Abstract
Objective
High expression alleles of macrophage migration inhibitory factor (MIF) are linked genetically to SLE disease severity. The U1-snRNP (snRNP) immune complex containing U1-snRNP and anti-U1-snRNP antibodies, which are found in SLE, activates the NLRP3 inflammasome comprised of NLRP3, ASC, and procaspase-1 in human monocytes, leading to the production of IL-1β. The role of the snRNP immune complex in upregulating the MIF and its interface with the NLRP3 inflammasome were investigated.
Methods
MIF, IL-1β, NLRP3, caspase-1, ASC, and MIF receptors were analyzed in human monocytes incubated with or without the snRNP immune complex by ELISA, Western blot, qPCR, and CyTOF. MIF pathway responses were probed with the novel small molecule antagonist MIF098.
Results
The snRNP immune complex induced the production of MIF and IL-1β from human monocytes. High-dimensional single cell CyTOF analysis established MIF regulation of inflammasome activation, including a quantitative relationship in MIF, its receptors, and IL-1β, in monocytes. MIF098, which blocks MIF binding to its cognate receptor, suppressed IL-1β production and NLRP3 upregulation, a rate-limiting step in activating the NLRP3 inflammasome, as well as caspase-1 activation in snRNP immune complex-stimulated human monocytes.
Conclusion
The U1-snRNP immune complex is a specific stimulus of MIF production in human monocytes, with MIF having an upstream role in defining the inflammatory characteristics of activated monocytes by regulating NLRP3 inflammasome activation and downstream IL-1β production. These findings provide mechanistic insight and a therapeutic rationale for targeting MIF in subgroups of lupus patients, such as high genotypic MIF expressers or those with anti-snRNP antibodies.
Keywords: macrophage migration inhibitory factor (MIF), systemic lupus erythematosus (SLE), NLRP3, inflammasome
Introduction
Macrophage migration inhibitory factor (MIF) produced primarily from activated monocytes and macrophages is an upstream activator of innate immune responses (1–3). In addition to its effect on inhibiting the migration of monocyte and macrophage mobility, MIF promotes inflammatory responses by counter-regulating the inhibitory effect of glucocorticoids on the production of the inflammatory cytokines from macrophages and suppressing p53-dependent cell death (4, 5). The MIF receptor complex is comprised of the transmembrane ligand-binding component CD74 and the CD44 signaling component (6, 7). MIF also competes with cognate ligands for CXCR4 and CXCR2, and directly binds to CXCR2 in a macromolecular receptor complex with CD74 (8). Human and animal studies have supported MIF’s role in the pathogenesis of infectious and inflammatory conditions including septic shock, malaria, rheumatoid arthritis, and systemic lupus erythematosus (SLE or lupus) (9–13). MIF is over-expressed in lupus-prone mice, and MIF-deficient MRL/lpr lupus-prone mice are protected from glomerular injury (14). The therapeutic efficacy of blocking MIF in lupus-prone mice also was previously demonstrated (11). An association between high expression MIF alleles with susceptibility and deep organ involvement has been reported (12, 15). In lupus patients, circulatory MIF levels are increased (15) and correlate with disease damage cross-sectionally and longitudinally (16, 17). These findings support the pathogenic role of MIF and the therapeutic value of targeting MIF-dependent pathways in lupus, which is currently under study with the clinical testing of anti-CD74 (18).
The pathologic hallmarks of SLE are altered immune responses to nuclear autoantigens with autoantibody production and subsequent tissue injury (19, 20). Experimental studies support the critical role of innate immunity, in addition to that of adaptive immunity, in the development of lupus and in the pathologic progression of disease. Plasmacytoid dendritic cells (pDCs) recognize lupus self-antigens via Toll-like receptors (TLRs), leading to the production of IFN-α, which is linked to lupus pathogenesis and its clinical manifestations (21–23). For instance, TLR7 and 8 recognize the ssRNA of the self-antigen U1-small nuclear ribonucleoprotein (U1-snRNP) that is targeted by anti-U1-snRNP antibodies (Abs) in lupus (21). In support of this pathway, TLR7-deficient lupus-prone mice have ameliorated disease (24). We recently showed the production of IL-1β from human monocytes in response to a combination of U1-snRNP and anti-U1-snRNP Ab-positive serum (referred to as snRNP immune complex) by activation of the NLRP3 inflammasome comprised of NLRP3, the adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD), and procaspase-1 (25). NLRP3 recruits ASC and procaspase-1, leading to the assembly of the NLRP3 inflammasome, which cleaves pro-IL-1β to mature IL-1β (26). NLRP3 appears to be a limiting step because the protein level of NLRP3 is relatively low in resting macrophages, a phenomenon observed in murine and immortalized human macrophages (27, 28). Of note, patients with SLE have increased activation of the NLRP3 inflammasome in monocytes, which may be related to exposure to IFN-α (29). Lupus-prone mice treated with an NLRP3 inflammasome inhibitor or deficient in caspase-1 also show reduced disease (30, 31), further supporting the pathogenic role of innate immunity and the NLRP3 inflammasome pathway.
Although genetic, clinical, and mouse modeling data implicate MIF and the NLRP3 inflammasome in the pathogenesis and clinical progression of lupus, little is known about the possible interface between the two pathways at the molecular level. Here we demonstrate the lupus snRNP immune complex as a specific stimulus of human MIF production and support the upstream regulatory role of MIF in activating the NLRP3 inflammasome and subsequent production of IL-1β. We also define the molecular characteristics of these activated monocyte populations.
Methods
Human monocytes and sera
Human peripheral blood was obtained from healthy adult donors after informed consent. Fresh monocytes were purified from blood using a negative cell purification kit (Stem cell Technologies Inc, Vancouver, BC, Canada). Anti–U1-snRNP Ab-positive sera were obtained from the L2 Diagnostic Laboratory. Anti-U1-snRNP Abs were measured by ELISA (DiaSorin, Stillwater, MN). Healthy control sera were obtained from the peripheral blood of healthy donors. This work was approved by the institutional review committee of Yale University.
