Abstract
Large-scale manufacturing of therapeutic cells requires bioreactor technologies with online feedback control enabled by monitoring of secreted biomolecular critical quality attributes (CQAs). Electrospray ionization mass spectrometry (ESI-MS) is a highly sensitive label-free method to detect and identify biomolecules, but requires extensive sample preparation before analysis, making online application of ESI-MS challenging. We present a microfabricated, monolithically integrated device capable of continuous sample collection, treatment, and direct infusion for ESI-MS detection of biomolecules in high-salt solutions. The dynamic mass spectrometry probe (DMSP) uses a microfluidic mass exchanger to rapidly condition samples for online MS analysis by removing interfering salts, while concurrently introducing MS signal enhancers to the sample for sensitive biomolecular detection. Exploiting this active conditioning capability increases MS signal intensity and signal-to-noise ratio. As a result, sensitivity for low-concentration biomolecules is significantly improved, and multiple proteins can be detected from chemically complex samples. Thus, the DMSP has significant potential to serve as an enabling portion of a novel analytical tool for discovery and monitoring of CQAs relevant to therapeutic cell manufacturing.
Keywords: biomarker monitoring and discovery, cell manufacturing, microfabricated sampling platform, online electrospray ionization mass spectrometry
1 |. INTRODUCTION
A recent review article made a clear case that the most significant bottleneck to the widespread clinical translation of cell therapies is the development of reproducible, large-scale production of therapeutically relevant cells. To achieve this goal, broadly applicable quality control methodologies and enabling monitoring technologies for online process control are essential (Aijaz et al., 2018). In this study, we present a new approach to dynamic electrospray ionization mass spectrometry (ESI-MS) sensing from complex mixtures, which has the potential to enable online monitoring of the cell secretome within the bioreactor. We demonstrate a microfabricated mass exchanger with an integrated ESI emitter, for solvent modification and ESI-MS detection of multiple proteins from high salt solutions.
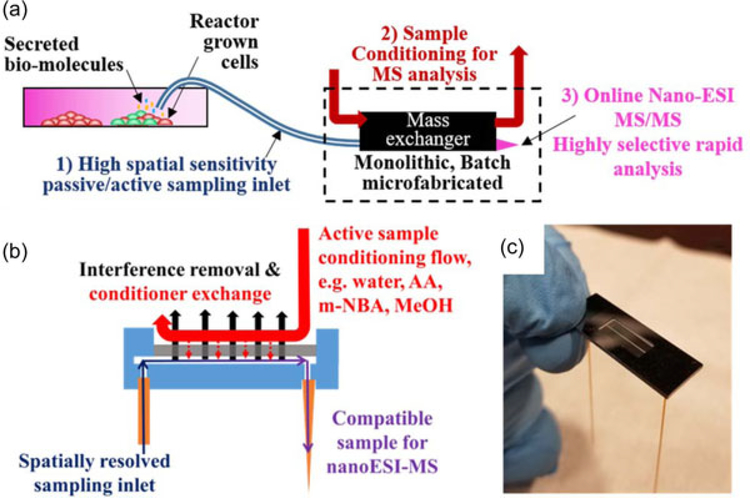
Emerging cell therapies have been shown to successfully treat a range of life-threatening illnesses and injuries. Technologies facilitating therapeutic cell production can also be used to develop new drugs, and can serve as models for in vitro studies, but are currently not widely available (Iyer, Wilems, & Sakiyama-Elbert Shelly, 2016; Kaiser et al., 2015; Kropp, Massai, & Zweigerdt, 2017; Maude, Teachey, Porter, & Grupp, 2015; Oettgen, 2006; Poulos, 2018; Simaria et al., 2013). To enable large-scale and cost-effective adoption of cell therapies, quality control methodologies and standards for therapeutic cell manufacturing need to be established (Aijaz et al., 2018; Albrecht et al., 2018; De Sousa et al., 2016; Du Moulin et al., 1994; Kaiser et al., 2015; Lipsitz, Timmins, & Zandstra, 2016). During the production of therapeutic cells, levels of metabolites (Mucida et al., 2007), cytokines (Agarwal & Rao, 1998; Coronel et al., 2001; Lai, Asthana, & Kisaalita, 2011), and other proteins (Albrecht et al., 2018) or biomolecules can be monitored as critical quality attributes (CQAs) directly related to cell health, efficacy, and differentiation. ESI-MS is an excellent candidate for biochemical analysis due to its broad molecular weight coverage and capacity for unlabeled biomolecule detection and discovery. ESIMS preserves the state of large biomolecules (“soft ionization”) with little fragmentation, requires no a priori labeling of biomolecules, and has limits of detection in the picomolar to even femtomolar range (Fenn, Mann, Meng, Wong, & Whitehouse, 1989; Hoofnagle & Wener, 2009; Yates, Ruse, & Nakorchevsky, 2009). However, direct online ESIMS of cell media is plagued by sample preparation challenges (Vaughn, Crockett, Lin, Lim, & Elenitoba-Johnson, 2006), demanding development of innovative approaches to the rapid in-flow conditioning of extracted samples in small volumes, such that continuous ESI-MS analysis becomes possible. Figure 1a depicts the key features necessary for a continuous bioreactor monitoring system for ESI-MS analysis. These include (a) small volume, localized sampling to capture spatial heterogeneities, and provide nondestructive analysis; (b) rapid sample conditioning to preserve temporal information (microfabrication enables treatment of small volumes with high throughput); and (c) online nano-ESI-MS/MS for identification.
FIGURE 1.
(a) A bioreactor monitoring system should ideally have the ability to continuously sample, either passively (no external pump, capillary filling, flow rates governed by electrospray physics) or actively (forced sampling, flow rates dependent on external pump), from spatially resolved locations within the heterogeneous bioreactor environment for critical quality attributes (CQAs), prepare the sample inline for analysis, and rapidly analyze with high sensitivity, specificity, and quantitation capability, thus providing real-time process control. Nano-ESIMS/MS is a promising analysis approach for untargeted CQA discovery and bioreactor monitoring. (b) Cross-sectional view of the dynamic mass spectrometry probe (DMSP), which enables direct sampling from a bioreactor via a fused silica inlet capillary, sample conditioning and direct online nano-ESI-MS analysis. Conditioning in the mass exchanger includes removal of salt and any other small interfering chemicals, and solvent exchange. Introduction of proton donating chemicals such as acetic acid (AA), protein denaturing solvents such as methanol (MeOH) and supercharging solutes such as 3-nitrobenzyl alcohol (m-NBA) in the mass exchanger can improve sensitivity. (c) Microfabricated monolithic DMSP without conditioning flow package to allow visualization of the sample channel through the semi-transparent nanoporous alumina mass exchange membrane. ESI-MS: electrospray ionization mass spectrometry [Color figure can be viewed at wileyonlinelibrary.com]
Current methods for continuous monitoring of therapeutic cell cultures, such as pH measurement, temperature measurement, off-gas mass spectrometry (i.e., MS monitoring of volatile compounds produced by cells in culture via mass spectrometry), infrared and near-infrared spectroscopy, and Raman spectroscopy noninvasively capture bulk characteristics, but are subpar in offering detailed information such as the secretome’s complete biochemical composition or spatial heterogeneity within the bioreactor (Abu-Absi et al., 2011; Biechele, Busse, Solle, Scheper, & Reardon, 2015; Forcinio, 2003; Zhao, Fu, Zhou, & Hu, 2015). Some progress has been made in the implementation of noninvasive technologies that deliver multidimensional information about the cell population in a bioreactor. For instance, advanced image processing to track and monitor cell count and distribution on microcarriers in a bioreactor (Odeleye, Castillo-Avila, Boon, Martin, & Coopman, 2017) or two-photon microscopy using endogenous fluorophores for monitoring stem cell differentiation (Rice, Kaplan, & Georgakoudi, 2010). As of yet, the level of quantitative detail required for high fidelity detection of CQAs has only seemed obtainable via offline methods such as LC-MS (Albrecht et al., 2018), microarrays (Wang et al., 2009), and enzymatic assays (Kirouac & Zandstra, 2008; Zhao et al., 2015), which have been the main workhorses in characterization of bioreactor processes and biomarker discovery but suffer from significant time delays and low throughput, which limits their utility for online reactor monitoring.
