
| HES4 | 5.22 | 0.09 | 6.83 | 0.35 | 0.0028 |
| IFIT1 | 9.06 | 0.50 | 12.90 | 0.93 | 0.0034 |
| OAS2 | 8.76 | 0.34 | 11.16 | 0.57 | 0.0036 |
| MX1 | 9.06 | 0.59 | 13.53 | 1.11 | 0.0039 |
| OTOF | 3.97 | 0.09 | 5.03 | 0.25 | 0.0053 |
| LY6E | 9.83 | 0.25 | 11.63 | 0.44 | 0.0058 |
| OAS3 | 10.31 | 0.40 | 12.94 | 0.62 | 0.0061 |
| IFI44L | 6.21 | 0.97 | 11.85s | 1.38 | 0.0087 |
| RSAD2 | 8.58 | 0.90 | 13.43 | 1.17 | 0.0090 |
| SERPING1 | 10.09 | 0.18 | 11.36 | 0.26 | 0.0318 |
| TRIM6 | 7.46 | 0.35 | 8.12 | 0.50 | 0.0837 |
Values, log2 (TPM); using P-values below 0.05/(# genes), the threshold for significance is P<0.0018.
Table 2.
hY3-treated macrophages do not demonstrate increased expression of housekeeping genes (21) compared to control macrophages in vitro.
| Gene Name | Control | SD | hY3 Rx | SD | p-value |
|---|---|---|---|---|---|
| CHMP2A | 11.24 | 0.15 | 10.92 | 0.09 | 0.0046 |
| PSMB2 | 12.16 | 0.03 | 12.50 | 0.08 | 0.0068 |
| NONO | 12.76 | 0.03 | 12.62 | 0.04 | 0.0072 |
| LDHA | 13.05 | 0.09 | 12.87 | 0.06 | 0.0170 |
| SNRPD3 | 11.91 | 0.04 | 11.77 | 0.06 | 0.0238 |
| C1orf43 | 12.44 | 0.04 | 12.36 | 0.03 | 0.0463 |
| UBC | 13.72 | 0.03 | 13.86 | 0.04 | 0.0519 |
| HPRT1 | 10.21 | 0.21 | 9.82 | 0.15 | 0.0546 |
| GAPDH | 13.66 | 0.03 | 13.42 | 0.05 | 0.0583 |
| GPI | 12.30 | 0.12 | 11.81 | 0.12 | 0.0673 |
| RPLP0 | 13.04 | 0.04 | 12.67 | 0.08 | 0.0758 |
| RPL13A | 13.82 | 0.02 | 13.70 | 0.03 | 0.0904 |
| RAB7A | 12.00 | 0.02 | 11.81 | 0.06 | 0.0945 |
| REEP5 | 12.22 | 0.09 | 11.84 | 0.09 | 0.1120 |
| TFRC | 13.17 | 0.05 | 13.05 | 0.03 | 0.1165 |
| PGK1 | 12.92 | 0.06 | 12.75 | 0.04 | 0.1340 |
| HSP90AB1 | 12.89 | 0.04 | 12.76 | 0.03 | 0.1425 |
| PPIH | 9.37 | 0.18 | 8.95 | 0.14 | 0.1613 |
| GUSB | 13.10 | 0.10 | 12.95 | 0.07 | 0.1635 |
| PPIA | 13.71 | 0.02 | 13.61 | 0.02 | 0.1766 |
| B2M | 13.74 | 0.04 | 13.77 | 0.04 | 0.5909 |
| PSMB4 | 13.05 | 0.04 | 13.03 | 0.02 | 0.6064 |
| VCP | 11.04 | 0.23 | 10.95 | 0.02 | 0.6697 |
| ACTB | 14.26 | 0.03 | 14.28 | 0.01 | 0.8151 |
Values, log2 (TPM); using P-values below 0.05/(# genes), the threshold for significance is P<0.0020.
Figure 1. Expression of targeted IFN-related genes in macrophages transfected with hY3 RNA.
In Panel A, an inspection of transcripts ranked by log2 (hY3-treated macrophages minus untreated macrophages alone) highlight an upregulation of type I IFN response genes (from Table 1) and SIGLEC1 but not housekeeping genes (from Table 2). Use of qPCR confirmed the increased expression of SIGLEC1 in macrophages transfected with hY3 versus macrophages treated solely with vehicle. Additionally, Panel B shows exposure of macrophages to IFNα resulted in significantly increased expression of SIGLEC1. The y axis represents 2-ΔΔCT values of SIGLEC1 normalized to 18S.