Monocyte stimulation
Purified monocytes (1 × 105) were resuspended in 200 μl of RPMI 1640 media supplemented with 10% FCS, penicillin, and streptomycin. Monocytes were treated for 30 min with the MIF antagonist 3-(3-hydroxybenzyl)-5-methylbenzooxazol-2-one, designated MIF098 (20 μM, at a dose determined by a dose-kinetic study, not shown) (32) followed by stimulation for 3, 7, or 18 hours with or without U1-snRNP (5 μg/ml, AroTec Diagnostics Limited, New Zealand) in the presence or absence of anti-U1-snRNP Ab+ or healthy serum (final concentration of 5 %) (25). Some cells were treated for 18 hours with U1-snRNP and anti-U1-snRNP Ab+ serum in the presence or absence of recombinant human MIF (40 μg/ml, R&D Systems, Minneapolis, MN)
ELISA, qPCR, flow cytometry and LDH-based cytotoxicity assay
IL-1β and MIF in culture supernatants were measured by sandwich enzyme-linked immunosorbant assay (ELISA) using a commercially available IL-1β kit (ebioscience, San Diego, CA) and specific antibodies for MIF (33), respectively. IL1B, NLRP3, MIF, MARCH7, and TRIM31 genes were determined by qPCR. Primer sequences for qPCR are shown in Supplementary Table 1. Total RNA was extracted from cells using the RNeasy Plus Midi kit (QIAGEN, Germantown, MD) and cDNA was synthesized. Each real-time PCR reaction was performed on a 10 μl reaction mixture containing cDNA, 2× Brilliant SYBR green master mix (Stratagene, San Diego, CA), and 3 μM of each primer. The reaction mixture was denatured for 10 min at 94°C and incubated for 40 cycles (denaturing for 15 s at 95°C and annealing and extending for 1 min at 60°C) using the Mx3005P QPCR system (Stratagene). GAPDH was amplified as an internal control. The relative RNA levels were calculated by the 2−ΔΔCT algorithm. Freshly isolated monocytes were stained with Abs to CD44-FITC, CD74-PE, CXCR2-FITC, or CXCR4-PE (all from Biolegend, San Diego, CA) and analyzed using an LSRII flow cytometer (BD Biosciences, Franklin Lakes, NJ) and FlowJo software (FlowJo, LLC, Ashland, Oregon). LDH-based cytotoxicity assay (Promega, Madison, WI) was performed on the culture supernatants of monocytes incubated for 18 hours with U1-snRNP (snRNP, 5 μg/ml) and anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration) in the presence or absence of the MIF antagonist MIF098 (20 μM) following the manufacturer’s instructions (34).
Western blotting
Protein extracts that were separated by SDS-PAGE and transferred onto PVDF membranes were probed with Abs against total NF-κB p65, phospho-NF-κB p65, caspase-1 p20, ASC (all from Cell signaling Technology, Danvers, MA), NLRP3 (Enzo life sciences, Farmingdale, NY), IL-1β (Santa Cruz Biotechnology, Santa Cruz, CA) and GAPDH (Santa Cruz Biotechnology). The probed membranes were washed and incubated with HRP-labeled secondary antibodies (Santa Cruz Biotechnology). The bands were visualized with the Pierce ECL Western blotting substrate (Thermo Scientific, Rockford, IL).
Immunofluorescence staining
Formalin-fixed paraffin-embedded sections from human acute cutaneous lupus and normal skin obtained from the Department of Pathology at Yale Medical School were de-waxed and rehydrated with serial ethanol treatments. Heat-induced antigen retrieval was performed. After blocking, the tissue slides were serially incubated overnight at 4°C with rabbit anti-CD14 (Invitrogen, Carlsbad, CA), mouse anti-CD74 (R&D Systems), and goat anti-NLRP3 Abs (R&D Systems) followed by staining with secondary Abs (Alexa594-donkey anti-rabbit, Alexa488-goat anti-mouse and Alexa647-rabbit anti-goat antibodies, Molecular Probes) and Hoechst 33342 (Immunochemistry). Some sections were incubated with mouse anti-MIF Abs (R&D Systems) and subsequently stained with secondary Abs (Alexa488-goat anti-mouse). Staining-positive cells were detected with the Leica DM6000 FS fluorescence microscope and LEICA 5.0 software (Leica Microsystems).
CyTOF Analysis
All mass cytometry reagents were purchased from Fluidigm, Inc (South San Francisco, CA) unless otherwise noted. Monocytes (5×105) were treated for 30 min with or without MIF098 followed by 5 hours of incubation with U1-snRNP (5 μg/ml) in the presence of anti-U1snRNP-Ab-positive serum (5% final concentration). Incubated cells were stained with a panel of metal-tagged Abs (Supplementary Table 2) and Cisplatin. For intracellular staining, cells were fixed and permeabilized with Maxpar Fix 1 buffer and Maxpar Perm-S buffer, respectively. Stained cells were washed and kept overnight in the MaxPar Fix & Perm Buffer containing intercalator-Ir. Cells were resuspended with MaxPar Water containing EQ Four Element Calibration Beads and acquired on a CyTOF system Helios (Fluidigm). All FCS files were normalized and analyzed using the CYT, an open source analytic tool for CyTOF data, and FlowJo software. PhenoGraph, t-distributed stochastic neighbor embedding (t-SNE), computational algorithms conditional-Density Resampled Estimate of Mutual Information (DREMI) and conditional-Density Rescaled Visualization (DREVI) were performed on gated cells (35, 36).
Statistical analysis
Data were statistically analyzed by the paired t-test and two-way ANOVA as appropriate using Microsoft Excel (Redmond, WA) and GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA), respectively. P values of less than 0.05 were considered statistically significant.