We have developed the dynamic mass spectrometry probe (DMSP; Figure 1b,c), which will be an enabling part of a complete system for online ESI-MS monitoring of cell bioreactors. Application of DMSP to samples that are unamenable to direct infusion ESI-MS analysis results in recovery of typical MS signals for protein samples in chemically simple (salt-free, aqueous) environments. We show that DMSP can use “active” sample treatment in a continuous flow format with a total response time of ~1 min for nano-ESI-MS analysis to enable simultaneous detection of multiple biomolecules in solutions representative of bioreactor environments. These results provide strong evidence that DMSP possesses key attributes to enable online monitoring of dynamic cellular processes, such us uptake and secretion of CQAs. DMSP, when combined with a sampling interface for localized uptake and gas flow assisted ion transfer to the mass spectrometer, will be part of an important contribution to the suite of quality control methods necessary to scale-up cell manufacturing (Aijaz et al., 2018; Garimella et al., 2012).
Continuous flow, passive sampling, spatially resolved biochemical detection have been demonstrated by Olivero, LaPlaca, and Kottke (2012) for a three-dimensional glial cell culture system using a device with inline microdialysis to desalt the sample for direct ESI-MS analysis. The inline salt removal device, which required hand assembly and used a cellulose dialysis membrane with high mass-transfer resistance, enabled detection of moderate concentrations of soluble proteins with a time response of ~1 min. Tibavinsky, Kottke, and Fedorov (2015) reduced online sample treatment time down to ~1 s with miniaturization via microfabrication of a mass exchanger with a monolithic ultrathin nanoporous alumina separation interface for microdialysis. In the DMSP, two important advances are made: First, a microfabricated mass exchanger is integrated with a sampling inlet and electrospray emitter, yielding a monolithic device capable of rapid biomolecule identification to meet the needs of online bioreactor monitoring; and second, a new operating mode with an active introduction of MS signal enhancing chemicals, in addition to sample desalting, is implemented to enable detection of a broader range of biomolecules. The DMSP comprises three elements (Figure 1b): (a) a sampling inlet; (b) a microfabricated mass exchanger for active sample treatment, which simultaneously removes compounds not amenable to MS analysis, such as inorganic salts, and introduces compounds that enhance MS analysis, for example, organic acids (Cech & Enke, 2001) and supercharging molecules (Lomeli, Peng, Yin, Ogorzalek Loo, & Loo, 2010); and (c) a nano-ESI emitter for direct infusion to the MS for analysis.
2 |. MATERIALS AND METHODS
2.1 |. Device description
Figure 1b shows a cross-sectional view of the microfabricated DMSP, as well as an isometric view (Figure 1c) of the fabricated and assembled device. The DMSP mass exchanger is nano/microfabricated and subsequently interfaced with an inlet capillary made from PEEK with a 360 μm outer diameter (OD) and a 150 μm inner diameter (ID) (IDEX Health and Science, Oak Harbor, WA) and an outlet (for ESI-MS) of fused silica with a 360-μm OD, 75-μm ID, and a 30-μm tapered outlet (New Objective, Inc., Woburn, MA). The microfabricated device consists of a 200-μm wide × a 5-μm tall sample channel that runs between the inlet/outlet capillaries. The sides of the sample channel are defined by SU-8 3005, a biocompatible photoresist (Nemani, Moodie, Brennick, Su, & Gimi, 2013). Above the sample channel is a nanoporous alumina membrane which selectively inhibits the diffusion of larger biomolecules of interest but allows free diffusion of low molecular weight MS interfering species, such as inorganic salts, into a high flow rate “active conditioning channel.” The bottom of the sample channel is defined by a 3-μm SiO2 layer deposited on top of the silicon base.