3.2. Transcriptome of human fetal leukocytes isolated from CHB and healthy hearts
Encouraged by the in vitro findings, in vivo studies leveraged an unprecedented opportunity to detail the transcriptomes of freshly isolated, cell-sorted leukocytes from three fetal hearts with CHB and three age-matched healthy hearts. A single-cell suspension was prepared and cells sorted after tissue processing of each fetal heart, with the application of sorting parameters as previously reported (16). The initial strategy for sorting leukocytes from CHB-1 and one healthy heart gated solely on CD45+ cells. For CHB-2 and CHB-3 and two additional healthy hearts, the CD45+ flow-sorted population gated on CD11c yielded two further subsets of leukocytes (CD45+CD11c- and CD45+CD11c+). For each population of flow sorted cells, the resultant RNA of the cell fractions was used to generate cDNA libraries for downstream RNA-seq analysis. Of note for the healthy control hearts, the yield of CD45+CD11c+ cells was below the threshold target for RNA-seq.
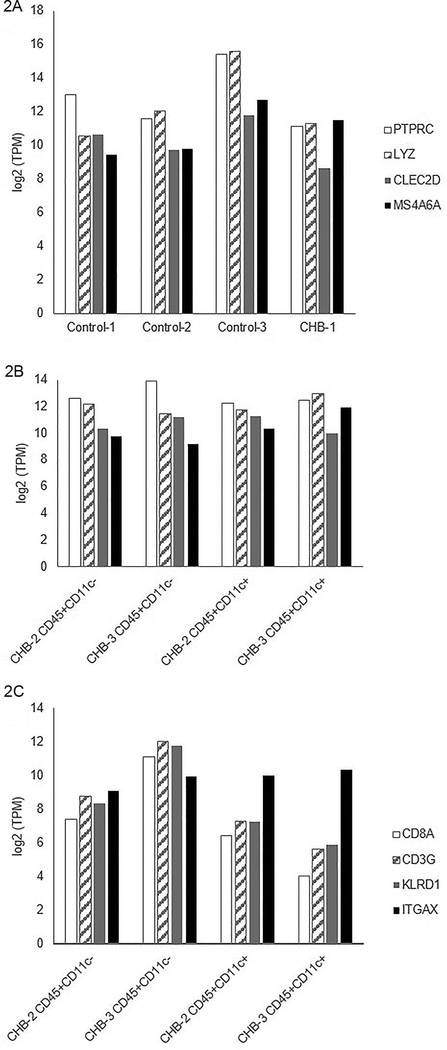
There were no differences in the CHB hearts compared to the controls when evaluating transcripts with utility to authenticate leukocytes, as shown in a comparison of the abundance of monocyte lineage markers within the transcriptome (PTPRC, LYZ, CLEC2D, MS4A6A, Figure 2A-B). Shown in Figure 2C are log2 transformed transcripts per million (TPM) of targeted transcripts (CD8A, CD3G, KLRD1, ITGAX) and leukocyte populations (CHB-1, CD45+CD11c- populations from CHB-2 and CHB-3, and CD45+CD11c+ populations from CHB-2 and CHB-3). Note that compared to ITGAX (a dendritic cell marker), there were lower levels of CD8A, CD3G and KLRD1 among CD45+CD11c+ flow-sorted leukocytes than among their CD45+CD11c- counterparts. While macrophages are versatile immune cells capable of polarizing into functional subsets depending on environmental stimulation (23), a barrier to facile perception of subsets was that, while NOS2 was evident in CD45+CD11c- cells, other markers, such as MRC1, MSR1 and STAB1, were found in CD45+CD11c+ cells (not shown).
Figure 2. Authentication of flow-sorted leukocytes based on expression of macrophage lineage transcripts.
Shown are log2 transformed transcripts per million (TPM) of myeloid lineage gene expression (PTPRC, LYZ, CLEC2D, MS4A6A) among leukocyte populations derived from control and CHB-afflicted hearts (Shown in Panel A are Control-1, Control-2, Control-3, and CHB-1; shown in Panel B are CD45+CD11c- populations from CHB-2 and CHB-3 and CD45+CD11c+ populations from CHB-2 and CHB-3). There were no differences in expression of myeloid lineage genes when comparing individual leukocyte populations with one another. Shown in Panel C are log2 transformed TPM of targeted transcripts (CD8A, CD3G, KLRD1, ITGAX) and leukocyte populations (CD45+CD11c- populations from CHB-2 and CHB-3 and CD45+CD11c+ populations from CHB-2 and CHB-3). Note that compared to ITGAX (a dendritic cell marker), there were lower levels of CD8A, CD3G and KLRD1 among CD45+CD11c+ flow-sorted leukocytes than among their CD45+CD11c- counterparts.