Results
The lupus immune complex of U1-snRNP and anti-U1-snRNP Ab-positive serum (snRNP immune complex) induced MIF from human monocytes, leading to promoting the production of IL-1β
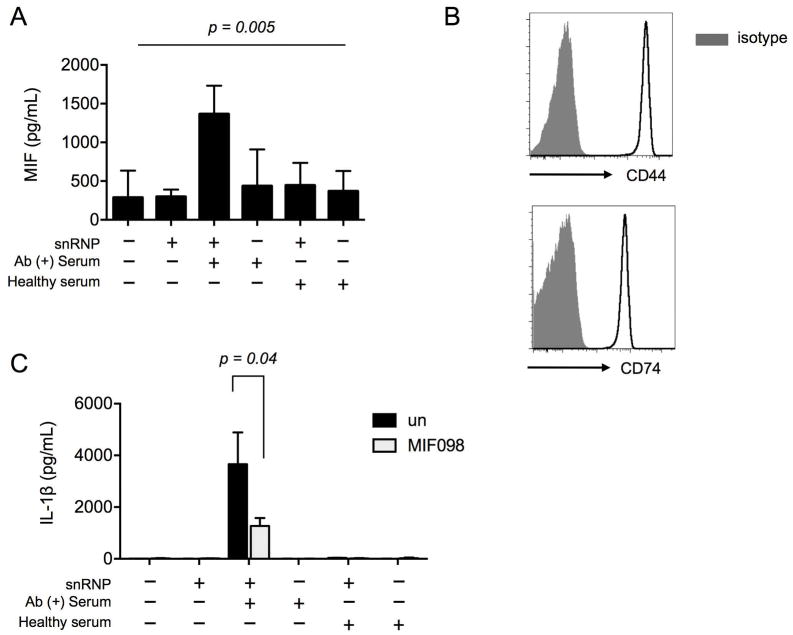
We explored whether MIF could be released from human monocytes in response to the snRNP immune complex and modulate the production of IL-1β. High levels of MIF were detected in the culture supernatants of monocytes incubated with the snRNP immune complex (Fig 1A). By contrast, U1-snRNP, anti-U1-snRNP Ab-positive serum, or a combination of U1-snRNP and serum from healthy donors induced relatively low levels of MIF. Given the evident co-expression of the MIF binding and signaling receptors, CD74 and CD44 in human monocytes (Fig 1B), we next determined whether the released MIF, by acting in an autocrine/paracrine manner, could affect the production of IL-1β. MIF098 is a potent and orally bioavailable small molecule that blocks MIF binding to the extracellular domain of CD74 (32, 37). Monocytes activated with the snRNP immune complex in the presence of MIF098 showed decreased production of IL-1β (Fig 1C). Adding recombinant human MIF to the snRNP immune complex showed a trend towards increased production of IL-1β although it was not statistically significant (Supplementary Fig 1). Monocytes treated with recombinant human MIF alone had no production of IL-1β (data not shown). Taken together, these findings support an upstream regulatory role for autocrine/paracrine MIF release in enabling high levels of IL-1β production from human monocytes stimulated with the snRNP immune complexes.
Figure 1. The lupus snRNP immune complex induces MIF release from human monocytes, leading to the promotion of IL-1β production.
(A) MIF ELISA at 18 hours from cell culture supernatants of human monocytes incubated with or without U1-snRNP (snRNP, 5 μg/ml) in the presence or absence of healthy serum or anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration). Bars and errors bars indicate mean and SEM, respectively (n = 15 donors). The P-value was obtained by ANOVA. (B) Flow cytometric analysis of CD44 and CD74 expression on monocytes freshly isolated from the peripheral blood of a healthy donor. Representative data from 2 donors. (C) IL-1β ELISA at 18 hours from cell culture supernatants of human monocytes incubated with or without U1-snRNP (snRNP, 5 μg/ml) and/or healthy serum or anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration) in the presence or absence of the MIF antagonist MIF098 (20 μM). Bars and errors bars indicate mean and SEM, respectively (n = 6 donors). The P-value was obtained by the paired t-test.
The up-regulation of NLRP3 in human monocytes in response to the snRNP immune complex was dependent on MIF
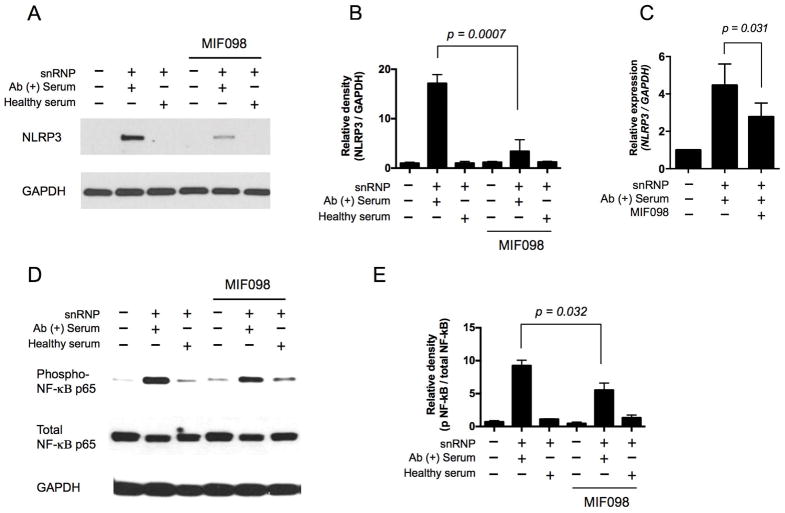
NLRP3 is a limiting step in NLRP3 inflammasome activation since the protein level of NLRP3 is low in resting murine and immortalized human macrophages (27, 28). Thus, we explored whether the decreased production of IL-1β from snRNP immune complex-activated monocytes by the MIF antagonist MIF098 was related to altered NLRP3 expression. Although unstimulated human monocytes had barely detectable levels of NLRP3 protein (Fig 2A), the snRNP immune complex induced high levels of NLRP3 protein expression as measured by Western blot. The expression of NLRP3 protein in these cells was substantially suppressed by MIF098 (Fig 2A–B). The expression levels of the NLRP3 gene that were upregulated by the snRNP immune complex also were decreased in the same cells by MIF098 (Fig 2C). It is known that MIF may contribute to the activation of NF-κB, which also up-regulates NLRP3 (28). The snRNP immune complex activated NF-κB in monocytes (Fig 2C), as previously reported (25), and MIF098 moderately reduced the activation of NF-κB in snRNP immune complex stimulated monocytes (Fig 2C–D). We also determined the levels of the E3 ubiquitin ligases membrane-associated ring-CH-type finger 7 (MARCH7) and tripartite motif containing 31 (TRIM31) in the same cells in that these molecules were reported to participate in degrading NLRP3 via ubiquitination (38, 39). We could not detect TRIM31, but noticed a trend towards increased levels of MARCH7 in monocytes stimulated with the snRNP immune complex, which was not affected by MIF098 (Supplementary Fig 2B, TRIM31 data not shown). Overall, these findings indicate the up-stream regulatory role of MIF in controlling the expression of NLRP3 in human monocytes in response to the snRNP immune complex.
Figure 2. The lupus snRNP immune complex induces NLRP3 expression and NF-κB activation in human monocytes that are suppressed by blocking MIF.
(A–B) Western blot analysis of NLRP3 in human monocytes that were incubated for 7 hours with or without U1-snRNP (snRNP, 5 μg/ml) and/or healthy serum or anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration) in the presence or absence of the MIF antagonist MIF098 (20 μM). (A) Representative data from 4 independent experiments with 4 donors. (B) Relative density of NLRP3/GAPDH Western blot analysis (n = 4 donors). (C) qPCR analysis of the NLRP3 gene in monocytes treated as in (A–B) (n = 4 donors). (D–E) Western blot analysis of phospho-NF-κB p65 and total NF-κB p65 in human monocytes that were incubated for 1 hour with or without U1-snRNP (snRNP, 5 μg/ml) and/or healthy serum or anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration) in the presence or absence of the MIF antagonist MIF098 (20 μM). (D) Representative data from 4 independent experiments with 4 donors. (E) Relative density of phospho-NF-κB p65/total NF-κB p65 Western blot analysis (n =4). Bars and error bars indicate mean ± and SEM, respectively. The P-values were obtained by the paired t-test.