2.2 |. Microfabrication process
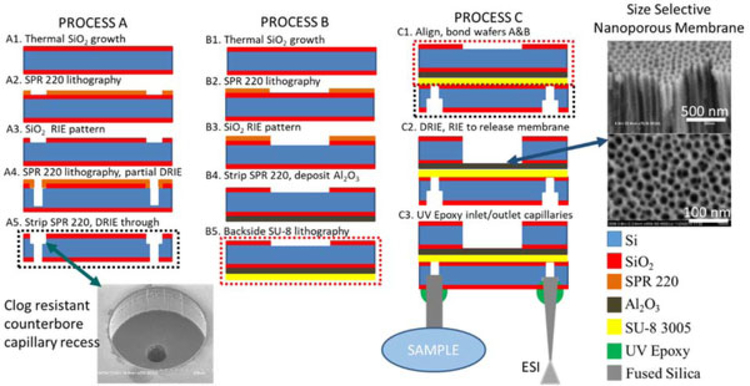
Fabrication of DMSP (Figure 2) is carried out in three process flows, the first two accomplished in parallel (processes A and B) and then a final process flow (process C) with steps carried out on the bonded two wafer stack, followed by incorporation of inlet capillary and outlet ESI emitter. The fabrication integrates both batch microfabrication and nanofabrication processes synergistically to produce a monolithic device. All processing except for the inlet/outlet integration is carried out in the cleanrooms at the Institute for Electronics and Nanotechnology at the Georgia Institute for Technology. Both processes A and B begin with the batch (25 wafers at once) thermal wet oxidation at 1,100°C of double side polished p-type 100 orientation silicon wafers (1–20 Ω-cm; Polishing Corp of America, Santa Clara, CA) resulting in a 3-μm thick silicon dioxide mask layer on both sides of each wafer (Figure 2A1,B1). Process A begins as a layer of Shipley’s SPR 220-7.0 (MicroChem, Westborough, MA) photoresist is spin coated at 1,000 rpm for 5 s, 2,500 rpm for 40 s followed by a 3 min soft bake at 110°C. Note that this SPR 220 recipe is used for all subsequent depositions of the photoresist. Next, 360-μm holes are patterned into the SPR 220 with darkfield photolithography using 405 nm wavelength at a 500 mJ/cm2 dose, followed by development in MF-319 (Figure 2A2). The hole pattern is transferred to through the silicon dioxide layer using a CHF3 reactiveion etching (RIE) step, followed by an acetone strip of the remaining SPR 220 layer (Figure 2A3). A new layer of SPR 220 is once again spun using the same recipe as above. Concentric 60 μm diameter holes in the SPR 220 are patterned using the same exposure as above, followed by a deep reactive-ion etching (DRIE) Bosch process carried out so that the 60-μm holes are etched for approximately 400 μm of depth (Figure 2A4). This Bosch process was characterized on test samples before this application so the etch rate was established on 50–100 μm etch depths. This etch rate was used to extrapolate the necessary amount of Bosch cycles to achieve 400 μm deep holes, since the high aspect ratio of these holes makes them difficult to characterize without using destructive cleaving methods to obtain cross-sectional measurements of the holes. The photoresist is stripped with a piranha bath (3:1, 96% H2SO4:30% H2O2 with no external heat source) and another DRIE process is carried out on the unmasked 360-μm patterned SiO2 layer with concentric partially etched 60-μm inlets until a through etch is achieved, which is verified when visible light is seen through the wafer, resulting in the clog-resistant counterbore capillary recess depicted on the inset image (Figure 2A5). Process B begins with the spin coating of an SPR 220 layer using the same parameters as above, followed by darkfield photolithography for 500 mJ/cm2 at 405 nm defining the active conditioning channel in the photoresist layer (Figure 5B2) followed by a subsequent CHF3 RIE process in the SiO2 masking layer (Figure 2B3). Afterwards, the wafer is cleaned with a piranha bath and a ~5-μm-thick film of aluminum is deposited on the side opposite the patterned SiO2. To deposit the thick aluminum layer, a 50-nm layer of titanium is deposited using an electron beam evaporator, followed by two depositions via electron beam evaporation of 2.5-μm thick layers of aluminum. During deposition, the thickness is monitored with the quartz crystalline microbalance incorporated into the evaporator. In addition, glass slides with a simple tape pattern are positioned near the wafers so that after deposition part of the aluminum film can be removed from the slide and profilometry measurements can be carried out on the resulting stepgeometry, ensuring that the thickness of the aluminum is correct. Next, the entire wafer is anodized using a two-step anodization process in 0.3 M oxalic acid, which creates a nanoporous alumina film with tunable pore diameters in the 10–50 nm range. For the purposes of this film, anodization was carried out at 40 V resulting in ~50 nm pores (Jessensky, Müller, & Gösele, 1998; Figure 2B4). Following the anodization, negative photoresist SU-8 3005 is spun at 500 rpm for 5 s, 3,000 rpm for 40 s, and 4,500 rpm for 2 s followed by a soft bake at 95°C for 2 min and 30 s. Backside photoresist patterning of SU-8 3005 is carried out using clearfield photolithography at 365 nm wavelength for 195 mJ/cm2 dosage, followed by a post exposure bake at 95°C for 2 min and 30 s. Development is carried out in SU-8 developer to define the sample channels (Figure 2B5). Process C begins as the two wafers are bonded at 130°C and 10 bar for 30 min using an Obducat Nanoimprinter (Obducat, Burlingame, CA) (Figure 2C1). A DRIE Bosch process is carried out on the patterned conditioning channels down to the layer of SiO2, followed by another CHF3 RIE process to remove the silicon dioxide layer and titanium adhesion layer on the backside of the porous alumina. Finally, a CF4 RIE process is used to the barrier layer in the nanoporous membrane to allow fluidic transfer between the sample and conditioning channels (Figure 2C2; Liang, Chik, Yin, & Xu, 2002). At this point, the batch fabricated devices are diced and isolated. Finally, using a DYMAX Light Welder PC-3D system (Dymax, Torrington, CT) and UV activated epoxy 9-3095-GEL, inlet and outlet capillaries are positioned and glued into place within each counterbore capillary recess (Figure 2C3).
FIGURE 2.
Dynamic mass spectrometry probe micro/nano-fabrication process diagram. ESI: electrospray ionization [Color figure can be viewed at wileyonlinelibrary.com]
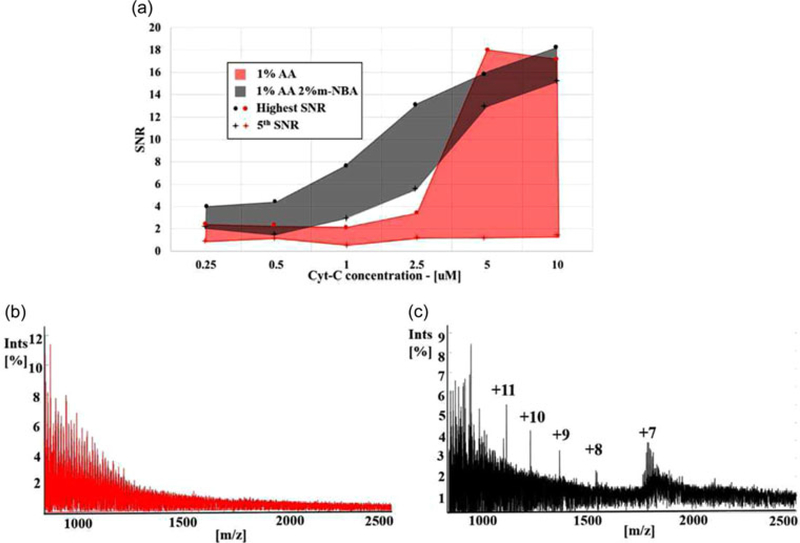
FIGURE 5.
(a) Highest and fifth highest signal-to-noise ratio (SNR) values of identified cytochrome-c (cyt-c) peaks within samples (50 mM KCl with: 0.25, 0.5, 1.0, 2.5, 5, and 10 μM cyt-c) treated with 1% AA versus 1% AA + 2% m-NBA. The m-NBA treatment reveals multiple charge states at all concentrations. (b) DMSP AA treatment ESI-MS of 50 mM KCl with 1 μM cyt-c shows no characteristic cyt-c spectral features—protein is unidentifiable after standard desalination without active sample conditioning. Normalization value 4,500 counts. (c) DMSP 1% AA + 2% m-NBA treatment-ESI-MS of 50 mM KCl with 1 μM cyt-c reveals multiple charge states for identification of the protein, which shows that the active sample conditioning can improve both SNR and sensitivity to low-concentration biomolecules. Normalization value 12,134 counts. AA: acetic acid; m-NBA: 3-nitrobenzyl alcohol [Color figure can be viewed at wileyonlinelibrary.com]
2.3 |. Fluidic connection package
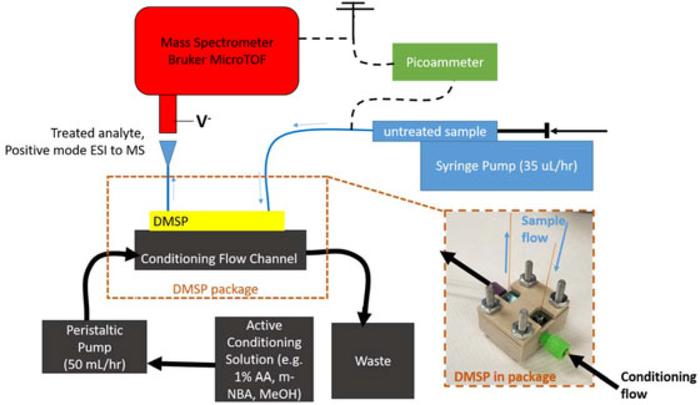
The device is packaged in a mechanically machined polycarbonate or PEEK package, which is designed to introduce a uniform flow of conditioning liquid across the top of the DMSP sample channel. The active conditioning flow direction is opposite that of the sample channel to enhance mass transfer efficiency. The conditioning flow is sealed around the device’s channel with a 1-mm-thick laser-cut self-adhesive gasket, which is fit into a machined groove in the package. Conditioning flow is continuously pumped through the package at 50 ml/hr using a peristaltic pump. An inset view of DMSP in the fluidic package is shown in Figure 3.
FIGURE 3.
Dynamic mass spectrometry probe (DMSP) experimental setup. Untreated sample is introduced into DMSP via syringe pump at 35 μl/hr. Conditioning solution is introduced to remove interfering material/introduce electrospray ionization mass spectrometry (ESI-MS) enhancements through a peristaltic pump at 50 ml/hr, which is sealed to DMSP through the package. Direct infusion ESI to the Bruker MicroTOF mass spectrometer is achieved by grounding the syringe, and applying a negative voltage to the mass spectrometer inlet. Consistent ESI characteristics are maintained during and between experiments by measuring the spray current with a picoammeter held at ground. AA: acetic acid; m-NBA: 3-nitrobenzyl alcohol; MeOH: methanol [Color figure can be viewed at wileyonlinelibrary.com]
2.4 |. Experimental setup
The DMSP’s experimental configuration is shown in Figure 3. A sample of known composition is drawn into a 50 μl syringe (Hamilton, Reno, NV) and infused with a syringe pump at 35 μl/hr through a length of fused silica capillary with a 75-μm ID and 360-μm OD, which is attached to the inlet capillary of the DMSP with Upchurch fittings (Oak Harbor, WA) in a Valco union (Houston, TX). On the tip of the syringe, an electrical connection is made to the probing end of a picoammeter. The ground end of the picoammeter is then tied to the ground of the mass spectrometer such that current in the ESI spray circuit can be measured at all times. The DMSP, in the fluidic package (Figure 3, inset), is then positioned 3–5-mm away from the Bruker micrOTOF mass spectrometer inlet (Billerica, MA), which is held at a negative bias between 1,000 and 1,500 V depending on the distance between the ESI emitter and the mass spectrometer inlet. During all experiments, spray current is kept between 10 and 20 nA at a steady value to ensure similar ESI conditions at the MS inlet.
Conditioning flow is delivered at 50 ml/hr with a peristaltic pump, which draws from a liquid reservoir through silicone tubing. Conditioning liquid then travels through PEEK 1/16″ OD tubing to the device package, where the connection is made with 1/4–28 Upchurch fluidic fittings into the DMSP package. Conditioning solution travels in the opposite direction of sample flow along the top side of the etched conditioning channel, separated from the sample channel within DMSP by the size-selective nanoporous membrane. The fluidic package is shown on the inset photograph in Figure 3. In this representation the DMSP is encased in a PEEK package, which is used to introduce chemicals that are incompatible with the transparent polycarbonate package (e.g., acetonitrile), but the polycarbonate package was used for the experimental results presented here. It is found that when changing between conditioning solutions, bubbles often are introduced. However, a polycarbonate package allows the visualization of bubbles or liquid dry out in the active conditioning channel during operation, and eliminating these bubbles ensures DMSP is operating properly during experiments.
3 |. RESULTS AND DISCUSSION
3.1 |. Salt removal and active sample conditioning
Inorganic salts interfere with positive mode electrospray mass spectrometry (ESI-MS) by suppressing the MS signal associated with analyte molecules. Removal of salts promotes charging of molecules with only protons, that is protonation, forming “nonadducted” ions where none of the charges is due to metal cation adduction. In addition to cation adduction, neutrals (e.g., KCl and NaCl with associated solvent molecules) can attach to the ionized analytes forming salt “clusters”. The signal at any nonadducted charge state associated with a given biomolecule becomes masked by the signal of the same or other molecules whose ions carry adducted salts and salt clusters, making it impossible to identify the biomolecule via MS (Jakubowski, Hatcher, & Sweedler, 2005; Keller, Sui, Young, & Whittal, 2008; Liu, Wu, Harms, & Smith, 1996; Olivero et al., 2012; Tibavinsky et al., 2015; Xiang, Lin, Wen, Matson, & Smith, 1999).
DMSP enhancement of ESI-MS analysis was explored through comparison of the effect of four different active conditioning solutions. Each conditioning solution contained 1% acetic acid (AA) to facilitate the protonation of compounds (Cech & Enke, 2001), and three of the conditioning solutions contained additional ESI-MS enhancing chemicals. Ammonium acetate has been reported to increase the acidity in the electrospray plume, enhancing protonation (Konerman, 2017), and to reduce the formation of salt adducts (Abbassi-Ghadi et al., 2016; Rush & Breemen Richard, 2016). A shift (increase) in the charge state distribution is due to enhanced protonation, which was expected to increase sensitivity in ESI-MS analysis. Supercharging molecule 3-nitrobenzyl alcohol (m-NBA) in concentrations from 1% to 20% has been shown to increase charge state distribution in nondenaturing solutions (Hogan, Ogorzalek Loo, Loo, & Mora, 2010). Although the mechanism is not completely settled, it is suggested that supercharging occurs due to the low volatility and low surface tension of the supercharging agent, a combination that promotes the Coulombic droplet fission process (Cassou & Williams, 2014; Lomeli et al., 2010). Finally, methanol (MeOH) can shift charge state distribution towards a lower m/z spectral range due to a denaturing of proteins, revealing more locations for protonation (Iavarone & Williams, 2003).
A solution of 100 mM KCl with 5 μM cytochrome-c (cyt-c) was used to carefully characterize the effect of active sample treatment. To maintain relevance to cell manufacturing, the salt level used is similar to that expected in a bioreactor (Stubblefield & Mueller, 1960), and cyt-c is an appropriate model protein in the mass range (~12 kDa) of signaling molecules secreted from cells, such as cytokines, that are expected to be CQAs for cell health (Agarwal & Rao, 1998; Coronel et al., 2001; Lai et al., 2011; Vaughn et al., 2006). When untreated, cyt-c signal is completely suppressed in the spectra obtained for ESI-MS of 100 mM KCl with 5 μM cyt-c (Figure 4a), indicating that the added KCl levels are high enough to render MS analysis incapable of detecting the protein—this is indicative of how the environment of a cell bioreactor will mask biomolecule signals in direct ESI-MS analysis. Salt concentrations need to be reduced to below about 1 mM for identification of cyt-c at a concentration of 5 μM, therefore at least 99% of the KCl in the solutions tested here was removed by DMSP (see supporting information). In cell monitoring applications, analyte concentrations could be lower than 5 μM which is why a highly selective mass exchanger, such as DMSP, is necessary to remove salt to a sufficient degree while retaining biomolecules of interest through size-selective separation.
FIGURE 4.
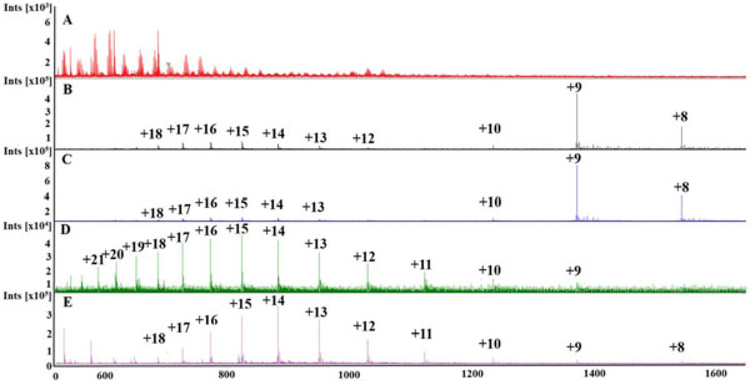
Resulting mass spectra from direct infusion electrospray ionization mass spectrometry (ESI-MS) through the dynamic mass spectrometry probe (DMSP) of a solution with 100 mM KCl and 5 μM cytochrome-c. A, Untreated, that is, empty conditioning channel—no characteristic cytochrome-c peaks are visible due to salt interference; B, 1% acetic acid (AA) reveals a strong cytochrome-c signal; C, Addition of ammonium acetate improves intensity of signal; D, Addition of 2% 3-nitrobenzyl alcohol (m-NBA) shifts to a higher charge state distribution; E, Addition of 50% methanol also increases the average charge state, but long-term operation is hindered due to increased inorganic salt precipitation, which can result in clogging of the device [Color figure can be viewed at wileyonlinelibrary.com]
The benefit of DMSP online active sample treatment can be seen by comparing the spectra of an untreated sample (Figure 4a), a sample with treatment (Figure 4b) using a 1% AA conditioning solution. Combined removal of salt (fewer salt adducts and clusters) and the introduction of AA for improved protonation results in a mass spectra characteristic of cyt-c (Figure 4b). The most intense peak (m/z ~1,374) is due to cyt-c molecules charged via addition of nine protons (called the +9 charge state). The appearance of cyt-c spectral peaks in the online analysis of a high salt solution occurs as a result of several mechanisms. First, the removal of salt reduces metal cation adduction of the protein, so that the protein is charged via protonation. Salt removal also eliminates the formation of cluster peaks, reducing charge competition and corresponding signal suppression. Finally, the addition of AA into the sample further increases the rate of protonation due to the increased proton concentration in acidified solution.
Adding 40 mM ammonium acetate to the conditioning solution increases the intensity of the largest nonadducted cyt-c associated peak (Figure 4b; 1% AA treatment) two-fold with no shift in charge state distribution (Figure 4c). While the addition of ammonium acetate further enhances protonation (Konerman, 2017) and mitigates salt adducts (Rush & Breemen Richard, 2016), ammonium acetate does not change the surface tension and has little to no denaturing effect on the protein so there is no apparent impact on the charge state.
In contrast, adding supercharging molecule m-NBA at 2% to the conditioning flow results in a significantly shifted charge state distribution (Figure 4d), so that the nonadducted peak with 15 protons attached (m/z ~824) to the biomolecule is most dominant. Although not producing the same maximum peak intensity as ammonium acetate, m-NBA causes a marked increase in the prevalence of peaks corresponding to the highly charged protein. This suggests the possibility of using m-NBA treatment of higher molecular weight proteins that would usually be outside the range of a given mass spectrometer. The Bruker micrOTOF time-of-flight mass spectrometer used in these experiments has a dynamic (m/z) range of 30–5,000. Thus increasing the maximum charge placed on proteins from 10 to 20, for example, would increase the size of the largest detectable protein from 50 to 100 kDa. The impact of supercharging is also seen in its potential for improved sensitivity. This is because tuning the ion-transfer voltages in a mass spectrometer to acquire signals over the entire available m/z range sacrifices sensitivity compared to that obtainable when tuned for a smaller m/z range. As seen in Figure 4d, super charging compresses the range of m/z values for cyt-c peaks enabling better use for the instrument capabilities for biomolecular detection.
Addition of 50% MeOH also has a significant effect on the charge state distribution (Figure 4e). This treatment results in a spectra in which the +14 protonation charge state (m/z ~883) is most abundant, which suggests that the effects of m-NBA and MeOH are not identical. Methanol denatures the protein, revealing more sites for protons to attach, resulting in a shift to a higher charge state distribution. Although MeOH, like m-NBA, has a lower surface tension than water, it is more volatile than water. Therefore it does not yield the same increase in droplet fission events m-NBA produces (Iavarone & Williams, 2003; Lomeli et al., 2010). Further, due to a lower solubility of inorganic salts in MeOH, continuous flow analysis is difficult using MeOH as an active conditioner (Pinho & Macedo, 2005). As MeOH is exchanged into the sample, salt precipitates form. These precipitates eventually clog the sample channel or the ESI emitter, causing the DMSP to cease operation.
3.2 |. Signal-to-noise ratio (SNR) and sensitivity enhancement
To quantify the impact of DMSP active sample treatment, SNR and sensitivity (i.e., ability to detect lower concentration biomolecules) were studied. Samples containing varying cyt-c concentrations (0.25, 0.5, 1.0, 2.5, 5, and 10 μM) in 50 mM KCl were treated with two conditioning solutions, 1% AA versus 1% AA + 2% m-NBA, to explore what effect active conditioning has on MS SNR and sensitivity. These experiments were designed to investigate how supercharging molecules specifically can enhance detection of proteins at low concentrations.
Figure 5a illustrates that the addition of m-NBA through active conditioning increases SNR across multiple charge states by plotting the highest and fifth highest SNR as a function of cyt-c concentration. Here, SNR is defined as SNR = Icyt-c/Iavg, where Icyt-c is the intensity (%) of an identified cyt-c peak and Iavg the averaged intensity (%) of all peaks within a window ± 0.3 m/z of the identified peak. This definition gives a local SNR value for every nonadducted cyt-c peak. An SNR above ~2.5 corresponds to a distinguishable, nonadducted cyt-c peak. These results demonstrate that m-NBA treatment improves the sensitivity to low-concentration biomolecules and increases the SNR for detected molecules across multiple charge states. This allows for easier identification of low-concentration biomolecules in complex mixtures.
Active treatment with m-NBA drastically improves the lowest concentration for which a peak associated with nonadducted cyt-c is detectable in 50 mM KCl by an order of magnitude, from 2.5 μM to 250 nM. Figures 5b,c show a comparison of spectra resulting from DMSP nano-ESI-MS analysis of 1 μM cyt-c in 50 mM KCl with 1% AA conditioning solution (Figure 5b) versus 1% AA + 2% m-NBA (Figure 5c) to highlight the significant effect addition of m-NBA has on revealing nonadducted cyt-c signal in a low-concentration (1 μM) mixture. With 1% AA treatment (Figure 5b), no distinguishable nonadducted cyt-c peaks are visible, while treatment with 1% AA + 2% m-NBA produces five distinct peaks from nonadducted species (Figure 5c). The concentration of some CQAs in bioreactors may be even lower than those explored here. Therefore, the ability to detect low-concentration biomolecules enabled by DMSP treatment is critical for MS application to cell manufacturing control, since a large range in signaling molecule concentrations is expected.
At 2.5 μM cyt-c, where both conditioning solution compositions successfully reveal cyt-c, m-NBA enhances SNR across multiple charge states far above the levels obtained without m-NBA. With very high SNR peaks across a range of multiply charged states, identification of biomolecules is easier, and applications which can benefit from biomolecule structural information are enabled through the use of tandem (MS/MS) mass spectrometry that performs best with highly charged species (Teo & Donald, 2014). Although for both 5.0 and 10.0 μM cyt-c concentrations, 1% AA treatment produces a very high SNR for the highest intensity peak, m-NBA treatment again creates higher average SNR values. These levels of cyt-c concentration (5 and 10 μM) are approaching the point where signal saturation was observed, such that MS measurements of abundance is not dependent on treatment type, muting the impact of active sample conditioning. These results show that DMSP active sample conditioning further enhances the detection of low-concentration biomolecules (i.e., <2.5 μM cyt-c), extending the potential utility of DMSP as part of an online bioreactor monitoring system.
3.3 |. Simultaneous identification of multiple proteins with DMSP
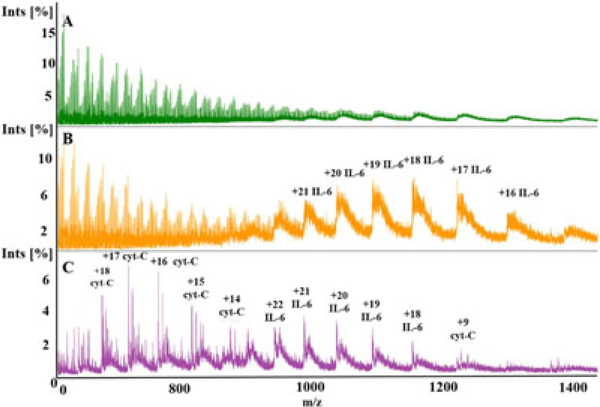
To assess the DMSP’s capability of multiple protein detection, we performed experiments with a cell buffer mixture, phosphate-buffered saline (1×PBS: 137 mM NaCl, 2.7 mM KCl, 1.0 mM Na2HPO4, and 1.8 mM KH2PO4), containing 5 μM of interleukin 6 (IL-6), interleukin 8 (IL-8), and cyt-c using two different active sample conditioning strategies previously described. With molecular weights of ~8 kDa (IL-8), ~12 kDa (cyt-C), and ~21 kDa (IL-6), the mixture of these molecules is a realistic proxy for cytokines, which are known to be indicative of cell health (Coronel et al., 2001; Lai et al., 2011), in a solution that has multiple inorganic compounds in concentrations representative of a bioreactor environment (Stubblefield & Mueller, 1960). As expected, ESI-MS of 5 μM IL-6, IL-8, and cyt-c in 1×PBS without DMSP treatment produces spectra in which all three biomolecules are undetectable (Figure 6a). When 1% AA treatment was used, only IL-6 was detected (Figure 6b), whereas treatment with 1% AA + 2% m-NBA yielded detection of both IL-6 and cyt-c (Figure 6C).
FIGURE 6.
Mass spectra produced via direct infusion electrospray ionization mass spectrometry through dynamic mass spectrometry probe (DMSP). Each spectra normalized by highest intensity peak. A, Untreated 1×PBS with 5 μM cytochrome-c (12 kDa), 5 μM IL-6 (21 kDa), and 5 μM IL-8 (8.4 kDa) shows no identifiable peaks associated with protonation of biomolecules. Normalization value 2,715 counts. B, 1% AA treatment reveals multiple charge states associated with IL-6 only. Normalization value 4,823 counts. C, 1% AA + 2% m-NBA treatment reveals nonadducted charge states of cytochrome-c and IL-6. In all cases, IL-8 is not visible due to unintended removal through the membrane being the smallest (lowest molecular weight) biomolecule in the protein mixture. Normalization value 34,180 counts. AA: acetic acid; m-NBA: 3-nitrobenzyl alcohol; IL: interleukin; phosphate-buffered saline [Color figure can be viewed at wileyonlinelibrary.com]
When 1% AA is used as the conditioning solution, removal of inorganic compounds and acidification of the sample in the DMSP reduces chemical noise associated with salt adducts and clusters, and IL-6 can be detected (Figure 6b). The highest concentration of salt in PBS is NaCl at 137 mM, which is drastically higher than the salt levels explored in the experiments described in earlier sections (100 mM, Figure 4; 50 mM, Figure 5). Still, based on prior experiments (Supporting Information) it can be estimated that the DMSP is removing over 99% of the salts present to reveal the signal associated with IL-6. However, the SNR is low and peaks formed via salt adducts are visible at higher m/z values along with each nonadduct peak, indicating not fully sufficient salt removal and pointing to the opportunities for additional DMSP optimization.
Treatment with 1% AA + 2% m-NBA reveals high SNR, nonadducted cyt-c and IL-6 peaks, but IL-8 is not observed (Figure 6c). As the smallest of the analytes, IL-8 may have suffered more parasitic loss in the DMSP mass exchanger than larger in size cyt-c and IL-6 (Tibavinsky et al., 2015). This result further emphasizes that active sample treatment with m-NBA is very useful for reducing chemical noise due to metal cation adduction and salt cluster formation. These promising results demonstrate that DMSP with active sample treatment enables the detection of multiple biomolecules online in a high salt content mixture, essential for cellhealth monitoring.
4 |. CONCLUSION
New quality control methods combined with in operando sensing inputs are critically needed to enable the industrialization of therapeutic cell manufacturing, leading to more widely available treatments for previously untreatable diseases, development of new drugs, and further innovations in cell therapies (Aijaz et al., 2018; De Sousa et al., 2016; Lipsitz et al., 2016; Simaria et al., 2013; Zhao et al., 2015). The DMSP is a monolithically integrated, microfabricated sample processing platform that enables the rapid (~1 min) ESI-MS detection of low-concentration biomolecules in chemically complex solutions, and is therefore an enabling part of a tool for online bioreactor monitoring. Modifications to DMSP’s geometry and flow configuration to enhance mass transfer are immediate points for improvement, as suggested by mass transfer modeling, which should help further increase salt removal capability, and allow more effective introduction of conditioning agents into the sample flow, resulting in an overall increase in the rapidity of the analysis. Further optimization will involve exploration of additional chemistries (e.g., acetonitrile) and a broader range of concentrations introduced via the active sample conditioning in DMSP, which should allow the detection of wider range biomolecules, and further enhance sensitivity to low-concentration biomolecules. Combination of DMSP with a sampling interface and gas assisted ion transfer will be the final steps necessary for demonstration of a DMSP based system for online bioreactor monitoring.
Supplementary Material
ACKNOWLEDGEMENTS
This material is based upon work supported by the National Science Foundation under Grant No. EEC-1648035. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Research was also supported by funds from the Marcus Foundation, The Georgia Research Alliance, and the Georgia Tech Foundation through their support of the Marcus Center for Therapeutic Cell Characterization and Manufacturing (MC3M) at Georgia Tech. Partial support (PAK and AGF) was also provided by Grant Number RO1GM112662 from the National Institute of General Medical Science (NIGMS), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. MC was in part supported by the Georgia Tech’s Renewable Bioproducts Institute Fellowship. Device micro-fabrication was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure, which is supported by the National Science Foundation (Grant ECCS1542174).
Funding information
National Science Foundation, Grant/Award Numbers: 1648035, ECCS-1542174; National Institute of General Medical Sciences, Grant/Award Number: RO1GM112662
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Abbassi-Ghadi N, Jones EA, Gomez-Romero M, Golf O, Kumar S, Huang J, … Takats Z (2016). A comparison of DESI-MS and LC-MS for the lipidomic profiling of human cancer tissue. Journal of the American Society for Mass Spectrometry, 27(2), 255–264. 10.1007/s13361-015-1278-8 [DOI] [PubMed] [Google Scholar]
- Abu-Absi NR, Kenty BM, Cuellar ME, Borys MC, Sakhamuri S, Strachan DJ, … Li ZJ (2011). Real time monitoring of multiple parameters in mammalian cell culture bioreactors using an in-line Raman spectroscopy probe. Biotechnology and Bioengineering, 108(5), 1215–1221. [DOI] [PubMed] [Google Scholar]
- Agarwal S, & Rao A (1998). Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity, 9(6), 765–775. 10.1016/S1074-7613(00)80642-1 [DOI] [PubMed] [Google Scholar]
- Aijaz A, Li M, Smith D, Khong D, LeBlon C, Fenton OS, … Parekkadan B (2018). Biomanufacturing for clinically advanced cell therapies. Nature Biomedical Engineering, 2(6), 362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht S, Kaisermayer C, Gallagher C, Farrell A, Lindeberg A, & Bones J (2018). Proteomics in biomanufacturing control: Protein dynamics of CHO-K1 cells and conditioned media during apoptosis and necrosis. Biotechnology and Bioengineering, 115(6), 1509–1520. 10.1002/bit.26563 [DOI] [PubMed] [Google Scholar]
- Biechele P, Busse C, Solle D, Scheper T, & Reardon K (2015). Sensor systems for bioprocess monitoring. Engineering in Life Sciences, 15(5), 469–488. [Google Scholar]
- Cassou CA, & Williams ER (2014). Desalting protein ions in native mass spectrometry using supercharging reagents. Analyst, 139(19), 4810–4819. 10.1039/C4AN01085J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech NB, & Enke CG (2001). Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrometry Reviews, 20(6), 362–387. 10.1002/mas.10008 [DOI] [PubMed] [Google Scholar]
- Coronel A, Boyer A, Franssen JD, Romet-Lemonne JL, Fridman WH, & Teillaud JL (2001). Cytokine production and T-cell activation by macrophage–dendritic cells generated for therapeutic use. British Journal of Haematology, 114(3), 671–680. 10.1046/j.1365-2141.2001.02982.x [DOI] [PubMed] [Google Scholar]
- Fenn J, Mann M, Meng C, Wong S, & Whitehouse C (1989). Electrospray ionization for mass-spectrometry of large biomolecules. Science, 246(4926), 64–71. 10.1126/science.2675315 [DOI] [PubMed] [Google Scholar]
- Forcinio H (2003). Pharmaceutical industry embraces NIR technology. Spectroscopy, 18(9), 16–19. [Google Scholar]
- Garimella S, Xu W, Huang G, Harper JD, Cooks RG, & Ouyang Z (2012). Gas-flow assisted ion transfer for mass spectrometry. Journal of Mass Spectrometry, 47(2), 201–207. 10.1002/jms.2955 [DOI] [PubMed] [Google Scholar]
- Hogan CJ Jr, Ogorzalek Loo RR, Loo JA, & Mora JF (2010). Ion mobility–mass spectrometry of phosphorylase B ions generated with supercharging reagents but in charge-reducing buffer. Physical Chemistry Chemical Physics, 12(41), 13476–13483. [DOI] [PubMed] [Google Scholar]
- Hoofnagle AN, & Wener MH (2009). The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. Journal of Immunological Methods, 347(1-2), 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone AT, & Williams ER (2003). Mechanism of charging and supercharging molecules in electrospray ionization. Journal of the American Chemical Society, 125(8), 2319–2327. 10.1021/ja021202t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NR, Wilems TS, & Sakiyama-Elbert SE (2016). Stem cells for spinal cord injury: Strategies to inform differentiation and transplantation. Biotechnology and Bioengineering, 114(2), 245–259. 10.1002/bit.26074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski JA, Hatcher NG, & Sweedler JV (2005). Online microdialysis-dynamic nanoelectrospray ionization-mass spectrometry for monitoring neuropeptide secretion. Journal of Mass Spectrometry, 40(7), 924–931. 10.1002/jms.869 [DOI] [PubMed] [Google Scholar]
- Jessensky O, Müller F, & Gösele U (1998). Self-organized formation of hexagonal pore arrays in anodic alumina. Applied Physics Letters, 72(10), 1173–1175. [Google Scholar]
- Kaiser AD, Assenmacher M, Schröder B, Meyer M, Orentas R, Bethke U, & Dropulic B (2015). Towards a commercial process for the manufacture of genetically modified T cells for therapy. Cancer Gene Therapy, 22, 72–78. 10.1038/cgt.2014.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller BO, Sui J, Young AB, & Whittal RM (2008). Interferences and contaminants encountered in modern mass spectrometry. Analytica Chimica Acta, 627(1), 71–81. 10.1016/j.aca.2008.04.043 [DOI] [PubMed] [Google Scholar]
- Kirouac DC, & Zandstra PW (2008). The systematic production of cells for cell therapies. Cell Stem Cell, 3(4), 369–381. 10.1016/j.stem.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Konermann L (2017). Addressing a common misconception: Ammonium acetate as neutral pH “buffer” for native electrospray mass spectrometry. Journal of the American Society for Mass Spectrometry, 28(9), 1827–1835. [DOI] [PubMed] [Google Scholar]
- Kropp C, Massai D, & Zweigerdt R (2017). Progress and challenges in large-scale expansion of human pluripotent stem cells. Process Biochemistry, 59, 244–254. 10.1016/j.procbio.2016.09.032 [DOI] [Google Scholar]
- Lai Y, Asthana A, & Kisaalita WS (2011). Biomarkers for simplifying HTS 3D cell culture platforms for drug discovery: The case for cytokines. Drug Discovery Today, 16(7), 293–297. 10.1016/j.drudis.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Liang J, Chik H, Yin A, & Xu J (2002). Two-dimensional lateral superlattices of nanostructures: Nonlithographic formation by anodic membrane template. Journal of Applied Physics, 91(4), 2544–2546. [Google Scholar]
- Lipsitz YY, Timmins NE, & Zandstra PW (2016). Quality cell therapy manufacturing by design. Nature Biotechnology, 34, 393–400. 10.1038/nbt.3525 [DOI] [PubMed] [Google Scholar]
- Liu C, Wu Q, Harms AC, & Smith RD (1996). On-line microdialysis sample cleanup for electrospray ionization mass spectrometry of nucleic acid samples. Analytical Chemistry, 68(18), 3295–3299. 10.1021/ac960286j [DOI] [PubMed] [Google Scholar]
- Lomeli SH, Peng IX, Yin S, Ogorzalek Loo RR, & Loo JA (2010). New reagents for increasing ESI multiple charging of proteins and protein complexes. Journal of the American Society for Mass Spectrometry, 21(1), 127–131. 10.1016/j.jasms.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Teachey DT, Porter DL, & Grupp SA (2015). CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood, 125(26), 4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Moulin GC, Stack J, Pitkin Z, Chew-Darke J, Cyr C, White A, … Liu V (1994). A 3-year experience of quality control and quality assurance in the multisite delivery of a lymphocyte-based cellular therapy for renal cell carcinoma. Biotechnology and Bioengineering, 43(8), 693–699. 10.1002/bit.260430804 [DOI] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, & Cheroutre H (2007). Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science, 317(5835), 256–260. [DOI] [PubMed] [Google Scholar]
- Nemani KV, Moodie KL, Brennick JB, Su A, & Gimi B (2013). In vitro and in vivo evaluation of SU-8 biocompatibility. Materials Science and Engineering: C, 33(7), 4453–4459. 10.1016/j.msec.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odeleye AOO, Castillo-Avila S, Boon M, Martin H, & Coopman K (2017). Development of an optical system for the non-invasive tracking of stem cell growth on microcarriers. Biotechnology and Bioengineering, 114(9), 2032–2042. 10.1002/bit.26328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettgen P, Boyle AJ, Schulman SP, & Hare JM (2006). Cardiac stem cell therapy. Circulation, 114(4), 353–358. [DOI] [PubMed] [Google Scholar]
- Olivero D, LaPlaca M, & Kottke PA (2012). Ambient nanoelectrospray ionization with in-line microdialysis for spatially resolved transient biochemical monitoring within cell culture environments. Analytical Chemistry, 84(4), 2072–2075. 10.1021/ac203009s [DOI] [PubMed] [Google Scholar]
- Pinho SP, & Macedo EA (2005). Solubility of NaCl, NaBr, and KCl in water, methanol, ethanol, and their mixed solvents. Journal of Chemical & Engineering Data, 50(1), 29–32. 10.1021/je049922y [DOI] [Google Scholar]
- Poulos J (2018). The limited application of stem cells in medicine: A review. Stem Cell Research & Therapy, 9(1), 1 10.1186/s13287-017-0735-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WL, Kaplan DL, & Georgakoudi I (2010). Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLOS One, 5(4), e10075 10.1371/journal.pone.0010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush MD, & Van breemen RB (2016). Role of ammonium in the ionization of phosphatidylcholines during electrospray mass spectrometry. Rapid Communications in Mass Spectrometry, 31(3), 264–268. 10.1002/rcm.7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simaria AS, Hassan S, Varadaraju H, Rowley J, Warren K, Vanek P, & Farid SS (2013). Allogeneic cell therapy bioprocess economics and optimization: Single-use cell expansion technologies. Biotechnology and Bioengineering, 111(1), 69–83. 10.1002/bit.25008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa PA, Downie JM, Tye BJ, Bruce K, Dand P, Dhanjal S, … Bateman M (2016). Development and production of good manufacturing practice grade human embryonic stem cell lines as source material for clinical application. Stem Cell Research, 17(2), 379–390. 10.1016/j.scr.2016.08.011 [DOI] [PubMed] [Google Scholar]
- Stubblefield E, & Mueller GC (1960). Effects of sodium chloride concentration on growth, biochemical composition, and metabolism of HeLa cells. Cancer Research, 20(11), 1646–1655. [Google Scholar]
- Teo CA, & Donald WA (2014). Solution additives for supercharging proteins beyond the theoretical maximum proton-transfer limit in electrospray ionization mass spectrometry. Analytical Chemistry, 86(9), 4455–4462. 10.1021/ac500304r [DOI] [PubMed] [Google Scholar]
- Tibavinsky IA, Kottke PA, & Fedorov AG (2015). Microfabricated ultrarapid desalting device for nanoelectrospray ionization mass spectrometry. Analytical Chemistry, 87(1), 351–356. 10.1021/ac5040083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn CP, Crockett DK, Lin Z, Lim MS, & Elenitoba-Johnson KSJ (2006). Identification of proteins released by follicular lymphoma-derived cells using a mass spectrometry-based approach. Proteomics, 6 (10), 3223–3230. 10.1002/pmic.200500539 [DOI] [PubMed] [Google Scholar]
- Wang M, Senger RS, Paredes C, Banik GG, Lin A, & Papoutsakis ET (2009). Microarray-based gene expression analysis as a process characterization tool to establish comparability of complex biological products: Scale-up of a whole-cell immunotherapy product. Biotechnology and Bioengineering, 104(4), 796–808. 10.1002/bit.22441 [DOI] [PubMed] [Google Scholar]
- Xiang F, Lin Y, Wen J, Matson DW, & Smith RD (1999). An integrated microfabricated device for dual microdialysis and on-line ESI-Ion trap mass spectrometry for analysis of complex biological samples. Analytical Chemistry, 71(8), 1485–1490. 10.1021/ac981400w [DOI] [PubMed] [Google Scholar]
- Yates JR, Ruse CI, & Nakorchevsky A (2009). Proteomics by mass spectrometry: Approaches, advances, and applications. Annual Review of Biomedical Engineering, 11, 49–79. [DOI] [PubMed] [Google Scholar]
- Zhao L, Fu HY, Zhou W, & Hu WS (2015). Advances in process monitoring tools for cell culture bioprocesses. