Abstract
Current advances in combined anti-retroviral therapy (cART) have rendered HIV infection a chronic, manageable disease; however, the problem of persistent immune activation still remains despite treatment. The immune cell receptor SLAMF7 has been shown to be upregulated in diseases characterized by chronic immune activation. Here, we studied the function of the SLAMF7 receptor in immune cells of HIV patients and the impacts of SLAMF7 signaling on peripheral immune activation. We observed increased frequencies of SLAMF7+ PBMCs in HIV+ individuals in a clinical phenotype-dependent manner, with discordant and long-term nonprogressor patients showing elevated SLAMF7 levels, and elite controllers showing levels comparable to healthy controls. We also noted that SLAMF7 was sensitive to IFN⍺ stimulation; a factor elevated during HIV infection. Further studies revealed SLAMF7 to be a potent inhibitor of the monocyte-derived proinflammatory chemokine CXCL10 (IP-10) and other CXCR3 ligands, except in a subset of HIV+ patients termed SLAMF7 silent (SF7S). Studies utilizing small molecule inhibitors revealed that the mechanism of CXCL10 inhibition is independent of known SLAMF7 binding partners. Furthermore, we determined that SLAMF7 activation on monocytes is able to decrease their susceptibility to HIV-1 infection in vitro via down-regulation of CCR5 and up-regulation of the CCL3L1 chemokine. Finally, we discovered that neutrophils do not express SLAMF7, are CXCL10+ at baseline, are able to secrete CXCL10 in response to IFN⍺ and LPS, and are non-responsive to SLAMF7 signaling. These findings implicate the SLAMF7 receptor as an important regulator of IFN⍺-driven innate immune responses during HIV infection.
Introduction:
HIV infection is now widely regarded as a manageable disease; however, cART, which controls viremia, fails to effectively control many secondary HIV-associated pathologies (1, 2). The universal mechanism believed to underlie the development of these diseases is chronic, global immune activation (3, 4). Known causes of which include increased gut permeability resulting in microbial translocation into systemic circulation (5) and constitutively elevated levels of proinflammatory cytokines and chemokines, including interferon alpha (IFN⍺) (6–9).
Sustained levels of detectable IFN⍺ in the plasma of cART-treated HIV patients results from persistent activation of plasmacytoid dendritic cells (pDCs) by HIV-antibody complexes (10) and causes a global type I interferon signature in circulating monocytes (11, 12). This induces monocyte transition from the classical subtype (CD14+CD16-) into the proinflammatory (CD14+CD16+) and non-classical (CD14lowCD16++) subtypes, as evidenced by upregulation of CD16 (13, 14). While beneficial in acute infections, these type I interferon-mediated effects on monocytes prove to be deleterious in chronic infections (15).
Some HIV+ individuals are known to have persistently elevated levels of CXCL10 (IP-10) and other proinflammatory cytokines and chemokines (9, 16). CXCL10 is produced primarily by monocytes and is of particular interest due to its ability to suppress T cell functions (17), induce neuronal apoptosis (18–20), and as a marker of systemic inflammation (21). During HIV infection some CD16+ monocytes become activated by IFN⍺, upregulate CCR5 (22), infected with HIV, and subsequently migrate across the blood-brain barrier where they set up a viral reservoir and are capable of inducing neuroinflammation and neuronal apoptosis via secretion of high levels of CXCL10, TNF⍺, IL-6, and IL-1β (22, 23). Here we investigated the role of an immune-modulatory receptor, signaling lymphocytic activation molecule family member 7 (SLAMF7) (a.k.a. CRACC, CS-1, CD319), in the context of HIV infection and immune activation.
SLAMF7 is a member of the signaling lymphocytic activation molecules (SLAM) family of receptors and is expressed on numerous immune cell types (24, 25). SLAMF7, and other SLAM family receptors (except 2B4), function as homotypic receptors that, upon activation, recruit SLAM-associated protein (SAP) family of adaptors or other SH2 domain-containing proteins to their cytoplasmic immunoreceptor tyrosine-based switch motifs (ITSMs) (24). SLAMF7 is unique among SLAM receptors in that it is only able to recruit a single SAP adaptor, EAT-2, to its tyrosine-phosphorylated ITSM (26). SLAMF7 receptor ligation in cells expressing EAT-2 results in activation of cellular immune responses (27), while SLAMF7 activation in the absence of EAT-2 results in cellular inhibition via recruitment of a number of inhibitory phosphatases (SHP1, SHP2, SHIP1, and csk) (26).
SLAMF7 is well known for being over-expressed on multiple myeloma (MM) cells (28), as an important regulator of NK cell function (25, 26, 29), and recently, as a critical factor in macrophage-mediated phagocytosis of tumor cells (30). Using a medium-sized cohort of middle-aged, HIV+ individuals, we sought to assess whether the SLAMF7 receptor was playing a role in the context of HIV infection in cART-treated patients. Our results implicate the SLAMF7 receptor as an important immunomodulatory receptor in human monocytes, and in the context of HIV-associated peripheral immune activation.
Materials and Methods:
Reagents used.
The following antibodies were used: CD14-FITC (61D3), CD16-BV510 (3G8), SLAMF7-PE (162), CD3-PE-Cy7 (OKT3), CD19-PE-Cy7 (SJ25C1), CD57-PE-Cy7 (TB01), CD66b-APC (G10F5), CXCL10-PerCp-eFluor710 (4NY8UN), EAT-2-APC (LS-C240730), YY1-Alexa647 (H-10), Blimp-1-DyLight650 (3H2-E8), CCR5-PerCp-eFluor710 (NP-6G4) and SLAMF7 (162.1) (used for cross-linking). The SLAMF7-Fc recombinant protein was designed and produced similar to mCRACC-Fc as previously described (25), with the following modifications: a human SLAMF7 extracellular domain was swapped for the murine SLAMF7 extracellular domain, the murine IgG Fc portion was switched to a human IgG4 domain to reduce Fc receptor binding and ADCC, and S228P and L235E mutations were made in the IgG4 domain to further reduce interactions with Fc receptors (31). Recombinant universal IFN⍺ (PBL Assay Bioscience) and Recombinant human IFNγ (ProSpec) were used at 100 IU/mL unless otherwise noted. SHP1/2 inhibitor (NSC87877) (Millipore Sigma) was used at 10μM. SHIP1 inhibitor (3AC) (Millipore Sigma) was used at 5μM. CD45 inhibitor (CAS 345630–40-2) (Millipore Sigma) was used at 1μM. Bortezomib (EMD Millipore) was used at 100nM. All HDACi’s (Selleck chemicals) were used at concentrations indicated in relevant figure legends.
HC and HIV blood sample collection.
HC PBMCs were obtained from either buffy coats purchased from Gulf Coast Regional Blood Center, Texas, or from whole blood samples purchased from Stanford Blood Center, California. HIV+ blood samples were collected from donors enrolled in the Mid-Michigan HIV Consortium. Plasma was stored at −80°C until use. PBMCs were isolated with Ficoll-Plaque Plus (GE Healthcare) as previously described (32).
In vitro cell culture and stimuli.
Cells were plated at 3×105 cells/well for PBMC and isolated monocyte experiments, and 1×105 cell/well for isolated neutrophil experiments, in 96-well plates. Cells were cultured in complete RPMI (RPMI 1640, 10% FBS, 1X PSF). For cross-linking experiments 10μg/mL anti-SLAMF7 mAb or SLAMF7-Fc was added to a sterile, high-binding 96-well cell culture plate O/N at 4°C. For all in vitro flow cytometry experiments cells were cultured in the presence of stimuli for 17–18h. In the monocyte maturation assay (Fig. 6E) the cells were stimulated with only 25 IU/mL IFN⍺ and for exactly 24h. Small molecule inhibitors were added 45–60min before IFN⍺ for all relevant experiments. For experiments involving intracellular staining, BD GolgiPlug was added for the final 5h. NK92 cells were purchased from ATCC and cultured as indicated by manufacturer.
Figure 6.
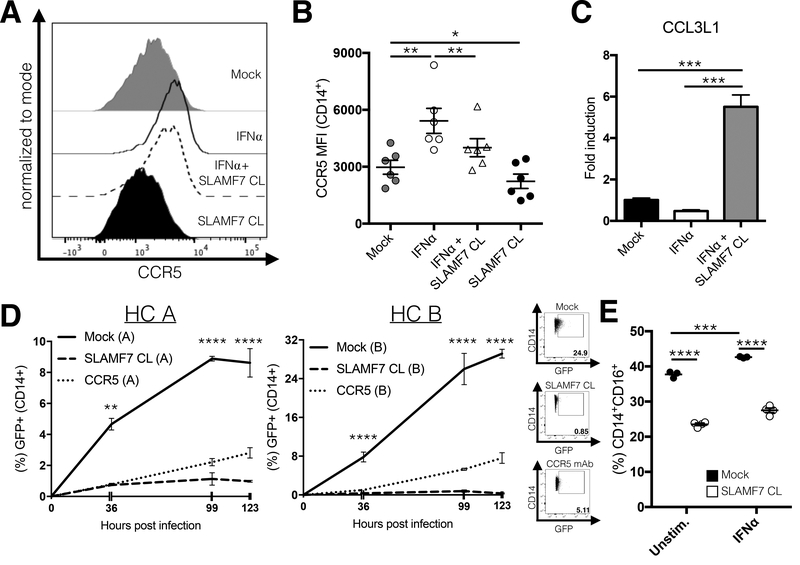
SLAMF7 activation inhibits monocyte infection with HIV-1 in vitro and down-regulates CD16. (A and B) PBMCs from HIV+ individuals (n=6) were stimulated in vitro with IFN⍺ and the SLAMF7 receptor was activated by cross-linking where indicated. Surface expression of CCR5 was measured by flow cytometry. (C) mRNA expression of CCL3L1 was assessed by qRT-PCR from the same samples in (Fig. 5A and B). (D) Isolated monocytes from 2 HCs (labeled “A” and “B”) were infected with HIV-1-Ba-L-GFP and infectivity was assessed at the indicated time points by FACS. Two technical replicates for each HC were analyzed. A CCR5 blocking mAb (10 μg/mL) was included as a positive control. (E) PBMCs from a single HC were stimulated in vitro as indicated for 24 hours before analysis by flow cytometry. This experiment used 25 IU/mL IFN⍺. Percent indicated on y-axis is from all cells in FSC-A/SSC-A “monocyte” gate. (B and C) Groups compared using a 1-way ANOVA with Tukey’s multiple comparison test. (D and E) Groups compared using a 2-way ANOVA with Tukey’s multiple comparison test. Data presented as mean ± SEM and are representative of 1 experiment (B and C) or 2 independent experiments showing similar results (D and E). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
BioPlex assay.
For analysis of cell culture supernatant and plasma samples 80μL of media or plasma was used, undiluted, and the assay was run per manufacturer’s instructions (Bio-Rad, Hercules, CA) via Luminex 100 technology. A 27-plex assay was run and analysis was performed on only those factors with detectable levels or contributing to the separation of Cluster 1 and Cluster 2.
Flow cytometry.
Cells were prepared and stained as previously described (25). Intracellular staining was performed using the BD fixation/permeabilization kit (BD Biosciences) per manufacturer’s instructions. Transcription factor staining was performed with the Transcription Factor Buffer Set (BD Pharmingen) per manufacturer’s instructions. Samples were analyzed on either a BD LSR II or BD FACSCanto. LIVE/DEAD staining with Aqua fixable stain (ThermoFisher) was included during initial experiments and cell viability was verified to be >90%. Monocyte and neutrophil gating strategy is shown in (Supplemental Fig. 1).
Monocyte and Neutrophil isolations.
CD14+ monocytes were isolated from PBMCs via positive selection using CD14 Microbeads (Miltenyi Biotec) per manufacturer’s instructions. Purity was consistently >90% as assessed by flow cytometry. Neutrophils were isolated from whole blood using the MACSxpress Neutrophil isolation kit (Miltenyi Biotec) per manufacturer’s instructions. Purity was consistently >98% as assessed by flow cytometry.
qRT-PCR experiments.
Isolated monocytes were plated in 96-well plates at 3×105 cells/well and stimulated as indicated for 4h. Cells were harvested and placed into Trizol (ThermoFisher) and RNA was isolated per manufacturer’s instructions. RNA was reverse transcribed with SuperStrand First Strand Synthesis Kit III (Invitrogen) per manufacturer’s instructions and analyzed on a QuantStudio7 system (ThermoFisher). GAPDH was used as a housekeeping gene and the ΔΔCt method was used for analysis.
In vitro HIV-1 infection of monocytes.
Isolated primary human monocytes from two HC’s were plated at 2×105 cells/well in a 96-well high-binding cell culture plate. SLAMF7 activation was induced by antibody cross-linking the same as in IFN⍺ stimulation experiments. Monocytes were isolated by positive selection with CD14 Microbeads (Miltenyi Biotec) per manufacturer’s instructions. Purity of monocyte samples used in (Fig. 6D) was 95.5% and 92.6% for donors A and B, respectively. Cells were cultured for 24h before addition of HIV-1Ba-L-GFP (a kind gift from Dr. Young-Hui Zheng, Michigan State University) at a final concentration of 190 pg/mL. HIV-1Ba-L-GFP virus stock concentration was determined by p24 ELISA (HIV-1 p24 Quantikine ELISA, R&D Systems) per manufacturer’s instructions. HIV-1Ba-L-GFP was generated by replacing the gene coding for nef with GFP (Young-Hui Zheng, unpublished, personal communication). Cells were washed 4h following infection and collected at indicated time points post-infection for assessment of infectivity via FACS analysis. Cells were stained for CD14, CD16, and viability (LIVE/DEAD Violet) and GFP+, living, CD14+ cells were analyzed with FlowJo. The CCR5 blocking mAb (2D7) (BD Biosciences) was added at a concentration of 10 μg/mL at the start of cell culture (24 hrs before HIV-1Ba-L-GFP infection) and was replenished at each media change.
Clustering and statistical analysis.
k-means and hierarchical clustering were performed using SPSS (IBM). Statistical analysis was performed in GraphPad Prism 7.0 as indicated. The heatmap from (Fig. 3B) was generated using the z-score normalized values of plasma biomarkers calculated in SPSS. The violin plots in (Fig. 1) were generated using the ggplot2 package in R. The gene expression heatmap from (Fig. 5B) was generated using the gplot package in R. MFI refers to “median fluorescence intensity” in all instances.
Figure 3.
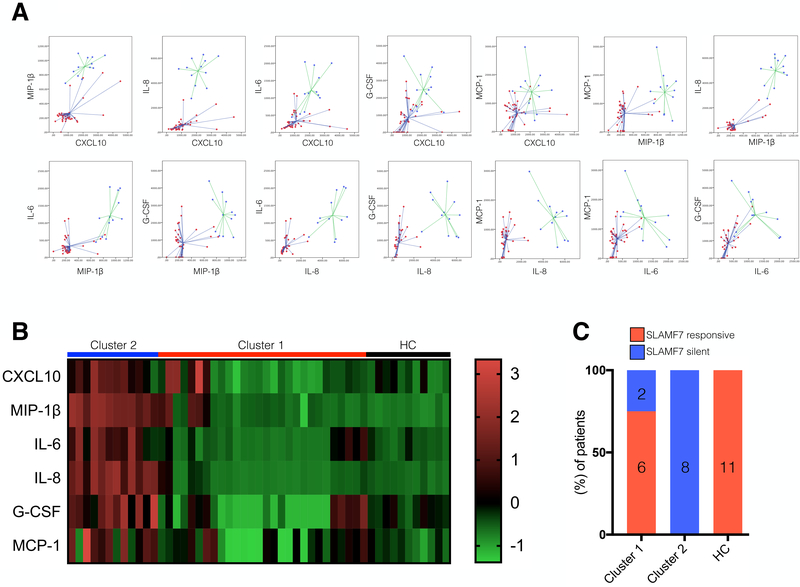
SLAMF7 silent patients have elevated plasma levels of proinflammatory factors and cluster distinctly from SLAMF7 responsive patients. Plasma from HIV patients and HCs was assessed by BioPlex assay for 6 proinflammatory cytokines and chemokines known to be involved in HIV-associated immune dysfunction (CXCL10, MIP-1β, IL-6, IL-8, G-CSF and MCP-1). (A) k-means clustering was performed with all 6 factors and plots depicting all relationships are shown. Previous hierarchal clustering identified two distinct clusters (data not shown). Red dots indicate cluster 1 (n=28) and blue dots indicate cluster 2 (n=12). (B) Heatmap of z-score normalized plasma cytokine and chemokine values. (C) Breakdown of percentages of SF7S and SLAMF7 responsive patients per cluster. Number of patients per group, per cluster indicated inside bars.
Figure 1.
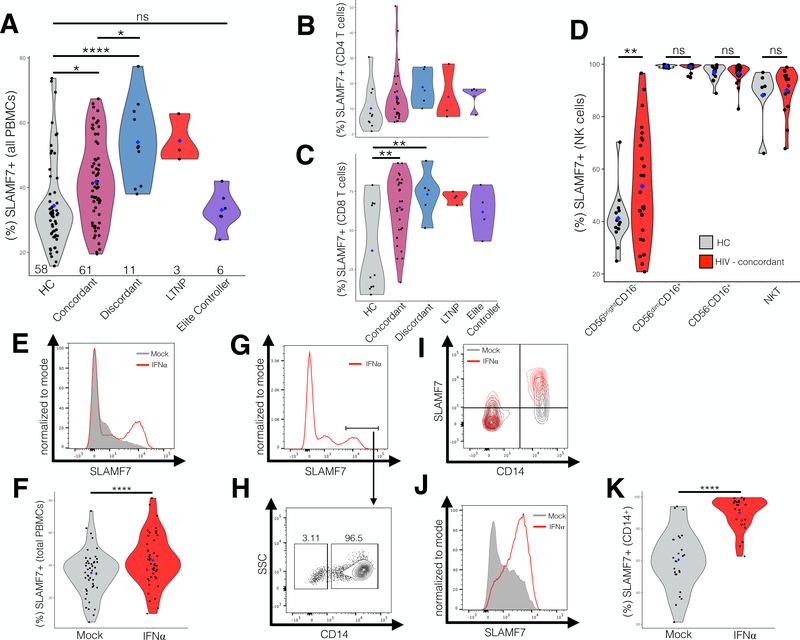
HIV+ individuals have increased expression of SLAMF7 on their PBMCs and SLAMF7 is upregulated by IFN⍺. HIV+ patients and healthy controls (HC) were screened for total SLAMF7 expression across all peripheral immune cell types (A). SLAMF7 expression was assessed on CD4 T cells (B), CD8 T cells (C), and NKT cells (D). The n is indicated along the x-axis (A) and the blue diamonds indicate the mean. The n for (B and C) is: HC: 10, Concordant: 28, Discordant: 5, LTNP: 3, EC: 4. The n for (D) is: HC: 14, HIV: 26. The effect of IFN⍺ (100 IU/mL) on SLAMF7 expression on total peripheral blood mononuclear cells (PBMCs) (E and F), and monocytes (J and K) was assessed in vitro. (F and K) n=46 and 26, respectively and data is pooled from 5 independent experiments. (G and H) CD14 expression on SLAMF7high cells from IFN⍺ stimulated total PBMCs. (I) Comparison of SLAMF7 expression on CD14+ and CD14- cells following IFN⍺ stimulation showing only CD14+ monocytes up-regulate SLAMF7. Groups compared using 1-way ANOVA with Tukey’s multiple comparison test (A) and paired two-tailed T test (C and E). *P<0.05, ****P<0.0001. LTNP: long-term nonprogressor, ns: not significant.
Figure 5.
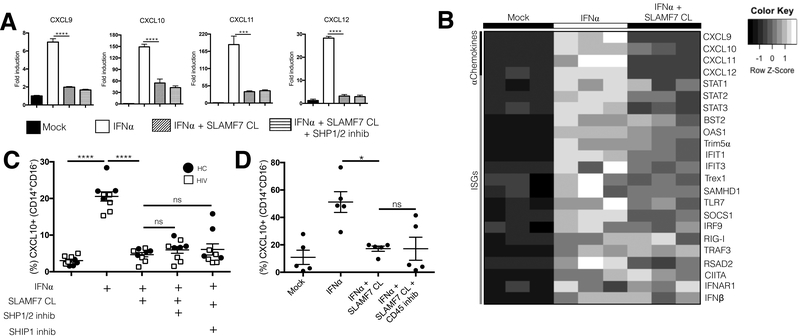
SLAMF7 activation on monocytes is selective for alpha chemokines and may not be mediated by any of the inhibitory phosphatases known to interact with SLAMF7. (A and B) We assessed the effects of SLAMF7-mediated inhibition of other alpha chemokines (A) and ISGs (B) in isolated monocytes from HCs at the mRNA level by qRT-PCR. (C) The effects of SHP1/2 and SHIP1 on SLAMF7-mediated inhibition of CXCL10 was assessed via small molecule inhibitors (SHP1/2: NSC87877, 10μM) (SHIP1: 3AC, 5μM). SHP1/2 effects on alpha chemokines were also assessed at the mRNA level (A). (D) The role of CD45 in SLAMF7-mediated inhibition of CXCL10 was assessed via a small molecule inhibitor (CAS 345630–40-2, 1μM). (A and B) representative of 4 independent experiments. Groups in (A, B, C, and D) were compared using 1-way ANOVA with Tukey’s multiple comparison test. Data presented as mean ± SEM. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Study approval.
Informed consent from all HIV patients was obtained prior to their enrollment in the study and approved by the MSU IRB (IRB#: 11–202). Patient data was de-identified and complies with all HIPAA regulations.
Results:
HIV+ individuals have increased expression of SLAMF7 on their peripheral blood mononuclear cells (PBMCs).
The SLAMF7 receptor functions as a self-ligand, therefore, global receptor levels across the complete spectrum of PBMCs will impact the function of all SLAMF7+ cells. To investigate if HIV infection alters SLAMF7 expression we screened 81 HIV patients and 58 healthy controls (HCs) for global expression of SLAMF7 across all peripheral immune cells (Fig. 1A). Subjects were grouped according to their clinical phenotype: cART concordant (CD4 count >250 with no decrease in CD4 count over 6 months), cART discordant (CD4 count <250 with undetectable viral load and no improvement in CD4 count over 6 months), long-term nonprogressor (LTNP), or elite controller (EC). We observed that SLAMF7 levels were minimally increased in concordant patients while discordant and LTNP patients had significantly higher SLAMF7 levels (Fig. 1A). Interestingly, elite controllers had global SLAMF7 levels comparable to HCs (Fig. 1A). Assessment of SLAMF7 expression on CD4 and CD8 T cells revealed SLAMF7 to be significantly up-regulated on CD8 T cells from concordant and discordant patients (Fig. 1B and C). Expression of SLAMF7 on 4 subsets of NK cells (CD56brightCD16-, CD56dimCD16+, CD56-CD16+, and NKT cells) showed comparable expression between HCs and HIV patients (Fig. 1D). Interestingly, HIV patients had significantly more SLAMF7+ cells in the CD56brightCD16- regulatory NK cell subset, as compared to other NK cell subsets (Fig. 1D).
SLAMF7 is upregulated in response to IFN⍺ in total PBMCs and monocytes from HIV+ individuals.
To examine whether SLAMF7 upregulation may be the result of chronic immune activation we stimulated PBMCs from HIV+ patients with IFN⍺; known to be chronically elevated in HIV+ patients (6). We observed a minor, but significant increase in the percent of SLAMF7+ PBMCs following IFN⍺ stimulation (Fig. 1E and F). To identify if a specific cell type was responsible for this increase in SLAMF7+ cells we gated on the SLAMF7high peak present in IFN⍺ samples and not in mock treated and looked at various cell markers. We observed that >95% of the cells in this SLAMF7high peak were CD14+ monocytes (Fig. 1G-I). Looking at just CD14+ monocytes we found they showed the most robust SLAMF7 response to IFN⍺ (Fig. 1J and K). Since monocytes play a critical role in type I interferon responses and are implicated in the pathogenesis of a number of secondary HIV-associated pathologies (22), we next assessed what role SLAMF7 may be playing in monocytes.
SLAMF7 activation on monocytes of HCs inhibits IFN⍺-mediated CXCL10 production.
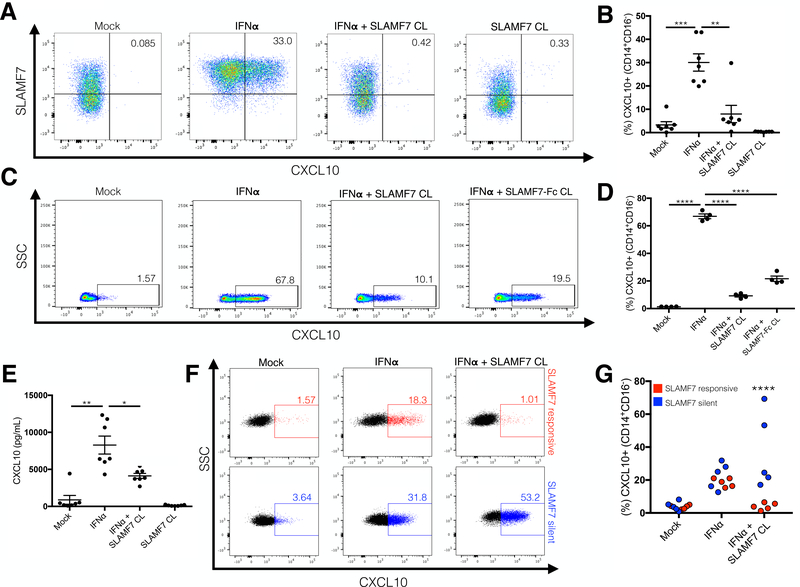
Monocytes are the primary source of CXCL10, especially in response to type I and II interferons (33). Therefore, we examined the effect of SLAMF7 signaling on IFN⍺ and IFNγ-stimulated PBMCs from HCs. Activation of SLAMF7 during IFN⍺ (Fig. 2A and B) and IFNγ-stimulation (Supplemental Fig. 2A) resulted in robust inhibition of CXCL10 production from monocytes. To confirm this effect was specific to monocytes we isolated primary CD14+ monocytes and repeated the previous experiment noting the same effect (Fig. 2C and D). To verify that CXCL10 inhibition was SLAMF7-specific we activated SLAMF7 via cross-linking with a recombinant version of the SLAMF7 extracellular domain, SLAMF7-Fc (Fig. 2C and D, final condition). Monocytes were also treated with soluble SLAMF7-Fc in vitro confirming that binding of cell surface SLAMF7 to an immobilized SLAMF7 extracellular domain was necessary for inhibition (Supplemental Fig. 2D and E). Furthermore, we confirmed that activation of SLAMF7 resulted in reduced secretion of CXCL10 from IFN⍺-stimulated PBMCs into the culture supernatant via BioPlex assay (Fig. 2E). However, it is important to note that SLAMF7 activation did not robustly reduce supernatant levels of CXCL10 to the degree we observed with flow cytometry, suggesting the putative involvement of another cell type(s).
Figure 2.
Activation of the SLAMF7 receptor on monocytes inhibits their IFN⍺-mediated production of CXCL10. (A and B) PBMCs from healthy controls were stimulated in vitro with IFN⍺ and the SLAMF7 receptor was activated by cross-linking where indicated. Expression of CXCL10 was measured by intracellular staining on flow cytometry. (C and D) The same experiment in (A) was carried out with isolated CD14+ monocytes. Cross-linking with a recombinant protein comprised of the extracellular domain of SLAMF7 fused to a modified IgG4 Fc fragment (SLAMF7-Fc) was performed to confirm that inhibition is SLAMF7 specific (C and D, last condition). (E) The levels of secreted CXCL10 in the supernatant from (A) was assessed by BioPlex assay. (F and G) PBMCs from HIV+ donors were isolated and the same experiment as in (A) was carried out. HIV patients failing to respond to SLAMF7 activation were classified as SLAMF7 silent (SF7S). (B and E n=7). (D) n=4 technical replicates from a single donor, representative of 3 independent experiments. (B and E) Data is presented as pooled results of 2 independent experiments, representative of 7 total experiments. (F and G) Data presented is pooled from 2 independent experiments. 3–4 HCs were run alongside HIV samples in all experiments to verify that assay worked. SF7S and SLAMF7 responsive groups are compared using 2-way ANOVA with Sidak’s multiple comparison test. Data in (B, D, and E) presented as mean ± SEM. Groups compared using 1-way ANOVA with Tukey’s multiple comparison test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
A subset of HIV+ patients are non-responsive to SLAMF7 inhibitory signaling.
We next examined if this effect was conserved in HIV+ individuals. The same experiment performed in (Fig. 2A and B) was repeated with freshly isolated PBMCs from HIV+ patients. We noted a dichotomous response in HIV+ patients with some responding the same as HCs (referred to hereafter as “SLAMF7 responsive”) and some showing a failure to inhibit CXCL10, which we termed “SLAMF7 silent” (SF7S) (Fig. 2F and G). As an internal control to verify that these differences were not due to experimental variability we included HCs alongside HIV+ samples in each experiment and noted in each case that all HCs did respond to SLAMF7 activation. Defining SLAMF7 as an inhibitory receptor in monocytes, and identifying a subset of HIV+ patients with a defect in SLAMF7 signaling led us to examine if there was any correlation to clinical biomarkers associated with chronic immune activation.
SLAMF7 responsive and SF7S patients have distinct peripheral immune activation signatures.
The plasma levels of six proinflammatory cytokines and chemokines implicated in chronic immune activation (3) during HIV infection were evaluated. Hierarchal and k-means clustering was used to identify patients with similar peripheral immune activation profiles. Hierarchal clustering identified two distinct clusters and (Fig. 3A) shows the results of k-means clustering. Cluster one was characterized by low levels of all six proinflammatory factors, while Cluster two showed patients with elevated levels of all six proinflammatory factors. A heatmap of plasma cytokines and chemokines shows Cluster one patients exhibiting a similar profile to that of HCs, while Cluster two patients have a markedly different cytokine profile (Fig. 3B). Comparison of demographics and clinical characteristics showed the two clusters to be otherwise well balanced (Table I). Interestingly, we found that Cluster 2 patients are all SF7S while Cluster 1 patients are predominantly SLAMF7 responsive (Fig. 3C). These results suggest that dysfunction of the SLAMF7 receptor may result in in vivo manifestations.
Table I.
Cluster characteristics
| Cluster 1 | Cluster 2 | |
|---|---|---|
| n | 28 | 12 |
| Median age, (IQR) | 51 (17.25) | 54 (6.25) |
| Median BMI, (IQR) | 28.5 (5.78) | 28.75 (12.9) |
| Race- n, (%) | ||
| Caucasian | 17 (60.7) | 7 (58.3) |
| African | 8 (28.6) | 3 (25) |
| Hispanic | 3 (10.7) | 1 (8.3) |
| Unknown | 0 | 1 (8.3) |
| Sex- n, (%) | ||
| Male | 21 (75) | 10 (83.3) |
| Female | 7 (25) | 2 (16.7) |
| Median CD4 count (cells/μL), (IQR) | 591.5 (500) | 688 (402) |
| Median CD4/CD8 ratio, (IQR) | 0.61 (0.96) | 0.45 (0.82) |
| CD4 nadir (cells/μL)-n, (%) | ||
| <50 | 7 (25) | 4 (33.3) |
| 50–100 | 5 (17.9) | 0 |
| 100–200 | 5 (17.9) | 4 (33.3) |
| 200–350 | 3 (10.7) | 1 (8.3) |
| 350–500 | 1 (3.6) | 2 (16.7) |
| >500 | 7 (25) | 0 |
| Unknown | 0 | 1 (8.3) |
| Clinical phenotype-n, (%) | ||
| Concordant | 20 (71.3) | 8 (66.7) |
| Discordant | 3 (10.7) | 3 (25) |
| Elite Controller | 4 (14.3) | 0 |
| LTNP | 1 (3.6) | 1 (8.3) |
| Viral load (copies/mL)-n, (%) | ||
| ND | 20 (71.3) | 8 (66.7) |
| 20–1000 | 5 (17.9) | 3 (25) |
| 1000–25000 | 2 (7.1) | 1 (8.3) |
| Unknown | 1 (3.6) | 0 |
| Median length of infection (years), (IQR) | 16.5 (17.25) | 14 (22.75) |
| Current MJ use- n, (%) | ||
| Yes | 9 (32.1) | 6 (50) |
| No | 18 (64.3) | 6 (50) |
| Unknown | 1 (3.6) | 0 |
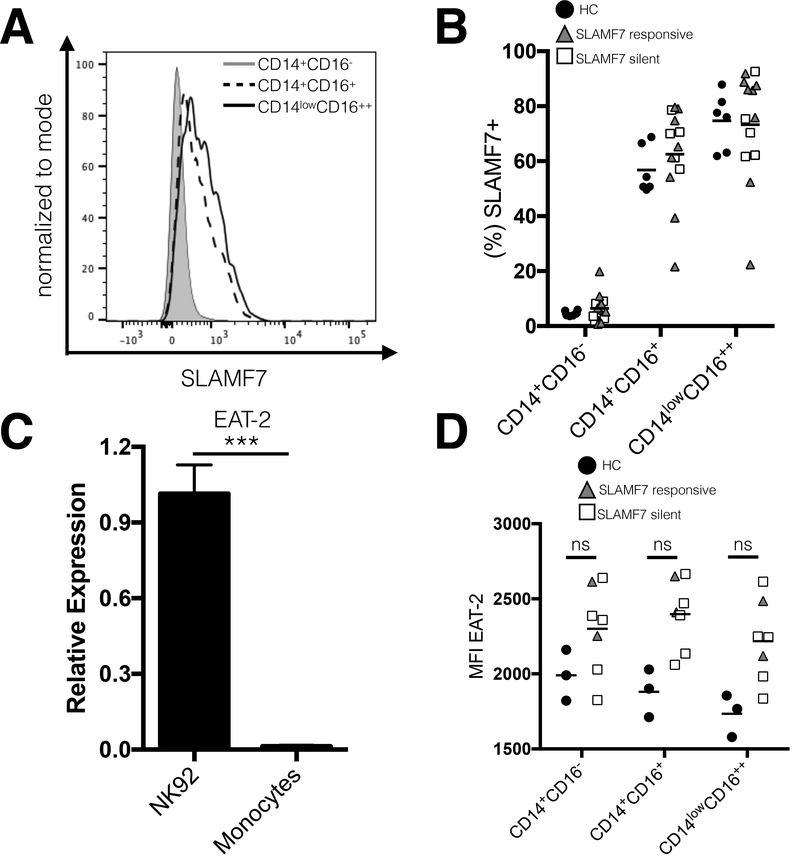
Monocytes from SF7S and SLAMF7 responsive patients do not differ in expression levels of the SLAMF7 receptor or EAT-2 adaptor.
To determine the mechanism behind this defective SLAMF7 function in SF7S patients we assessed levels of SLAMF7 expression across all monocyte subsets in HCs, SF7S, and SLAMF7 responsive patients. We noted that a small percentage of classical monocytes (~5%) express SLAMF7 at baseline, while a significantly higher percentage of both intermediate (~60%) and non-classical monocytes (~70%) express SLAMF7 (Fig. 4A and B). Critically, we did not notice a difference in SLAMF7 expression between SF7S and SLAMF7 responsive patients (Fig. 4B). Since it has been well established that the presence or absence of the SLAM family adaptor EAT-2 can dramatically alter SLAMF7 signaling and govern the activating function of SLAMF7 (26), we next looked to the levels of EAT-2 across all three monocyte subsets in HCs, SF7S, and SLAMF7 responsive individuals (Fig. 4D). However, we first examined whether monocytes from HCs express any EAT-2 at all. Comparison of EAT-2 mRNA levels between monocytes and a human NK cell line known to express EAT-2 (NK92), showed that monocytes have little to no EAT-2 (Fig. 4C). This result confirms previous findings (34) and partially explains why SLAMF7 acts in a purely inhibitory manner in monocytes. Comparison of EAT-2 at the protein level via flow cytometry showed that all HIV+ individuals have slightly increased EAT-2 expression over HCs (Fig. 4D). We did not, however, observe any differences in EAT-2 expression between SF7S and SLAMF7 responsive patients. Together, these results suggest that the mechanism underlying the loss of SLAMF7 inhibitory activity in monocytes may be independent of SLAMF7 and EAT-2 expression levels.
Figure 4.
Monocytes from SF7S and SLAMF7 responsive patients do not differ in expression levels of SLAMF7 or EAT-2. (A) Expression of SLAMF7 on monocyte subsets. (B) Comparison of SLAMF7 expression between SF7S, SLAMF7 responsive, and HCs on all monocyte subsets. (C) We assessed the levels of EAT-2, a known SLAM family receptor adaptor, in a human cell line known to express EAT-2 (NK92) and isolated primary monocytes by qRT-PCR. (D) Analysis of EAT-2 protein levels in monocytes between SF7S and SLAMF7 responsive individuals by intracellular staining on flow cytometry. (C) Results are technical replicates from a single donor compared with unpaired 2-tailed T test. Groups in (B and D) compared using 1-way ANOVA with Tukey’s multiple comparison test as well as a 2-way ANOVA with Sidak’s multiple comparison. (B) n=6 for HCs, 8 for SLAMF7 responsive, and 5 for SF7S. (D) n=3 for HCs, 2 for SLAMF7 responsive, and 5 for SF7S. Data presented as mean ± SEM. ***P<0.001.
SLAMF7 signaling in monocytes selectively inhibits IFN⍺-mediated production of alpha chemokines over other interferon-stimulated genes (ISGs) and host restriction factors.
We then investigated if the robust inhibition of CXCL10 was conserved across other alpha chemokines and ISGs. We noted that the inhibition of CXCL10 was conserved at the mRNA level and that this effect was consistent across CXCL9, CXCL11, and CXCL12 (Fig. 5A). Surprisingly, a number of other interferon stimulated genes and HIV-associated, interferon-modulated, host restriction factors including: BST2, OAS1, Trex1, and STAT1 showed only minor, or no inhibition following SLAMF7 activation (Fig. 5B).
SLAMF7-mediated inhibition of CXCL10 production in monocytes may be independent of SHP1, SHP2, SHIP1, or CD45.
To better understand the mechanism behind SLAMF7’s inhibitory functions in monocytes, we evaluated a number of inhibitory phosphatases previously shown to interact with SLAMF7 and mediate its inhibitory functions (26, 35). In an effort to continue using primary human monocytes, we tested a number of small molecule inhibitors targeting SHP1, SHP2, and SHIP1. Pre-treatment with either a small molecule inhibitor of SHP1/2 (NSC87877) (Fig. 5A and C) or SHIP1 (3AC) (Fig. 5C) did not result in recovery of CXCL10 expression in monocytes stimulated with IFN⍺ and SLAMF7 cross-linking. We then hypothesized that SLAMF7’s inhibitory function might be mediated by CD45, as CD45 has been previously implicated in SLAMF7’s ability to propagate inhibitory signals in NK and multiple myeloma cells (35). Pre-treatment with a small molecule inhibitor specific for CD45 (CAS 345630–40-2) in the presence of IFN⍺ and SLAMF7 cross-linking also failed to rescue CXCL10 production (Fig. 5D). Finally, we speculated that SLAMF7 might prevent CXCL10 production by a mechanism that involves CXCL10 proteasomal degradation. Addition of the proteasome inhibitor Bortezomib did not result in recovery of CXCL10 expression in SLAMF7-stimulated cells, and actually further decreased CXCL10 production (Supplemental Fig. 2B). Interestingly, we also noted that Bortezomib acted synergistically with IFN⍺ to upregulate SLAMF7 expression on monocytes (Supplemental Fig. 2C).
SLAMF7 inhibits monocyte infection with HIV-1 in vitro.
HIV+ patients are known to have increased levels of circulating inflammatory monocytes (CD14+CD16+ and CD14lowCD16++) which promote peripheral immune activation (36, 37), can become infected with HIV virus (22), and induce HIV-associated neurocognitive disorder (HAND). We next looked to see if SLAMF7 activation could affect the ability of monocytes to transition into pro-inflammatory subtypes (CD16+), as well as monocyte susceptibility to HIV virus infection. Monocyte infection with HIV occurs via CCR5 (38, 39), which is known to be upregulated following IFN⍺ stimulation (40). We discovered that activation of SLAMF7 on monocytes from HIV+ individuals could down-regulate CCR5 both in the presence and absence of IFN⍺ (Fig. 6A and B). Further supporting SLAMF7’s utility as a means to prevent monocyte infection by HIV-1, we noted that activation of SLAMF7 in the presence of IFN⍺ significantly upregulated the CCL3L1 chemokine (Fig. 6C), a chemokine that binds to CCR5 and directly prevents HIV infection of monocytes (41, 42). Based on these findings we hypothesized that SLAMF7 activation in human monocytes might prevent HIV infection. Therefore, we performed an in vitro infection of isolated primary human monocytes from 2 HCs to determine if SLAMF7 activation could prevent HIV-1 infection. As a control, we also included a CCR5 blocking antibody. As expected, pre-treatment of human monocytes with anti-CCR5 mAb prevented HIV infection (Fig. 6D). Critically, infection of monocytes with HIV-1-Ba-L-GFP in the presence of SLAMF7 cross-linking resulted in dramatically reduced percentages of HIV-1 infected cells (Fig. 6D). To determine if SLAMF7 signaling affected CD16 expression on monocytes we utilized a previously described, IFN⍺-driven monocyte maturation assay (13). We observed a significant down-regulation of CD16 on monocytes with SLAMF7 activation both in the presence and absence of IFN⍺ stimulation (Fig. 6E) suggesting that SLAMF7 signaling can prevent induction of inflammatory monocyte subsets.
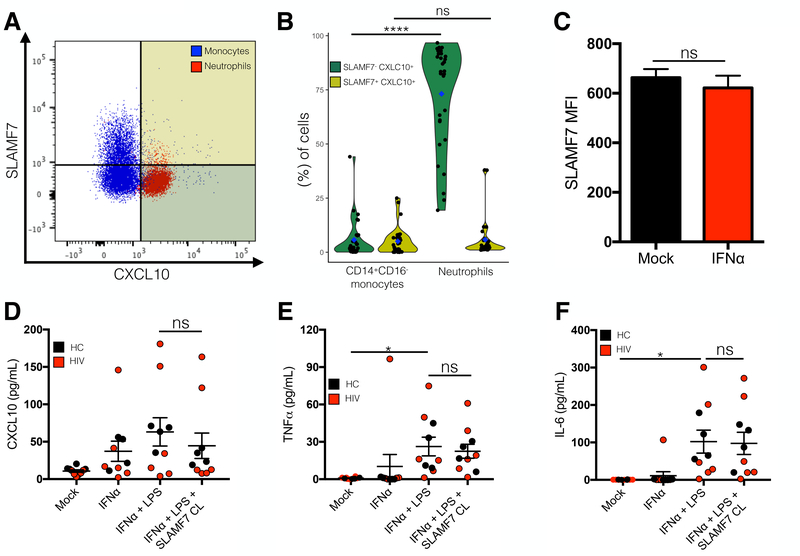
Neutrophils constitutively express CXCL10 and do not express SLAMF7.
Upon further review of our in vitro data we noticed a peculiar discrepancy between our flow cytometry and BioPlex results. Comparing (Fig. 2B and E) we noted that while we observed near complete inhibition of CXCL10 from IFN⍺-stimulated monocytes following SLAMF7 activation on FACS analysis, we saw just an ~50% reduction in CXCL10 concentration in the culture supernatant. While this could have been from residual buildup of CXCL10 in the supernatant that occurs before SLAMF7 inhibition takes effect, we had determined that SLAMF7 begins to exert its inhibitory effects within a very short time frame (<4h), based on isolated monocyte experiments (Fig. 2D). This suggested that another cell type may also be responsible for CXCL10 production and may not be responding to SLAMF7 activation. We identified neutrophils (CD66b+ CD3- CD19- CD57- CD16++ CD14low/-) as being SLAMF7- and CXCL10+ at baseline in cART treated HIV individuals (Fig. 7A and B).
Figure 7.
Neutrophils are SLAMF7- and CXCL10+ and do not respond to SLAMF7 activation. Baseline SLAMF7 and CXCL10 expression was examined on CD14+CD16- monocytes and neutrophils by flow cytometry (A). (B) Quantification of results from (A) focusing on CXCL10+ cells. Color-coded quadrants in (A) correspond to dot colors in (B). (C) Neutrophils do not increase SLAMF7 expression in response to IFN⍺ stimulation. (D-F) Isolated neutrophils from HCs and HIV patients were stimulated in vitro with IFN⍺, LPS, and SLAMF7 as indicated and supernatants were assessed by BioPlex assay for indicated cytokines/chemokines. (B) n=30. (C) n=3 independent donors from a single experiment. (D-F) Data presented is pooled from 2 independent experiments with n=10 donors. (B) Groups are compared using 2-way ANOVA with Sidak’s multiple comparison test. Groups in (C) compared using 2-tailed T test. Groups in (D-F) compared using 1-way ANOVA with Tukey’s multiple comparison test. Data presented as mean ± SEM. *P<0.05, ****P<0.0001.
Neutrophils are unable to upregulate SLAMF7 in response to IFNα and SLAMF7 activation does not inhibit proinflammatory cytokine and chemokine release from neutrophils.
SLAMF7 expression on neutrophils in the presence and absence of IFN⍺ stimulation was assessed by flow cytometry. We observed that neutrophils are unable to upregulate SLAMF7 in response to IFN⍺ (Fig. 7C). This, combined with the knowledge that they are CXCL10+ at baseline, suggested that following appropriate stimulation, they should constitutively release CXCL10 regardless of the presence of SLAMF7 activating mAbs. We tested this via BioPlex analysis utilizing freshly isolated neutrophils from both HCs and HIV+ patients. We discovered that IFN⍺ by itself can induce CXCL10 release from neutrophils, that this effect is enhanced by addition of LPS, and that addition of SLAMF7 cross-linking mAbs very minimally decreases CXCL10 release (Fig. 7D). Consistent with this result, we noted a similar pattern in regards to TNF⍺ and IL-6 release from IFN⍺ and LPS co-stimulated neutrophils (Fig. 7E and F, respectively).
Discussion:
The discovery that global levels of SLAMF7+ PBMCs are increased in HIV infected patients in a clinical phenotype-dependent manner suggests that SLAMF7 receptor functions may play a role in modulating peripheral immune activation. Supporting the idea that SLAMF7 is a marker of elevated type I interferons are reports showing SLAMF7 to be upregulated in systemic lupus erythematosus (43), rheumatoid arthritis (44), and multiple sclerosis (45). While we focused on the effects of IFN⍺ on SLAMF7 expression, it is well known that LPS can also up-regulate SLAMF7 through a NF-κB-dependent mechanism (34). HIV+ patients are known to have elevated levels of LPS in their blood as a result of a “leaky gut” (5); we cannot exclude this as an additional possible reason for elevated SLAMF7 levels. The findings that discordant patients have extremely elevated SLAMF7 levels and that elite controllers have levels of SLAMF7 comparable to healthy controls supports the idea that assessing SLAMF7 expression levels on peripheral immune cells could be an effective gauge of a patients’ immune-activation status.
The finding that SLAMF7 activation can specifically inhibit CXCL10 and other alpha chemokines in monocytes is consistent with a previous study showing SLAMF7 has inhibitory effects in LPS-stimulated monocytes (34). Interestingly, SLAMF7 activation minimally reduced HIV restriction factors, thus pharmacological modulation of SLAMF7 in the context of HIV infection could be beneficial in reducing peripheral immune activation, and preventing monocyte infection without affecting important viral restriction factors. However, this approach may only be effective in some HIV patients, since we identified a subset of HIV+ individuals, SF7S, who show a lack of response (or inverse response) to SLAMF7 activation. It is possible that the underlying mechanism to this paradoxical lack of SLAMF7 response is due to either genetic differences between patients, differential alterations in circulating cytokines or chemokines, or from specific interactions with HIV viral proteins.
Our small molecule studies suggest that there are yet other, unidentified inhibitory factor(s) which can interact with SLAMF7, or SLAMF7 may propagate its signals through ITSM-independent mechanisms (30). Supporting this is the fact that most of the studies regarding the interaction of SLAMF7 with inhibitory phosphatases were performed in mice or human cell lines (26, 27, 35), thus it is possible that some of those findings are not entirely translatable to primary human cells. Regardless, SLAMF7 activation in monocytes may prove to be a useful method of preventing pathological activation of the CXCR3 receptor since it inhibits all CXCR3 ligands.
Both clinical and pre-clinical attempts at CXCL10 neutralization and/or CXCR3 receptor blockade have largely failed thus far (46–50). Reasons for this include: failure of CXCL10- specific mAbs to inhibit all CXCR3 ligands, inability of anti-CXCL10 mAbs to compete with the high synthesis and turnover rate of CXCL10, and inability of some anti-CXCL10 mAbs to bind the glycosaminoglycan (GAG) bound form of CXCL10 (the active form of CXCL10). Importantly, SLAMF7-mediated inhibition of alpha chemokines has the ability to overcome all of these limitations and may find itself to be a useful therapeutic modality in diseases where over-expression of CXCL10 has been linked to pathogenesis including: rheumatoid arthritis (51), type I diabetes mellitus (52), systemic lupus erythematous (53), multiple sclerosis (54), ulcerative colitis (55), and primary biliary cirrhosis (56).
It is well established that patients with HIV or a number of other diseases (57–59) have elevated levels of CD16+ monocytes and that these pro-inflammatory cells have been implicated in the pathogenesis of these conditions. To our knowledge there are currently no methods to reduce the number of circulating CD16+ monocytes in these individuals, except for a single in vitro report studying one of the components of cannabis, THC (13). It would be interesting to see if SLAMF7-mediated down-regulation of CD16 is consistent in vivo and, if so, if there is any resultant effect on pathology in pre-clinical animal models.
Similar to its effect on CD16, the down-regulation of CCR5 on monocytes by SLAMF7 signaling could have potential benefits in the setting of HIV infection (60). While there have been numerous attempts at either blocking/down-regulating CCR5 (notably, Maraviroc) or increasing the levels of chemokines, such as CCL3L1, (or engineered chemokines) specific for CCR5 for the treatment of HIV infection (61), there have not been any attempts to simultaneously apply both approaches. Activation of SLAMF7 on monocytes is unique in that it can accomplish this and potentially inhibit HIV-1 infection of monocytes and other CCR5-expressing immune cells.
We found that neutrophils are SLAMF7- and CXCL10+ at baseline and are able to respond to stimulation by type I interferons, but not to SLAMF7 activation. The role of neutrophils in the context of HIV infection is complex and understudied (62). Specifically, whether or not neutrophils contribute to peripheral immune activation in cART-treated individuals is unclear (62). Our results suggest that neutrophils likely do contribute to peripheral immune activation. Additionally, neutrophils are also known to play an important role in neuroinflammation (63) and have recently been discovered to be present in high levels in the brain at steady state (64). While the role of neutrophils in the CNS of HIV+ individuals and in HAND is unknown, our results suggest that if they do play a role, it is likely one that cannot be modulated through the SLAMF7 receptor. The discovery that neutrophils were able to secrete CXCL10 following IFN⍺ and LPS stimulation highlights the need to consider the effects neutrophils are playing in diseases characterized by chronic type I interferon activation.
In summary, we determined that SLAMF7 is up-regulated globally in HIV patients who have high levels of peripheral immune activation and that SLAMF7 functions to both prevent HIV viral infection of monocytes, and inhibit CXCL10 from monocytes stimulated with type I and II interferons, except in a subset of HIV patients. We also discovered that neutrophils fail to express SLAMF7, constitutively express CXCL10, and are non-responsive to SLAMF7 activation, implicating them as potential propagators of chronic, peripheral immune activation in type I interferon-mediated diseases.
Supplementary Material
Acknowledgments:
We thank the MSU flow cytometry core and Dr. Louis King for assistance with flow cytometry, the MSU genomics core for assistance with qRT-PCR, the MSU Center for Statistical Training and Consulting (CSTAT) and Hope Akaeze for advice on clustering, MSU Clinical and Translational Sciences Institute (MSU-CTSI) for support with patient recruitment and sample collection, Candace Savonen for assistance with R, and to all of the patients for graciously volunteering to assist with this study. Thanks to Joseph Henriquez for assistance with sample preparation and advice on assay design. Special thanks to Dr. Young-Hui Zheng for providing HIV-1-Ba-L-GFP virus.
Funding: This work was supported by the US National Institute of Health grant:1R21AI122808–01 (to YAA).
Abreviations:
- cART
combined anti-retroviral therapy
- SF7S
SLAMF7 silent
- pDCs
plasmacytoid dendritic cells
- SLAM
signaling lymphocytic activation molecules
- ITSMs
immunoreceptor tyrosine-based switch motifs
- SAP
SLAM-associated protein
- ADCC
antibody-dependent cell-mediated cytotoxicity
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- PSF
penicillin, streptomycin and fungizone
- LTNP
long-term nonprogressor
- EC
elite controller
- HC
healthy control
- ISGs,
interferon stimulated genes
- HAND
HIV-associated neurocognitive disorder
- MM
multiple myeloma
Footnotes
Conflicts of interest: YAA and AA hold a patent on the SLAMF7-Fc fusion protein. YAA and PO have a patent pending regarding the modulation of SLAMF7 signaling to prevent inflammation and HIV viral infection. The authors declare no additional conflicts of interest.
References
- 1.Katlama C, Deeks SG, Autran B, Martinez-Picado J, van Lunzen J, Rouzioux C, Miller M, Vella S, Schmitz JE, Ahlers J, Richman DD, and Sekaly RP 2013. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 381: 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardoso SW, Torres TS, Santini-Oliveira M, Marins LM, Veloso VG, and Grinsztejn B 2013. Aging with HIV: a practical review. Braz J Infect Dis 17: 464–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paiardini M, and Muller-Trutwin M 2013. HIV-associated chronic immune activation. Immunol Rev 254: 78–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sereti I, and Altfeld M 2016. Immune activation and HIV: an enduring relationship. Curr Opin HIV AIDS 11: 129–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong S, and Banks WA 2015. Role of the immune system in HIV-associated neuroinflammation and neurocognitive implications. Brain Behav Immun 45: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha L, Berry CM, Nolan D, Castley A, Fernandez S, and French MA 2014. Interferon-alpha, immune activation and immune dysfunction in treated HIV infection. Clin Transl Immunology 3: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rho MB, Wesselingh S, Glass JD, McArthur JC, Choi S, Griffin J, and Tyor WR 1995. A potential role for interferon-alpha in the pathogenesis of HIV-associated dementia. Brain Behav Immun 9: 366–377. [DOI] [PubMed] [Google Scholar]
- 8.Sas AR, Bimonte-Nelson H, Smothers CT, Woodward J, and Tyor WR 2009. Interferon-alpha causes neuronal dysfunction in encephalitis. J Neurosci 29: 3948–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French MA, Cozzi-Lepri A, Arduino RC, Johnson M, Achhra AC, Landay A, and Group ISS 2015. Plasma levels of cytokines and chemokines and the risk of mortality in HIV-infected individuals: a case-control analysis nested in a large clinical trial. AIDS 29: 847–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veenhuis RT, Freeman ZT, Korleski J, Cohen LK, Massaccesi G, Tomasi A, Boesch AW, Ackerman ME, Margolick JB, Blankson JN, Chattergoon MA, and Cox AL 2017. HIV-antibody complexes enhance production of type I interferon by plasmacytoid dendritic cells. J Clin Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulliam L, Rempel H, Sun B, Abadjian L, Calosing C, and Meyerhoff DJ 2011. A peripheral monocyte interferon phenotype in HIV infection correlates with a decrease in magnetic resonance spectroscopy metabolite concentrations. AIDS 25: 1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rempel H, Sun B, Calosing C, Pillai SK, and Pulliam L 2010. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS 24: 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizzo MD, Crawford RB, Henriquez JE, Aldhamen YA, Gulick P, Amalfitano A, and Kaminski NE 2018. HIV-infected cannabis users have lower circulating CD16+ monocytes and IFN-gamma-inducible protein 10 levels compared with nonusing HIV patients. AIDS 32: 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arico E, Castiello L, Urbani F, Rizza P, Panelli MC, Wang E, Marincola FM, and Belardelli F 2011. Concomitant detection of IFNalpha signature and activated monocyte/dendritic cell precursors in the peripheral blood of IFNalpha-treated subjects at early times after repeated local cytokine treatments. J Transl Med 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNab F, Mayer-Barber K, Sher A, Wack A, and O’Garra A 2015. Type I interferons in infectious disease. Nat Rev Immunol 15: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ploquin MJ, Madec Y, Casrouge A, Huot N, Passaes C, Lecuroux C, Essat A, Boufassa F, Jacquelin B, Jochems SP, Petitjean G, Angin M, Gartner K, Garcia-Tellez T, Noel N, Booiman T, Boeser-Nunnink BD, Roques P, Saez-Cirion A, Vaslin B, Dereudre-Bosquet N, Barre-Sinoussi F, Ghislain M, Rouzioux C, Lambotte O, Albert ML, Goujard C, Kootstra N, Meyer L, and Muller-Trutwin MC 2016. Elevated Basal Pre-infection CXCL10 in Plasma and in the Small Intestine after Infection Are Associated with More Rapid HIV/SIV Disease Onset. PLoS Pathog 12: e1005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez LA, Arango TA, Thompson E, Naji M, Tebas P, and Boyer JD 2014. High IP-10 levels decrease T cell function in HIV-1-infected individuals on ART. J Leukoc Biol 96: 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehla R, Bivalkar-Mehla S, Nagarkatti M, and Chauhan A 2012. Programming of neurotoxic cofactor CXCL-10 in HIV-1-associated dementia: abrogation of CXCL-10-induced neuro-glial toxicity in vitro by PKC activator. J Neuroinflammation 9: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Marle G, Henry S, Todoruk T, Sullivan A, Silva C, Rourke SB, Holden J, McArthur JC, Gill MJ, and Power C 2004. Human immunodeficiency virus type 1 Nef protein mediates neural cell death: a neurotoxic role for IP-10. Virology 329: 302–318. [DOI] [PubMed] [Google Scholar]
- 20.Sui Y, Stehno-Bittel L, Li S, Loganathan R, Dhillon NK, Pinson D, Nath A, Kolson D, Narayan O, and Buch S 2006. CXCL10-induced cell death in neurons: role of calcium dysregulation. Eur J Neurosci 23: 957–964. [DOI] [PubMed] [Google Scholar]
- 21.Kamat A, Misra V, Cassol E, Ancuta P, Yan Z, Li C, Morgello S, and Gabuzda D 2012. A plasma biomarker signature of immune activation in HIV patients on antiretroviral therapy. PLoS One 7: e30881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams DW, Veenstra M, Gaskill PJ, Morgello S, Calderon TM, and Berman JW 2014. Monocytes mediate HIV neuropathogenesis: mechanisms that contribute to HIV associated neurocognitive disorders. Curr HIV Res 12: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M, Suk K, Kang Y, McGeer E, and McGeer PL 2011. Neurotoxic factors released by stimulated human monocytes and THP-1 cells. Brain Res 1400: 99–111. [DOI] [PubMed] [Google Scholar]
- 24.Cannons JL, Tangye SG, and Schwartzberg PL 2011. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol 29: 665–705. [DOI] [PubMed] [Google Scholar]
- 25.Aldhamen YA, Rastall DP, Chen W, Seregin SS, Pereira-Hicks C, Godbehere S, Kaminski NE, and Amalfitano A 2016. CRACC-targeting Fc-fusion protein induces activation of NK cells and DCs and improves T cell immune responses to antigenic targets. Vaccine 34: 3109–3118. [DOI] [PubMed] [Google Scholar]
- 26.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, and Veillette A 2009. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat Immunol 10: 297–305. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Quintero LA, Roncagalli R, Guo H, Latour S, Davidson D, and Veillette A 2014. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cgamma, Ca++, and Erk, leading to granule polarization. J Exp Med 211: 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, Huseni M, Powers D, Nanisetti A, Zhang Y, Rice AG, van Abbema A, Wong M, Liu G, Zhan F, Dillon M, Chen S, Rhodes S, Fuh F, Tsurushita N, Kumar S, Vexler V, Shaughnessy JD Jr., Barlogie B, van Rhee F, Hussein M, Afar DE, and Williams MB 2008. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin Cancer Res 14: 2775–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tassi I, and Colonna M 2005. The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase Cgamma signaling pathways in human NK cells. Journal of immunology 175: 7996–8002. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Zhong MC, Guo H, Davidson D, Mishel S, Lu Y, Rhee I, Perez-Quintero LA, Zhang S, Cruz-Munoz ME, Wu N, Vinh DC, Sinha M, Calderon V, Lowell CA, Danska JS, and Veillette A 2017. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature 544: 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Thudium KB, Han M, Wang XT, Huang H, Feingersh D, Garcia C, Wu Y, Kuhne M, Srinivasan M, Singh S, Wong S, Garner N, Leblanc H, Bunch RT, Blanset D, Selby MJ, and Korman AJ 2014. In vitro characterization of the anti-PD-1 antibody nivolumab, BMS-936558, and in vivo toxicology in non-human primates. Cancer Immunol Res 2: 846–856. [DOI] [PubMed] [Google Scholar]
- 32.Henriquez JE, Rizzo MD, Schulz MA, Crawford RB, Gulick P, and Kaminski NE 2017. Delta9-Tetrahydrocannabinol Suppresses Secretion of IFNalpha by Plasmacytoid Dendritic Cells From Healthy and HIV-Infected Individuals. J Acquir Immune Defic Syndr 75: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons RP, Scully EP, Groden EE, Arnold KB, Chang JJ, Lane K, Lifson J, Rosenberg E, Lauffenburger DA, and Altfeld M 2013. HIV-1 infection induces strong production of IP-10 through TLR7/9-dependent pathways. AIDS 27: 2505–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JR, Horton NC, Mathew SO, and Mathew PA 2013. CS1 (SLAMF7) inhibits production of proinflammatory cytokines by activated monocytes. Inflamm Res 62: 765–772. [DOI] [PubMed] [Google Scholar]
- 35.Guo H, Cruz-Munoz ME, Wu N, Robbins M, and Veillette A 2015. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol Cell Biol 35: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong KL, Yeap WH, Tai JJ, Ong SM, Dang TM, and Wong SC 2012. The three human monocyte subsets: implications for health and disease. Immunol Res 53: 41–57. [DOI] [PubMed] [Google Scholar]
- 37.Ziegler-Heitbrock L 2007. The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol 81: 584–592. [DOI] [PubMed] [Google Scholar]
- 38.Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, Sonza S, and Crowe SM 2007. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. Journal of immunology 178: 6581–6589. [DOI] [PubMed] [Google Scholar]
- 39.Tuttle DL, Harrison JK, Anders C, Sleasman JW, and Goodenow MM 1998. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol 72: 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoddart CA, Keir ME, and McCune JM 2010. IFN-alpha-induced upregulation of CCR5 leads to expanded HIV tropism in vivo. PLoS Pathog 6: e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell R J, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, and Ahuja SK 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307: 1434–1440. [DOI] [PubMed] [Google Scholar]
- 42.Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, Anaya JM, Miura T, Hecht FM, Mamtani M, Pereyra F, Marconi V, Mangano A, Sen L, Bologna R, Clark RA, Anderson SA, Delmar J, O’Connell RJ, Lloyd A, Martin J, Ahuja SS, Agan BK, Walker BD, Deeks SG, and Ahuja SK 2007. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol 8: 1324–1336. [DOI] [PubMed] [Google Scholar]
- 43.Niklas Hagberg JT, Alm Gunnar V, Eloranta Maija-Leena, Bryceson Yenan and Rönnblom Lars 2012. SLE immune complexes upregulate the expression of slamf7 (cd319) on plasmacytoid dendritic cells. Annals of Rheumatic Disease 71: A3. [Google Scholar]
- 44.Walsh AM, Wechalekar MD, Guo Y, Yin X, Weedon H, Proudman SM, Smith MD, and Nagpal S 2017. Triple DMARD treatment in early rheumatoid arthritis modulates synovial T cell activation and plasmablast/plasma cell differentiation pathways. PLoS One 12: e0183928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jordan S Mizrahi JJP, Masvaker Ruturaj and Bielekova Bibi 2017. Role of SLAMF immunoregulatory molecules in MS disease processes. The journal of immunology 198: 55.27913631 [Google Scholar]
- 46.de Graaf L, Marie KHK-V; Nicolas Fischer 2012. Therapeutic Targeting of Chemokines with Monoclonal Antibodies. Current Immunology Reviews 8: 141–148. [Google Scholar]
- 47.Yellin M, Paliienko I, Balanescu A, Ter-Vartanian S, Tseluyko V, Xu LA, Tao X, Cardarelli PM, Leblanc H, Nichol G, Ancuta C, Chirieac R, and Luo A 2012. A phase II, randomized, double-blind, placebo-controlled study evaluating the efficacy and safety of MDX-1100, a fully human anti-CXCL10 monoclonal antibody, in combination with methotrexate in patients with rheumatoid arthritis. Arthritis Rheum 64: 1730–1739. [DOI] [PubMed] [Google Scholar]
- 48.Mayer L, Sandborn WJ, Stepanov Y, Geboes K, Hardi R, Yellin M, Tao X, Xu LA, Salter-Cid L, Gujrathi S, Aranda R, and Luo AY 2014. Anti-IP-10 antibody (BMS-936557) for ulcerative colitis: a phase II randomised study. Gut 63: 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solari R, Pease JE, and Begg M 2015. “Chemokine receptors as therapeutic targets: Why aren’t there more drugs?”. Eur J Pharmacol 746: 363–367. [DOI] [PubMed] [Google Scholar]
- 50.Schall TJ, and Proudfoot AE 2011. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat Rev Immunol 11: 355–363. [DOI] [PubMed] [Google Scholar]
- 51.Laragione T, Brenner M, Sherry B, and Gulko PS 2011. CXCL10 and its receptor CXCR3 regulate synovial fibroblast invasion in rheumatoid arthritis. Arthritis Rheum 63: 3274–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimada A, Oikawa Y, Yamada Y, Okubo Y, and Narumi S 2009. The role of the CXCL10/CXCR3 system in type 1 diabetes. Rev Diabet Stud 6: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okamoto H, Katsumata Y, Nishimura K, and Kamatani N 2004. Interferon-inducible protein 10/CXCL10 is increased in the cerebrospinal fluid of patients with central nervous system lupus. Arthritis Rheum 50: 3731–3732. [DOI] [PubMed] [Google Scholar]
- 54.Bonechi E, Aldinucci A, Mazzanti B, di Gioia M, Repice AM, Manuelli C, Saccardi R, Massacesi L, and Ballerini C 2014. Increased CXCL10 expression in MS MSCs and monocytes is unaffected by AHSCT. Ann Clin Transl Neurol 1: 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Q, Kim T, Pang J, Sun W, Yang X, Wang J, Song Y, Zhang H, Sun H, Rangan V, Deshpande S, Tang H, Cvijic ME, Westhouse R, Olah T, Xie J, Struthers M, and Salter-Cid L 2017. A novel function of CXCL10 in mediating monocyte production of proinflammatory cytokines. J Leukoc Biol 102: 1271–1280. [DOI] [PubMed] [Google Scholar]
- 56.Zhang W, Fei Y, Gao J, Liu B, and Zhang F 2011. The role of CXCR3 in the induction of primary biliary cirrhosis. Clin Dev Immunol 2011: 564062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng AL, Zhu JK, Sun JT, Yang MX, Neckenig MR, Wang XW, Shao QQ, Song BF, Yang QF, Kong BH, and Qu X 2011. CD16+ monocytes in breast cancer patients: expanded by monocyte chemoattractant protein-1 and may be useful for early diagnosis. Clin Exp Immunol 164: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukherjee R, Kanti Barman P, Kumar Thatoi P, Tripathy R, Kumar Das B, and Ravindran B 2015. Non-Classical monocytes display inflammatory features: Validation in Sepsis and Systemic Lupus Erythematous. Sci Rep 5: 13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chuluundorj D, Harding SA, Abernethy D, and La Flamme AC 2014. Expansion and preferential activation of the CD14(+)CD16(+) monocyte subset during multiple sclerosis. Immunol Cell Biol 92: 509–517. [DOI] [PubMed] [Google Scholar]
- 60.Giri MS, Nebozyhn M, Raymond A, Gekonge B, Hancock A, Creer S, Nicols C, Yousef M, Foulkes AS, Mounzer K, Shull J, Silvestri G, Kostman J, Collman RG, Showe L, and Montaner LJ 2009. Circulating monocytes in HIV-1-infected viremic subjects exhibit an antiapoptosis gene signature and virus- and host-mediated apoptosis resistance. Journal of immunology 182: 4459–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopalco L 2010. CCR5: From Natural Resistance to a New Anti-HIV Strategy. Viruses 2: 574–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hensley-McBain T, and Klatt NR 2018. The Dual Role of Neutrophils in HIV Infection. Curr HIV/AIDS Rep 15: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makinde HM, Cuda CM, Just TB, Perlman HR, and Schwulst SJ 2017. Nonclassical Monocytes Mediate Secondary Injury, Neurocognitive Outcome, and Neutrophil Infiltration after Traumatic Brain Injury. Journal of immunology 199: 3583–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, Lelios I, Heppner FL, Kipnis J, Merkler D, Greter M, and Becher B 2018. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 48: 380–395 e386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.