The activation of caspase-1 in human monocytes in response to the snRNP immune complex was decreased by MIF antagonism
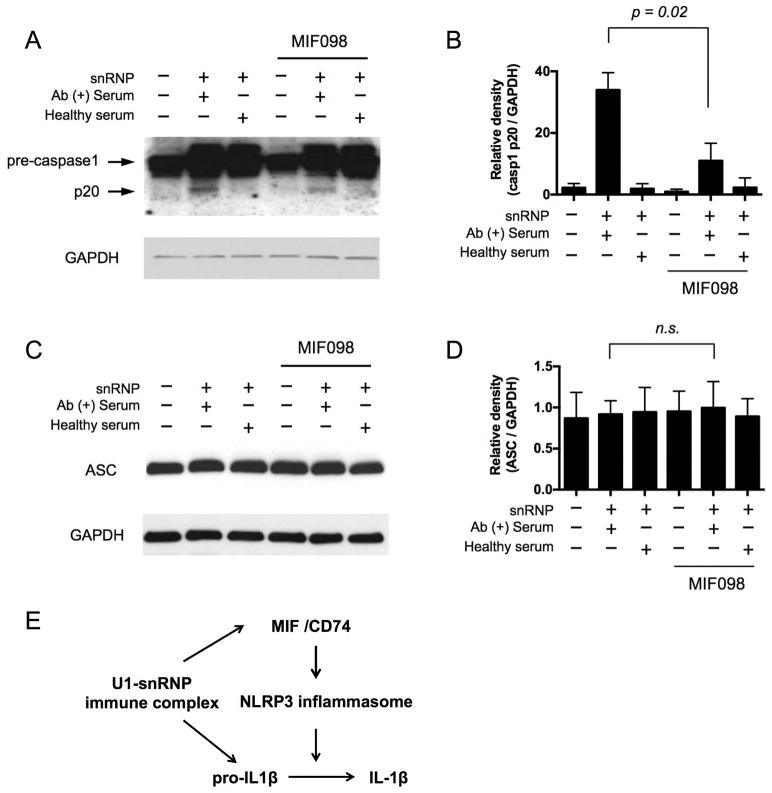
We next determined whether antagonizing MIF suppresses the activation of the NLRP3 inflammasome component caspase-1 by reducing NLRP3 expression. Monocytes were incubated with the snRNP immune complex and the generation of the caspase-1 p20 subunit, an indicator of the activation of caspase-1, was decreased by MIF098 (Fig 3A–B). Also, the mature form of IL-1β, which is processed from the immature form pro-IL-1β by activated caspase-1, was decreased in monocytes incubated in the same condition including the snRNP immune complex and MIF098 (Supplementary Fig 3). We explored whether the activation of the caspase-1 in monocytes by the snRNP immune complex induced pyropotosis which is a form of cell death mediated by the activation of caspase-1 (40). We noticed modest levels of cell death (about 20%) in monocytes incubated with the snRNP immune complex, which was not affected by antagonizing MIF (Supplementary Figure 4). We also analyzed the adaptor molecule ASC, which is a component of the NLRP3 inflammasome, in human monocytes stimulated with or without the snRNP immune complex in the presence or absence of MIF098. Unstimulated human monocytes showed substantial expression of ASC, which was not affected by snRNP immune complex stimulation and/or incubation with MIF098 (Fig 3C–D). These findings suggest that MIF-mediated upregulation of the rate-limiting molecule NLRP3 is essential for activating the NLRP3 inflammasome in monocytes upon snRNP immune complex stimulation (Fig 3E).
Figure 3. The activation of caspase-1 in human monocytes in response to the lupus snRNP immune complex is decreased by blocking MIF.
(A–D) Western blot analysis of pro-caspase1, caspase1 p20 (A) and ASC (C) in human monocytes that were incubated for 18 hours with or without U1-snRNP (snRNP, 5 μg/ml) and/or healthy serum or anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration) in the presence or absence of the MIF antagonist MIF098 (20 μM). Representative data from 4 independent experiments with 4 donors. (B, D) Relative density of caspase1 p20/GAPDH (B) and ASC/GAPDH (D) Western blot analysis (n = 4). Bars and error bars indicate mean ± and SEM, respectively. The P-values were obtained by the paired t-test. NS, not significant. (E) A model showing the possible role of MIF in the production of IL-1β from human monocytes upon U1-snRNP immune complex stimulation. The U1-snRNP immune complex induces the secretion of MIF. The secreted MIF binds the MIF receptor CD74 on monocytes, leading to the activation of the NLRP3 inflammasome by promoting NLRP3 gene and protein expression. The activated NLRP3 inflammasome cleaves pro-IL-1β into IL-1β.
NLRP3 and CD74 were expressed by CD14+ cells in human acute cutaneous lupus lesion
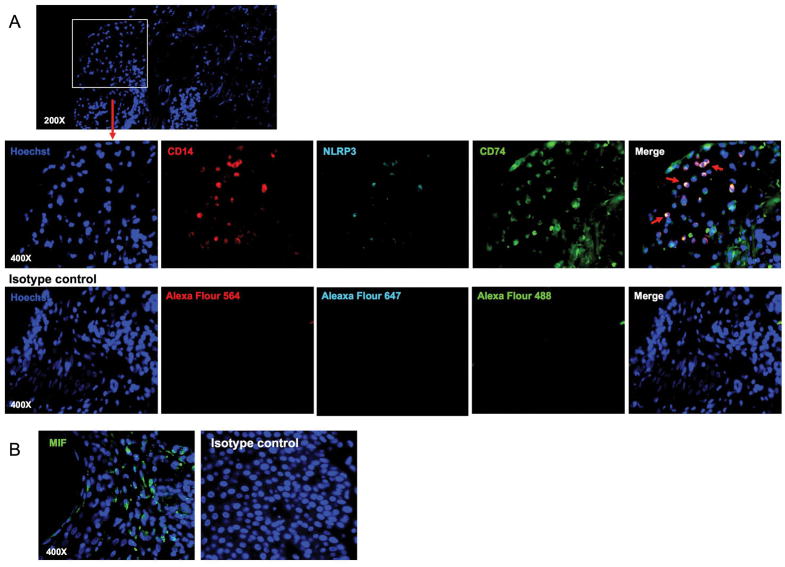
Increased levels of MIF are detected in the kidney tissues of lupus proliferative glomerulonephritis as well as in both skin and kidney lesions from the lupus-prone MRL/lpr mice (14, 41). However, the relationship of the MIF receptor CD74 and NLRP3 expression in lupus skin lesions has not been explored. Thus, we measured the expression of CD74 and NLRP3 by CD14+ cells as well as MIF in human acute cutaneous lupus lesion using immunofluorescence staining. We noticed the presence of CD14+ cells like monocytes, which expressed CD74 and NLRP3 in the lupus skin lesion (Fig. 4 and Supplementary Fig 5).
Figure 4. NLRP3 and CD74 are expressed by CD14+ cells in human acute cutaneous lupus lesion.
(A) Immunofluorescent staining of human acute cutaneous lupus lesion with antibodies to CD14 (red), NLRP3 (cyan) and CD74 (green) or control IgG. All nuclei were counterstained with Hoechst 33342. The upper panel shows nucleus staining (original magnification, ×200). The lower panels show fluorescent images for CD14, NLRP3, CD74 or control IgG staining in the areas indicated by the rectangle in the upper panels (original magnification, ×400). Arrows indicate triple-stained cells for CD14, NLRP3, and CD74. (B) Immunofluorescent staining of human acute cutaneous lupus lesion with antibodies to MIF (green) or control IgG. All nuclei were counterstained with Hoechst 33342. Representative data from 2 independent experiments.
High-dimensional single cell analysis supports the unique cellular traits of monocytes stimulated with the snRNP immune complex that were altered by antagonizing MIF
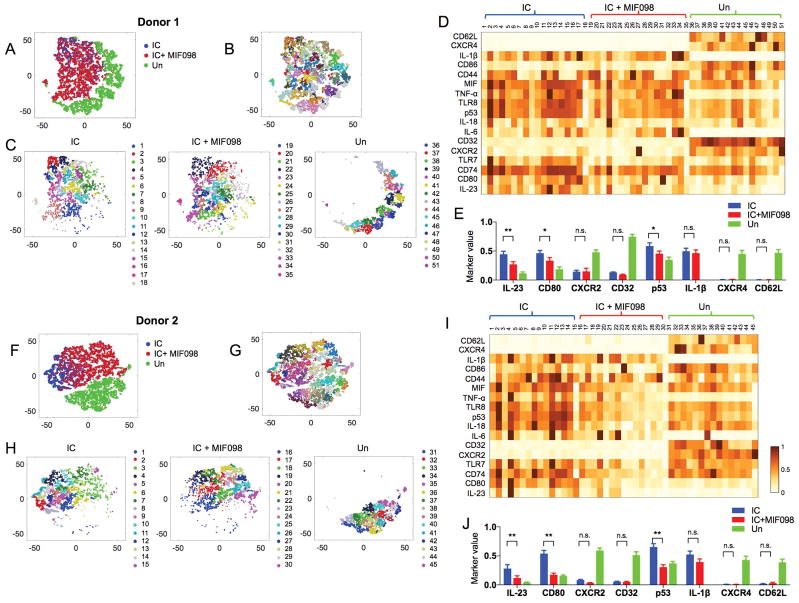
We explored whether monocytes stimulated with the snRNP immune complex develop unique cellular traits by using mass cytometry or Cytometry by Time-Of-Flight (CyTOF) together with high-dimensional computational analysis at the single cell level. CyTOF utilizes heavy metal ions and mass spectrometry as labels and a readout, respectively, which allows the measurement of multiple molecules in a single analysis (42). High-dimensional CyTOF data can be analyzed to demonstrate the multidimensional relationships of molecules expressed by single cells using computational methods such as the nonlinear dimensionality-reduction tool t-distributed stochastic neighbor embedding (t-SNE). The latter can be utilized in combination with PhenoGraph clustering analysis to robustly identify distinct cellular subsets (35, 43). t-SNE dimensionality reduction analysis showed a segregation of snRNP immune complex-stimulated monocytes from unstimulated monocytes based on the expression of 17 molecules including MIF, CD74, CD44, CXCR4, CXCR2, and IL-1β (Fig 5A and 5F). Phenograph clustering revealed subsets of cells within monocytes incubated with or without snRNP immune complex, and in the presence or absence of MIF098 (Fig 5B–D and F–I). Of note, unstimulated cells expressed high levels of CXCR2, CD32, CXCR4, and CD62L compared to the stimulated cells (Fig 5D–E and I–J), while the latter cells had higher levels of intracellular cytokines including IL-1β and the activation marker CD80. A group of monocytes stimulated with the snRNP immune complex in the presence of MIF098 were segregated from the same stimulated cells in the absence of MIF098. Such cell clusters had decreased expression levels of IL-23, CD80, and p53 (Fig 5D–E and I–J). Some unstimulated monocytes expressed MIF, indicating the constitutive expression of MIF, as reported previously (44). IL-1β was not detected in unstimulated monocytes and the expression levels of intracellular IL-1β, including both pro- and active forms of IL-1β, appeared largely similar in monocytes activated with the snRNP immune complex in the presence and absence of MIF098 (Fig 5D–E and I–J). This finding, which is consistent with the results of IL1B gene expression analysis in the same cells (Supplementary Fig 2C), supports the conclusion that the suppressive effect of MIF098 on the production of IL-1β is mediated primarily by decreasing activation of the NLRP3 inflammasome and the subsequent generation of the active form IL-1β.
Figure 5. High-dimensional single cell analysis shows the unique cellular traits of monocytes stimulated with the snRNP immune complex (IC) that were altered by antagonizing MIF.
Monocytes were incubated for 5 hours with or without IC (U1-snRNP (5 μg/ml)/anti-U1-snRNP serum, 5% final concentration) alone or with MIF098 (20 μM) followed by CyTOF analysis. The PhenoGraph clustering was performed on monocytes based on the expression of 17 molecules (column labels in D and I). (A–C, F–H) t-SNE plots show a landscape of subsets and their relationships in the incubated monocytes. C and H show subsets identified by PhenoGraph clustering on monocytes incubated in indicated conditions. Numbers and matched color dots indicate individual cell subsets. D and I show mean expression levels of 17 molecules (rows) by the individual cell subsets (columns) identified in C and H. Values are scaled between 0 and 1 for each molecule. (E, J) Bar graphs show the intensity of each molecule expressed by individual subsets of the incubated monocytes identified in C and H. Bars and error bars indicate mean ± and SEM, respectively. P < *0.05, **0.005 by two-way ANOVA (multiple comparison controlled by the Benjamin, Krieger and Yekutieli method, FDR 0.05). N.S., not significant.
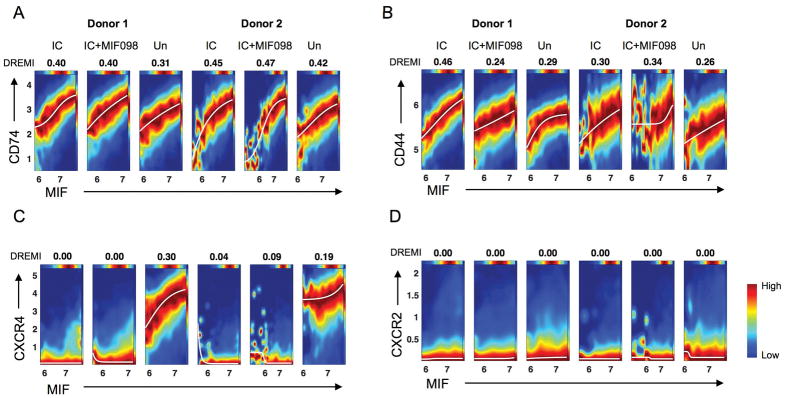
We determined how the expression levels of MIF and MIF receptors changed at the single cell level using the computational algorithms conditional-Density Resampled Estimate of Mutual Information (DREMI) and conditional-Density Rescaled Visualization (DREVI) (36). DREMI computes mutual information that describes how the state of Y alters with different states of X (36), while DREVI visualizes the function underlying such interactions (36). DREMI scores show the strength of the statistical dependency between two molecules. CD74 and CD44 increased as the expression levels of MIF increased in monocytes stimulated with or without the snRNP immune complex (Fig 6A–B), supporting the auto and paracrine effects of MIF on monocyte activation. A similar relationship with MIF was noticed with CXCR4, but not with CXCR2 (Fig 6C–D).
Figure 6. MIF has a robust quantitative relationship with CD74, CD44, and CD44 at the single cell level in monocytes stimulated with the lupus snRNP immune complex.
Monocytes purified from healthy donors were incubated for 5 hours with or without U1-snRNP (snRNP, 5 μg/ml) and anti-U1-snRNP antibody-positive (Ab+) serum (5% final concentration) (referred to as immune complex, IC) in the presence or absence of the MIF antagonist MIF098 (20 μM), stained with a set of antibodies, and run on a Helios CyTOF as in Fig 5. The computational algorithms conditional-Density Resampled Estimate of Mutual Information (DREMI) and conditional-Density Rescaled Visualization (DREVI) were performed on the incubated monocytes. DREVI plots show the quantitative relationship of MIF with CD74 (A), CD44 (B), CXCR4 (C), and CXCR2 (D). DREMI scores indicating the strength of the statistical dependency between two molecules are shown above the DREVI plots. Representative data from 4 independent experiments with 4 donors.
Discussion
The present study identifies the snRNP immune complex as an up-regulator of MIF production in human monocytes that is relevant to innate immune activation in lupus, and provides the first evidence for an upstream role of MIF in promoting NLRP3 expression, the rate-limiting step in NLRP3 inflammasome formation. Previous studies indicate that MIF-deficient mice express decreased IL-1β (45), supporting the upstream regulatory role of MIF in inducing this cytokine. In accordance with this finding, we noticed decreased production of IL-1β from snRNP immune complex-stimulated human monocytes in the presence of the MIF antagonist MIF098. Our findings of decreased NLRP3 expression and NLRP3 inflammasome activation in the same cells indicate that MIF likely functions upstream by enabling NLRP3 expression and subsequent formation of the NLRP3 inflammasome, which cleaves pro-IL-1β into bioactive IL-1β. Of interest, the expression levels of ASC, which was highly expressed at the basal level, were not different between monocytes stimulated and unstimulated with the snRNP immune complex. MIF antagonism did not alter the expression of ASC, further supporting the role of MIF in regulating the NLRP3 inflammasome by specific control of the expression of NRLP3, a rate-limiting molecule in forming the NLRP3 inflammasome in monocytes.
NF-κB is known to promote the expression of the NLRP3 gene (28). We noticed the suppression of NF-κB activation and NLRP3 gene expression in snRNP immune complex-stimulated monocytes by antagonizing MIF with MIF098. Previous studies reported the activation of NF-κB in murine B cells by MIF in a CD74-CD44-dependent manner (46) as well as in HEK-293 cells transfected with human CD74 (47). The latter findings support the autocrine and paracrine activation effect of MIF on the upregulation of NLRP3 in snRNP immune complex-stimulated monocytes through the CD74/CD44 receptor complex and subsequent NF-κB activation. IL1B gene expression also decreased modestly in monocytes activated with the snRNP immune complex in the presence of MIF098, although such a decrease was not statistically significant. It is possible that MIF regulates the NLRP3 expression through mechanism(s) redundant to those modulating NF-κB activation. We noticed no changes in the expression of MARCH7 and TRIM31 that were reported to be involved in degrading NLRP3 in snRNP immune complex-stimulated monocytes (38, 39). Our findings imply that the effect of MIF on IL-1β production is in part through NF-κB-mediated regulation of the NLRP3 gene and subsequent expression of the NLRP3 protein.
We explored how the cellular phenotype of monocytes, especially those molecules related to MIF, changed upon stimulation with the snRNP immune complex at the single cell level using high-dimensional CyTOF analysis. The dimensional reduction analysis t-SNE showed a segregation of unstimulated monocytes from stimulated monocytes based on 17 cytokines, chemokine receptors, and activation markers. Monocytes stimulated with the snRNP immune complex in the presence of the MIF antagonist MIF098 were segregated from both stimulated and unstimulated monocytes. These findings support the interpretation that MIF action on monocytes upon stimulation with the snRNP immune complex is more extensive than simply on the NLRP3 pathway and IL-1β production. Furthermore, the effect of MIF on individual monocytes and molecules expressed by them is not uniform, as distinct unstimulated and stimulated monocyte subsets can be identified with diverse characteristics, including expression of the MIF cognate (CD74/CD44) and non-cognate receptors (CXCR2/4). Analysis using the DREMI and DREVI algorithms further support the autocrine and paracrine activation effect of MIF through the CD74/CD44 receptor complex in stimulated and resting monocytes at the single cell level.
The results of this study identify the snRNP immune complex as a specific trigger of MIF production and NLRP3 inflammasome activation in human monocytes, with downstream biological significance evidenced by a decrease in the activation of caspase-1 and IL-1β production by MIF receptor blockade. Genetic deficiency or pharmacologic MIF antagonism has been shown previously to reduces functional and histological indices of glomerulonephritis and inflammatory cytokine and chemokine expression in lupus-prone MRL/lpr or NZB/NZW F1 mice (11, 14). In pristane-induced murine lupus, genetic caspase-1 deficiency also improves diseases (48), while hyperactivation of the NLRP3 inflammasome produces more severe renal disease and increased mortality (49). Inhibiting the NLRP3/ASC/caspase-1 pathway also suppressed nephritis in MRL/lpr lupus mice (30), although genetic lack of NLRP3 or ASC appeared paradoxically to trigger lupus-like disease in C57BL/6-lpr/lpr mice (50).
Of note, MIF was one of the decreased 77 molecules in ultraviolet B (UVB)-irradiated keratinocytes in the presence of the caspase-1 inhibitor YVAD as measured by a mass spectrometry-based method (51). However, this phenomenon is likely through an indirect mechanism since MIF does not require nor has a caspase-cleavage site necessary for secretion unlike IL-1β. A possible molecular link between snRNP immune complex and MIF production could exist in the TLR7 pathway in that snRNP can activate this pathway (52). Previous studies reported the post-translational modification of N-terminal proline, which is targeted by MIF098, in MIF in vitro by dietary isothiocyanates or myeloperoxidase-derived oxidants of neutrophils (reviewed in (53)). This modification impaired tautomerase activity but not immunomodulatory activity of MIF (54). However, here we studied only monocytes without adding these molecules. MIF098 targets the region encompassing the N-terminal proline in MIF, which mediates its tautomerase activity and also participates in binding to the MIF receptor CD74 (32, 37). Our data suggest that MIF098 primarily blocks extracellular MIF since MIF098 does not inhibit MIF tautomerase activity intracellularly, in contrast to the previously described MIF inhibitor 4-IPP (55) (Bucala et al, unpublished observation).
Given long-standing observations that anti-snRNP autoantibody responses are associated with distinct inflammatory sequelae (e.g., a mixed connective tissue disease phenotype), these observations suggest a rationale for specific targeting of the NLRP3 inflammasome or MIF signaling, potentially in high genotypic MIF expressers with SLE. High-dimensional CyTOF analysis also could be applied to identify patient subsets with relevant monocyte populations (e.g., high MIF or MIF receptor expression) that may suggest responsiveness to MIF or inflammasome directed therapies.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (R21AI1266042, R56AG0280691,1R01AG055362 to IK, and 1R01AR049610 to RB; the Yale CTSA grant UL1TR000142) and Connecticut Innovations (Regenerative Medicine Research Fund, 14-SCC-Yale-01 to IK). The authors thank Dr. Ala Nassar and Ms. Shelly Ren of the Yale CyTOF Core.
Footnotes
Conflict of Interest: Dr. Bucala is an inventor of MIF antagonists and has received past licensing royalties from Baxter Healthcare and Debiopharm SA.
References
- 1.Lang T, Foote A, Lee JP, Morand EF, Harris J. MIF: Implications in the Pathoetiology of Systemic Lupus Erythematosus. Front Immunol. 2015;6:577. doi: 10.3389/fimmu.2015.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucala R. MIF, MIF alleles, and prospects for therapeutic intervention in autoimmunity. J Clin Immunol. 2013;33(Suppl 1):S72–8. doi: 10.1007/s10875-012-9781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 5.Fingerle-Rowson G, Petrenko O, Metz CN, Forsthuber TG, Mitchell R, Huss R, et al. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci U S A. 2003;100(16):9354–9. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197(11):1467–76. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi X, Leng L, Wang T, Wang W, Du X, Li J, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25(4):595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–96. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 9.Ayoub S, Hickey MJ, Morand EF. Mechanisms of disease: macrophage migration inhibitory factor in SLE, RA and atherosclerosis. Nat Clin Pract Rheumatol. 2008;4(2):98–105. doi: 10.1038/ncprheum0701. [DOI] [PubMed] [Google Scholar]
- 10.McDevitt MA, Xie J, Ganapathy-Kanniappan S, Griffith J, Liu A, McDonald C, et al. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J Exp Med. 2006;203(5):1185–96. doi: 10.1084/jem.20052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leng L, Chen L, Fan J, Greven D, Arjona A, Du X, et al. A small-molecule macrophage migration inhibitory factor antagonist protects against glomerulonephritis in lupus-prone NZB/NZW F1 and MRL/lpr mice. J Immunol. 2010;186(1):527–38. doi: 10.4049/jimmunol.1001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sreih A, Ezzeddine R, Leng L, LaChance A, Yu G, Mizue Y, et al. Dual effect of the macrophage migration inhibitory factor gene on the development and severity of human systemic lupus erythematosus. Arthritis Rheum. 2011;63(12):3942–51. doi: 10.1002/art.30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calandra T, Echtenacher B, Roy DL, Pugin J, Metz CN, Hultner L, et al. Protection from septic shock by neutralization of macrophage migration inhibitory factor. Nat Med. 2000;6(2):164–70. doi: 10.1038/72262. [DOI] [PubMed] [Google Scholar]
- 14.Hoi AY, Hickey MJ, Hall P, Yamana J, O’Sullivan KM, Santos LL, et al. Macrophage migration inhibitory factor deficiency attenuates macrophage recruitment, glomerulonephritis, and lethality in MRL/lpr mice. J Immunol. 2006;177(8):5687–96. doi: 10.4049/jimmunol.177.8.5687. [DOI] [PubMed] [Google Scholar]
- 15.De la Cruz-Mosso U, Bucala R, Palafox-Sanchez CA, Parra-Rojas I, Padilla-Gutierrez JR, Pereira-Suarez AL, et al. Macrophage migration inhibitory factor: association of -794 CATT5-8 and -173 G>C polymorphisms with TNF-alpha in systemic lupus erythematosus. Hum Immunol. 2014;75(5):433–9. doi: 10.1016/j.humimm.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foote A, Briganti EM, Kipen Y, Santos L, Leech M, Morand EF. Macrophage migration inhibitory factor in systemic lupus erythematosus. J Rheumatol. 2004;31(2):268–73. [PubMed] [Google Scholar]
- 17.Connelly KL, Kandane-Rathnayake R, Hoi A, Nikpour M, Morand EF. Association of MIF, but not type I interferon-induced chemokines, with increased disease activity in Asian patients with systemic lupus erythematosus. Sci Rep. 2016;6:29909. doi: 10.1038/srep29909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace DJ, Weisman MH, Wegener WA, Horne H, Goldenberg DM. THU0288 IMMU-115 (Humanized Anti-CD74 Antibody) for Subcutaneous (SC) Administration: A Phase Ib Study in Patients with Systemic Lupus Erythematosus (SLE) Annals of the Rheumatic Diseases. 2016;75(Suppl 2):291. [Google Scholar]
- 19.Shin MS, Lee N, Kang I. Effector T-cell subsets in systemic lupus erythematosus: update focusing on Th17 cells. Curr Opin Rheumatol. 2011;23(5):444–8. doi: 10.1097/BOR.0b013e328349a255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 21.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6(11):823–35. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–36. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 23.Guiducci C, Gong M, Xu Z, Gill M, Chaussabel D, Meeker T, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465(7300):937–41. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25(3):417–28. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Shin MS, Kang Y, Lee N, Kim SH, Kang KS, Lazova R, et al. U1-small nuclear ribonucleoprotein activates the NLRP3 inflammasome in human monocytes. J Immunol. 2012;188(10):4769–75. doi: 10.4049/jimmunol.1103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10(3):210–5. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 27.Haneklaus M, O’Neill LA, Coll RC. Modulatory mechanisms controlling the NLRP3 inflammasome in inflammation: recent developments. Curr Opin Immunol. 2013;25(1):40–5. doi: 10.1016/j.coi.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183(2):787–91. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Berthier CC, Kahlenberg JM. Enhanced Inflammasome Activity in Systemic Lupus Erythematosus Is Mediated via Type I Interferon-Induced Up-Regulation of Interferon Regulatory Factor 1. Arthritis Rheumatol. 2017;69(9):1840–9. doi: 10.1002/art.40166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Wang H, Dai C, Wang H, Zhang H, Huang Y, et al. P2X7 blockade attenuates murine lupus nephritis by inhibiting activation of the NLRP3/ASC/caspase 1 pathway. Arthritis Rheum. 2013;65(12):3176–85. doi: 10.1002/art.38174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol. 2013;190(3):1217–26. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo SA, Leng L, Kim BJ, Du X, Tilstam PV, Kim KH, et al. MIF allele-dependent regulation of the MIF coreceptor CD44 and role in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2016;113(49):E7917–E26. doi: 10.1073/pnas.1612717113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grigorenko EL, Han SS, Yrigollen CM, Leng L, Mizue Y, Anderson GM, et al. Macrophage migration inhibitory factor and autism spectrum disorders. Pediatrics. 2008;122(2):e438–45. doi: 10.1542/peds.2007-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect Immun. 2014;82(1):112–23. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162(1):184–97. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science. 2014;346(6213):1250689. doi: 10.1126/science.1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauler M, Zhang Y, Min JN, Leng L, Shan P, Roberts S, et al. Endothelial CD74 mediates macrophage migration inhibitory factor protection in hyperoxic lung injury. FASEB J. 2015 doi: 10.1096/fj.14-260299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160(1–2):62–73. doi: 10.1016/j.cell.2014.11.047. [DOI] [PubMed] [Google Scholar]
- 39.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:13727. doi: 10.1038/ncomms13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73(4):1907–16. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan HY, Yang N, Nikolic-Paterson DJ, Yu XQ, Mu W, Isbel NM, et al. Expression of macrophage migration inhibitory factor in human glomerulonephritis. Kidney Int. 2000;57(2):499–509. doi: 10.1046/j.1523-1755.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 42.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–52. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci U S A. 2015;112(7):E607–15. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stosic-Grujicic S, Stojanovic I, Maksimovic-Ivanic D, Momcilovic M, Popadic D, Harhaji L, et al. Macrophage migration inhibitory factor (MIF) is necessary for progression of autoimmune diabetes mellitus. J Cell Physiol. 2008;215(3):665–75. doi: 10.1002/jcp.21346. [DOI] [PubMed] [Google Scholar]
- 46.Gore Y, Starlets D, Maharshak N, Becker-Herman S, Kaneyuki U, Leng L, et al. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008;283(5):2784–92. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- 47.Binsky I, Haran M, Starlets D, Gore Y, Lantner F, Harpaz N, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci U S A. 2007;104(33):13408–13. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahlenberg JM, Yalavarthi S, Zhao W, Hodgin JB, Reed TJ, Tsuji NM, et al. An essential role of caspase 1 in the induction of murine lupus and its associated vascular damage. Arthritis Rheumatol. 2014;66(1):152–62. doi: 10.1002/art.38225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu A, Li H, Niu J, Wu S, Xue G, Yao X, et al. Hyperactivation of the NLRP3 Inflammasome in Myeloid Cells Leads to Severe Organ Damage in Experimental Lupus. J Immunol. 2017;198(3):1119–29. doi: 10.4049/jimmunol.1600659. [DOI] [PubMed] [Google Scholar]
- 50.Lech M, Lorenz G, Kulkarni OP, Grosser MO, Stigrot N, Darisipudi MN, et al. NLRP3 and ASC suppress lupus-like autoimmunity by driving the immunosuppressive effects of TGF-beta receptor signalling. Ann Rheum Dis. 2014 doi: 10.1136/annrheumdis-2014-205496. [DOI] [PubMed] [Google Scholar]
- 51.Keller M, Ruegg A, Werner S, Beer HD. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132(5):818–31. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 52.Savarese E, Chae OW, Trowitzsch S, Weber G, Kastner B, Akira S, et al. U1 small nuclear ribonucleoprotein immune complexes induce type I interferon in plasmacytoid dendritic cells through TLR7. Blood. 2006;107(8):3229–34. doi: 10.1182/blood-2005-07-2650. [DOI] [PubMed] [Google Scholar]
- 53.Schindler L, Dickerhof N, Hampton MB, Bernhagen J. Post-translational regulation of macrophage migration inhibitory factor: Basis for functional fine-tuning. Redox Biol. 2018;15:135–42. doi: 10.1016/j.redox.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickerhof N, Schindler L, Bernhagen J, Kettle AJ, Hampton MB. Macrophage migration inhibitory factor (MIF) is rendered enzymatically inactive by myeloperoxidase-derived oxidants but retains its immunomodulatory function. Free Radic Biol Med. 2015;89:498–511. doi: 10.1016/j.freeradbiomed.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Winner M, Meier J, Zierow S, Rendon BE, Crichlow GV, Riggs R, et al. A novel, macrophage migration inhibitory factor suicide substrate inhibits motility and growth of lung cancer cells. Cancer Res. 2008;68(18):7253–7. doi: 10.1158/0008-5472.CAN-07-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.