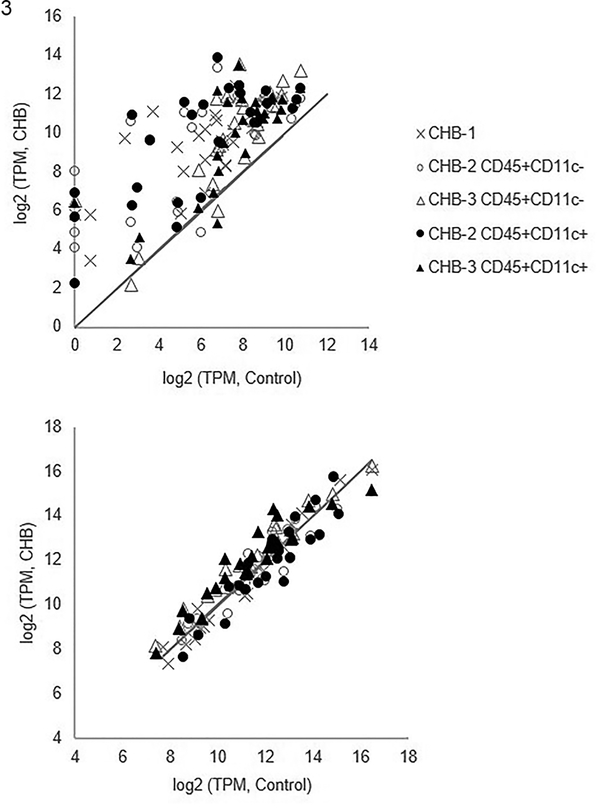
Next, flow-sorted leukocyte transcripts were evaluated applying the IFN and housekeeping modules (same gene modules as described in Tables 1–2). Transcripts within the IFN module plus SIGLEC1, but not the housekeeping module, were significantly elevated in CHB leukocytes compared to controls (Figure 3).
Figure 3. Expression of targeted candidate genes of flow-sorted human leukocytes derived from CHB and healthy fetal hearts.
Shown are log2 transformed transcripts per million (TPM) of CHB leukocyte populations (y-axis) and log2 transformed TPM of the same genes within control leukocyte populations (x-axis) using values of CHB-1, CHB-2, and CHB-3 with corresponding control values (Control-1, Control-2, and Control-3, respectively). The panels show IFN genes (Upper panel, genes listed in Table 1 plus SIGLEC1) and housekeeping genes (Lower panel, genes listed in Table 2). The solid line in both panels represents equivalent expression between CHB and control leukocytes. Notably, the transcripts within the IFN module plus SIGLEC1, but not the housekeeping module, are significantly elevated in CHB leukocytes compared to controls.
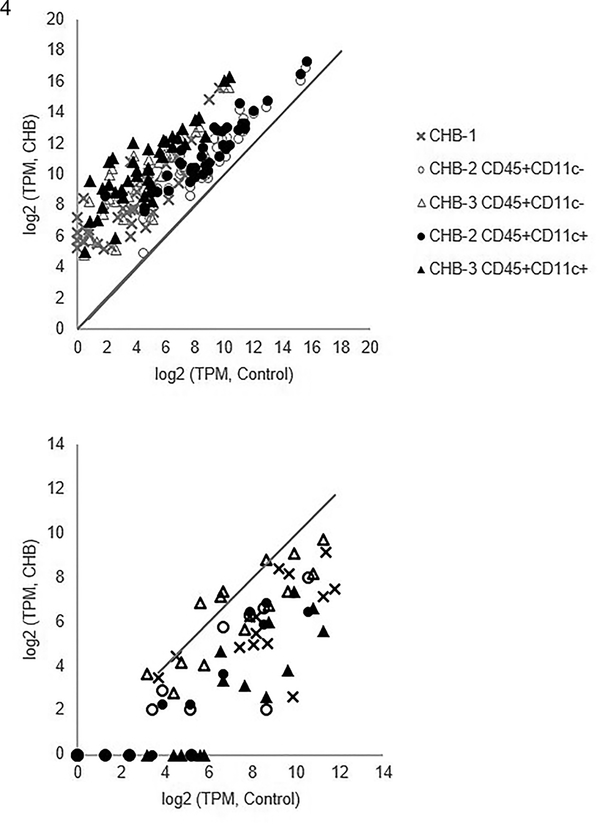
Based on an agnostic bioinformatics evaluation of CHB leukocytes, inspection of the top 500 transcripts ranked by fold change in expression revealed the enrichment of five gene ontology (GO) categories including Extracellular region, Extracellular matrix organization, Cytokine activity, and BMP signaling pathway (Table 3). The transcripts within the top GO module, Extracellular matrix, are significantly elevated in CHB leukocytes compared to controls (Figure 4, upper panel). Among the 39 overexpressed genes within this category (Figure 4), EDN1 and MMP2 have been associated with CHB in prior studies (6, 24). Of the top ten upregulated genes in the CHB leukocytes as ranked by fold increase of expression (Table 4), half were reported within this GO category (MMP2, EPHB2, MFAP5, NOV, and SIGLEC1). Conversely, the transcripts within the GO module Integral component of plasma membrane are significantly lowered in CHB leukocytes compared to control (Figure 4, lower panel). Three of the ten most steeply down-regulated genes in the CHB leukocytes as ranked by fold decrease in expression (Table 5, PTGDR2, CD1C and CD1E) are among the 14 underexpressed genes within this category (Figure 4).
Table 3.
Association of enriched transcripts derived from CHB leukocytes and GO categories from DAVID bioinformatics.
| GO Category | P-value |
|---|---|
| Extracellular matrix | 3.9 × 10−25 |
| Platelet-derived growth factor binding | 5.2 × 10−5 |
| Type 1 interferon signaling pathway | 6.2 × 10−5 |
| Negative chemotaxis | 7.2 × 10−4 |
| Cellular response to interleukin 1 | 0.004 |
Figure 4. Agnostic survey of genes of flow-sorted human leukocytes derived from CHB and healthy fetal hearts.
Shown are log2 transformed transcripts per million (TPM) of CHB leukocyte populations (y-axis) and log2 transformed TPM of the same genes within control leukocyte populations (x-axis) using values of CHB-1, CHB-2, and CHB-3 with corresponding control values (Control-1, Control-2, and Control-3, respectively). The upper and lower panels correspond to genes within the GO categories Extracellular region and Integral component of plasma membrane, respectively. The solid line in both panels represents equivalent expression between CHB and control leukocytes. In the upper panel, the transcripts within the Extracellular region GO module are significantly elevated in CHB leukocytes compared to control. Among the 39 upregulated genes within this category (ELN, EDN1, CX3CL1, MMP2, EPHB2, NOV, OLFML1, ACE, BCHE, PAPPA, FNDC1, GDF6, COL27A1, SERPINE1, COL6A3, CFH, COL6A1, AGRN, IGFBP5, BMP6, COL8A1, NRG1, THBS2, BMP4, SVEP1, STC2, EFEMP1, HSPG2, COL5A3, SLIT3, PRELP, SIGLEC1, VEGFC, BGN, COL1A2, COL1A1, ADAMTS2, MFAP5, and PLA2G), note that EDN1, MMP2, and SERPINE1 have been associated with CHB in prior studies (6, 24). Of the top ten upregulated genes in the CHB leukocytes as ranked by fold increase of expression (Table 4), half were reported within this GO category (MMP2, EPHB2, MFAP5, NOV, and SIGLEC1). For the lower panel, the genes within the Integral component of plasma membrane GO module are significantly lowered in CHB leukocytes compared to control. Three of the ten most steeply down-regulated genes in the CHB leukocytes as ranked by fold decrease in expression (Table 5, PTGDR2, CD1C and CD1E) are among the 14 underexpressed genes within this category (FCER1A, LTK, CXCR5, CD40LG, CD8B, ST14, CD1C, MS4A2, MAL, CD1A, PTGDR2, CD1E, CD1D, and SLC14A2).
Table 4.
Upregulated genes for CHB vs healthy flow-sorted leukocytes.
| Rank | Gene symbol | Gene Name | Comments | Fold increase* |
|---|---|---|---|---|
| 1 | NOV | Nephroblastoma overexpressed | Wound healing | 74 |
| 2 | NRK | Nik related kinase | Inflammation | 69 |
| 3 | IFI27 | Interferon alpha inducible gene 27 | Interferon | 68 |
| 4 | EPHB2 | EPH Receptor B2 | Fibrosis | 68 |
| 5 | MFAP5 | Microfibrillar-associated protein 5 | Inflammation | 67 |
| 6 | DLK1 | Delta Like non-canonical Notch Ligand 1 | Unknown | 55 |
| 7 | CDH19 | Cadherin 19 | Unknown | 53 |
| 8 | MMP2 | Matrix Metalloproteinase 2 | Fibrosis | 49 |
| 9 | SIGLEC1 | Sialic Acid Binding Ig Like Lectin 1 | Interferon | 47 |
| 10 | SVEP1 | Sushi, Von Willebrand Factor Type A, EGF And Pentraxin Domain Containing 1 | Fibrosis | 44 |
TPM of CHB flow-sorted leukocytes/TPM of healthy flow-sorted leukocytes
Table 5.
Downregulated genes for CHB vs healthy flow-sorted leukocytes.
| Rank | Gene symbol | Gene Name | Comments | Fold decrease* |
|---|---|---|---|---|
| 1 | CHRNA3 | Cholinergic Receptor Nicotinic Alpha 3 | Cholinergic (nicotinic) anti-inflammatory pathway | 35 |
| 2 | GSTM1 | Glutathione S- Transferase Mu 1 | Cell-metabolism | 28 |
| 3 | PTGDR2 | Prostaglandin D2 Receptor 2 | Promotes macrophage activation | 17 |
| 4 | LINC00526 | Long Intergenic Non-Protein Coding RNA 526 | Non-coding RNA | 16 |
| 5 | FCER1A | Fc Fragment Of IgE Receptor Ia | Lineage marker of mast cell | 16 |
| 6 | EDAR | Ectodysplasin A Receptor | Unknown | 14 |
| 7 | SNAI3 | SNAI3 Antisense RNA 1 | Non-coding RNA | 13 |
| 8 | CD1C | T-Cell Surface Glycoprotein CD1c | Activation marker of T cells | 13 |
| 9 | CD1E | T-Cell Surface Glycoprotein CD1e | Activation marker of T cells | 12 |
| 10 | TPSAB1 | Tryptase Alpha/Beta 1 | Lineage marker of mast cell | 11 |
1/(TPM of CHB flow-sorted leukocytes/TPM of healthy flow-sorted leukocytes)
3.3. Immunohistologic evidence implicating IFN in the pathogenesis of CHB
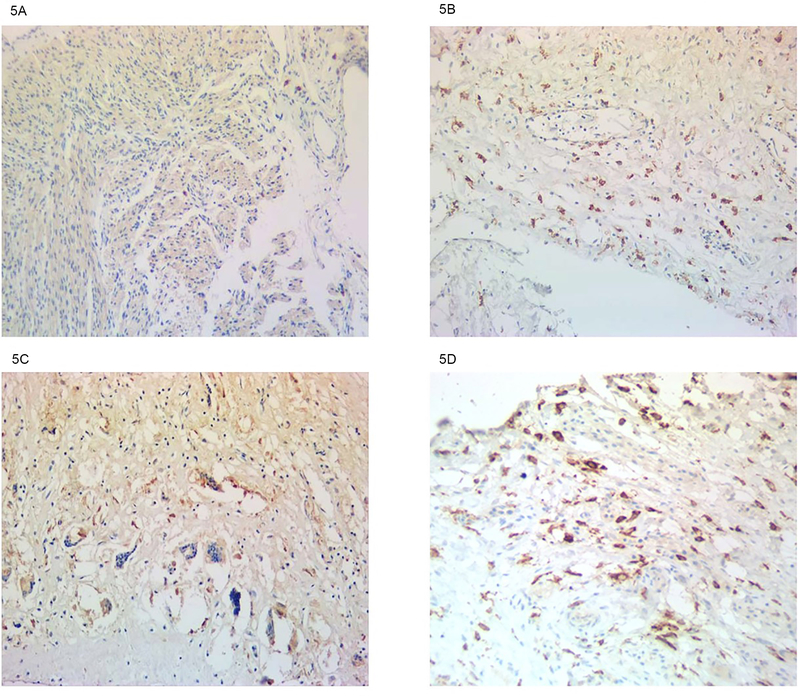
To directly address the potential contribution of an activated type I IFN system to conduction injury mediated by anti-SSA/Ro antibodies, available cardiac tissue from several cases of CHB and an age-matched healthy heart were interrogated for Siglec-1 expression. Assessing longitudinal sections through the septa of three CHB hearts, there was diffuse staining of Siglec-1, which was observed in cells proximal to areas of injury (Figure 5). Based on morphology, the two positive cell types expressing Siglec-1 were macrophages and dendritic cells. In contrast, Siglec-1 stained cells were rare in the healthy age-matched hearts (Figure 5).
Figure 5. Detection of Siglec-1-expressing macrophages in healthy control and CHB fetal hearts.
A normal 22-week-old fetal heart is shown in A, whereas CHB fetal hearts at ages 22, 20 and 25 weeks are shown in B, C, and D respectively. Longitudinal septal sections of cases (see Methods, CHB autopsies) and control were stained with anti-Siglec-1 antibody along with a counterstain of hematoxylin.
4.0. Discussion
Numerous studies focusing on anti-SSA/Ro injury and macrophage recruitment demonstrate that stimulated macrophages are strongly associated with inflammation and fibrosis. Using an agnostic in vitro approach, TLR ligation of macrophages with a Ro60-associated ssRNA led to the upregulation of a panel of IFN transcripts, including SIGLEC1, a result which was corroborated using qPCR. Transcriptome analysis of CD45+ flow-sorted cells, representing in vivo leukocytes, provided further evidence. Utilizing independent and agnostic bioinformatics approaches, leukocytes flow-sorted from three hearts of fetuses dying with CHB highly expressed type I IFN response genes inclusive of SIGLEC1. Furthermore, Siglec-1 was identified in the septal region of several affected fetal hearts. In sum, these data support a connection of Siglec-1 as a disease-provoking factor leading to a pathological, persistently activated macrophage.
Over the last few decades of research into the pathogenesis of CHB, immunohistochemistry of the affected tissue has provided important clues. In a seminal article reported in 1985, Litsey described a mononuclear cell infiltration in the myocardium of an anti-SSA/Ro-exposed fetus dying in utero at 18 weeks of gestation (25). In 1987, Lee reported patchy lymphoid aggregates throughout the myocardium of an infant delivered at 30 weeks and dying in the immediate postnatal period (26). It was later observed that macrophages and multinucleated giant cells were frequently seen proximal to IgG and apoptotic cells in several hearts dying with CHB (3). Llanos and coworkers identified mononuclear infiltrates as a consistent feature, most apparent in the youngest fetuses (15).
The current study extends our in vitro model in which macrophages have centered prominently as a cellular link between anti-SSA/Ro-mediated injury and fibroblast transdifferentiation and scar formation. In line with our working hypothesis that macrophage uptake of apoptotic fetal cardiocytes bound by immune complexes containing ssRNA results in the secretion of inflammatory and profibrosing mediators (6, 7, 27), treatment of macrophages with hY3 resulted in the increased expression of SIGLEC1 and other IFN-related genes. Based on microarray, mRNA expression of both IFNα and IFNβ are increased in macrophages treated with hY3 (17). Thus, macrophages could provide a source and feedforward loop of IFN and subsequent signaling through an IFN receptor. In parallel with the macrophage response to IFN as reflected by Siglec-1, we recently reported a further bystander effect of IFN enrichment in the transcriptome of fibroblasts isolated from a CHB heart (16). In aligning these data with our previous observations of enhanced TNFα in situ (5), it is of note that Yarilina et al (28) reported that the binding of TNF and TNF receptors as occurs in rheumatoid arthritis synovium results in a secretion of Type I IFN. In our CHB model, macrophage-derived TNFα may be a driver of the release of IFN by fibroblasts; then, in a paracrine manner the IFN and IFN receptors on macrophages may result in upregulation of SIGLEC1 or as stated above in an autocrine manner by the macrophages themselves.
Highly relevant to the affected CHB hearts studied herein, it was recently reported that in CD14+ monocytes, levels of Siglec-1 were higher in anti-SSA/Ro-positive women carrying a fetus with CHB compared to anti-SSA/Ro-positive women carrying a healthy fetus. This finding was independent of maternal disease burden (11). Moreover, mothers of affected children have significantly higher levels of circulating IFNα. While the investigative team speculated that IFNα influences the development of CHB via maternal transplacental passage, our results suggest that there may also be a fetal source.
Increased Siglec-1 expression has also been demonstrated in other diseases in which there is fibrosis (29). Using flow cytometry, Siglec-1-positive cells were shown to be increased in patients compared to controls, noting also that the major cell type was CD14+ monocytes. In parallel with our cardiac results, Siglec-1 cells were prevalent in the dermis of lesional sclerotic skin while absent in skin of healthy controls. To explore pathways yielding Siglec-1-expressing macrophages, York and co-workers demonstrated a significant upregulation of Siglec-1 by PBMCs treated with IFNα, a TLR7 agonist, and a TLR3 agonist, but not a TLR2 agonist. Blockage of IFN by B18R (a vaccinia virus-encoded receptor, which specifically binds IFNα) attenuated Siglec-1 expression. Furthermore, Siglec-1 expression seems to have a pathological contribution in a murine model of colitis. Prior to chemical induction of colitis, Siglec-1 cells were located adjacent to the vascular-rich sub-mucosa of intestinal tissue (30). Chemical injury resulted in the contact of Siglec-1cells with dying/dead cells, which triggered a cascade of events including the synthesis and release of CCL8, a chemokine that recruits additional macrophages to the site of injury. The co-administration of anti-CCL8 antibody did not completely suppress the symptoms of chemically-induced colitis as compared with the depletion of CD169 macrophages. In addition to recruitment, there is also the speculation that Siglec-1 might regulate monocyte interactions with other mononuclear cells in the circulation or in the tissues (13). A similar pathway may involve an axis of Siglec-1/CCL8 with recruitment of lymphocytes in other diseases such as atopic dermatitis (31).
Several limitations of this study are acknowledged. While the analysis of only three affected fetuses precludes generalizability to all affected cases, immunohistochemistry from three additional CHB cases provided supportive data showing diffuse expression of Siglec-1 in the septal region. Certainly, the opportunity to harvest cells in short temporal proximity to an electively terminated CHB fetus is quite rare. Although it is acknowledged that leukocytes were isolated from the whole diseased heart and not the specific area of injury, it is well described that injury can extend beyond the AV node (15, 32), which indeed it did in two of these cases as evidenced by echocardiograms that revealed involvement of the myocardium with endocardial fibroelastosis and extremely poor overall cardiac functioning. For in vivo assessments, the pool of flow-sorted leukocytes may contain contaminating cells of non-leukocyte origin. However, there was enrichment of lineage markers, reflecting an isolate within acceptable limits of flow sorting. Finally, our premise that macrophages undergo persistent activation is a concept challenging to affirm in human studies because procurement was obtained at early third term gestation.
In summary, data generated directly from the diseased CHB hearts support a role for IFNα and positions Siglec-1-positive macrophages as integral to the pathologic process. These cells may have the potential to amplify the burden of disease through recruitment of other mononuclear cells and biological properties related to matrix regulation. Thus, a reasonable prediction is that IFNs may contribute to both susceptibility and regulatory factors, each of which could influence the fate of anti-SSA/Ro exposure and whether the proposed pathologic cascade eventuates in irreversible third degree heart block.
Acknowledgements
The authors are grateful to Ben Wainwright for excellent editorial assistance.
This work was supported by a National Institutes of Health Merit Award (R37 AR042455, R37 AR042455–21S1, R37 AR042455–21S2 (J.P.B.)), the Research Registry for Neonatal Lupus (N01 AR-4–2220 (J.P.B.)), a Lupus Foundation of America Lifeline Grant (J.P.B.) and the National Institutes of Health (R03 HD069986 and R01 HD079951–01A1 (J.P.B.)). For the genomics facility, RNA sequencing relied on a shared resource, which is partially supported by a Cancer Center Support Grant (P30 CA016087).
Literature Cited.
- 1.Buyon JP, Clancy RM, and Friedman DM. 2009. Autoimmune associated congenital heart block: integration of clinical and research clues in the management of the maternal / foetal dyad at risk. J Intern Med 265: 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buyon JP, Hiebert R, Copel J, Craft J, Friedman D, Katholi M, Lee LA, Provost TT, Reichlin M, Rider L, Rupel A, Saleeb S, Weston WL, and Skovron ML. 1998. Autoimmune-associated congenital heart block: demographics, mortality, morbidity and recurrence rates obtained from a national neonatal lupus registry. J Am Coll Cardiol 31: 1658–1666. [DOI] [PubMed] [Google Scholar]
- 3.Clancy RM, Kapur RP, Molad Y, Askanase AD, and Buyon JP. 2004. Immunohistologic evidence supports apoptosis, IgG deposition, and novel macrophage/fibroblast crosstalk in the pathologic cascade leading to congenital heart block. Arthritis Rheum 50: 173–182. [DOI] [PubMed] [Google Scholar]
- 4.Clancy RM, Askanase AD, Kapur RP, Chiopelas E, Azar N, Miranda-Carus ME, and Buyon JP. 2002. Transdifferentiation of cardiac fibroblasts, a fetal factor in anti-SSA/Ro-SSB/La antibody-mediated congenital heart block. J Immunol 169: 2156–2163. [DOI] [PubMed] [Google Scholar]
- 5.Clancy RM, Backer CB, Yin X, Kapur RP, Molad Y, and Buyon JP. 2003. Cytokine polymorphisms and histologic expression in autopsy studies: contribution of TNF-alpha and TGF-beta 1 to the pathogenesis of autoimmune-associated congenital heart block. J Immunol 171: 3253–3261. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez D, Briassouli P, Clancy RM, Zavadil J, Reed JH, Abellar RG, Halushka M, Fox-Talbot K, Barrat FJ, and Buyon JP. 2011. A novel role of endothelin-1 in linking Toll-like receptor 7-mediated inflammation to fibrosis in congenital heart block. J Biol Chem 286: 30444–30454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy RM, Alvarez D, Komissarova E, Barrat FJ, Swartz J, and Buyon JP. 2010. Ro60-associated single-stranded RNA links inflammation with fetal cardiac fibrosis via ligation of TLRs: a novel pathway to autoimmune-associated heart block. J Immunol 184: 2148–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranda-Carus ME, Askanase AD, Clancy RM, Di Donato F, Chou TM, Libera MR, Chan EK, and Buyon JP. 2000. Anti-SSA/Ro and anti-SSB/La autoantibodies bind the surface of apoptotic fetal cardiocytes and promote secretion of TNF-alpha by macrophages. J Immunol 165: 5345–5351. [DOI] [PubMed] [Google Scholar]
- 9.Emamian ES, Leon JM, Lessard CJ, Grandits M, Baechler EC, Gaffney PM, Segal B, Rhodus NL, and Moser KL. 2009. Peripheral blood gene expression profiling in Sjogren’s syndrome. Genes Immun 10: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, and Behrens TW. 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100: 2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lisney AR, Szelinski F, Reiter K, Burmester GR, Rose T, and Dorner T. 2017. High maternal expression of SIGLEC1 on monocytes as a surrogate marker of a type I interferon signature is a risk factor for the development of autoimmune congenital heart block. Ann Rheum Dis 76: 1476–1480. [DOI] [PubMed] [Google Scholar]
- 12.Hartnell A, Steel J, Turley H, Jones M, Jackson DG, and Crocker PR. 2001. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood 97: 288–296. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg TK, Nath D, Ziltener HJ, Vestweber D, Fukuda M, van Die I, and Crocker PR. 2001. Cutting edge: CD43 functions as a T cell counterreceptor for the macrophage adhesion receptor sialoadhesin (Siglec-1). J Immunol 166: 3637–3640. [DOI] [PubMed] [Google Scholar]
- 14.Biesen R, Demir C, Barkhudarova F, Grun JR, Steinbrich-Zollner M, Backhaus M, Haupl T, Rudwaleit M, Riemekasten G, Radbruch A, Hiepe F, Burmester GR, and Grutzkau A. 2008. Sialic acid-binding Ig-like lectin 1 expression in inflammatory and resident monocytes is a potential biomarker for monitoring disease activity and success of therapy in systemic lupus erythematosus. Arthritis Rheum 58: 1136–1145. [DOI] [PubMed] [Google Scholar]
- 15.Llanos C, Friedman DM, Saxena A, Izmirly PM, Tseng CE, Dische R, Abellar RG, Halushka M, Clancy RM, and Buyon JP. 2012. Anatomical and pathological findings in hearts from fetuses and infants with cardiac manifestations of neonatal lupus. Rheumatology (Oxford) 51: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy RM, Markham AJ, Jackson T, Rasmussen SE, Blumenberg M, and Buyon JP. 2017. Cardiac Fibroblast Transcriptome Analyses Support a Role for Interferogenic, Profibrotic and Inflammatory Genes in Anti-SSA/Ro-Associated Congenital Heart Block. Am J Physiol Heart Circ Physiol: ajpheart 00256 02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clancy RM, Markham AJ, Reed JH, Blumenberg M, Halushka MK, and Buyon JP. 2016. Targeting downstream transcription factors and epigenetic modifications following Toll-like receptor 7/8 ligation to forestall tissue injury in anti-Ro60 associated heart block. J Autoimmun 67: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, and Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology 4: P3. [PubMed] [Google Scholar]
- 19.Brusic V, Gottardo R, Kleinstein SH, Davis MM, and H. s. committee. 2014. Computational resources for high-dimensional immune analysis from the Human Immunology Project Consortium. Nat Biotechnol 32: 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, Bennett L, Allantaz F, Mejias A, Ardura M, Kaizer E, Monnet L, Allman W, Randall H, Johnson D, Lanier A, Punaro M, Wittkowski KM, White P, Fay J, Klintmalm G, Ramilo O, Palucka AK, Banchereau J, and Pascual V. 2008. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 29: 150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiche L, Jourde-Chiche N, Whalen E, Presnell S, Gersuk V, Dang K, Anguiano E, Quinn C, Burtey S, Berland Y, Kaplanski G, Harle JR, Pascual V, and Chaussabel D. 2014. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol 66: 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, Wang T, Jordan N, Bornkamp N, Nwaukoni J, Martinez J, Goilav B, Buyon JP, Tuschl T, and Putterman C. 2017. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohensinner PJ, Baumgartner J, Kral-Pointner JB, Uhrin P, Ebenbauer B, Thaler B, Doberer K, Stojkovic S, Demyanets S, Fischer MB, Huber K, Schabbauer G, Speidl WS, and Wojta J. 2017. PAI-1 (Plasminogen Activator Inhibitor-1) Expression Renders Alternatively Activated Human Macrophages Proteolytically Quiescent. Arterioscler Thromb Vasc Biol 37: 1913–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena A, Izmirly PM, Han SW, Briassouli P, Rivera TL, Zhong H, Friedman DM, Clancy RM, and Buyon JP. 2015. Serum Biomarkers of Inflammation, Fibrosis, and Cardiac Function in Facilitating Diagnosis, Prognosis, and Treatment of Anti-SSA/Ro-Associated Cardiac Neonatal Lupus. J Am Coll Cardiol 66: 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litsey SE, Noonan JA, O’Connor WN, Cottrill CM, and Mitchell B. 1985. Maternal connective tissue disease and congenital heart block. Demonstration of immunoglobulin in cardiac tissue. N Engl J Med 312: 98–100. [DOI] [PubMed] [Google Scholar]
- 26.Lee LA, Coulter S, Erner S, and Chu H. 1987. Cardiac immunoglobulin deposition in congenital heart block associated with maternal anti-Ro autoantibodies. Am J Med 83: 793–796. [DOI] [PubMed] [Google Scholar]
- 27.Clancy RM, Markham AJ, and Buyon JP. 2016. Endosomal Toll-like receptors in clinically overt and silent autoimmunity. Immunol Rev 269: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarilina A, Park-Min KH, Antoniv T, Hu X, and Ivashkiv LB. 2008. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol 9: 378–387. [DOI] [PubMed] [Google Scholar]
- 29.York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, and Lafyatis R. 2007. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum 56: 1010–1020. [DOI] [PubMed] [Google Scholar]
- 30.Asano K, Takahashi N, Ushiki M, Monya M, Aihara F, Kuboki E, Moriyama S, Iida M, Kitamura H, Qiu CH, Watanabe T, and Tanaka M. 2015. Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes. Nat Commun 6: 7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, Lira SA, Charo IF, and Luster AD. 2011. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat Immunol 12: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuneo BF, Fruitman D, Benson DW, Ngan BY, Liske MR, Wahren-Herlineus M, Ho SY, and Jaeggi E. 2011. Spontaneous rupture of atrioventricular valve tensor apparatus as late manifestation of anti-Ro/SSA antibody-mediated cardiac disease. Am J Cardiol 107: 761–766. [DOI] [PubMed] [Google Scholar]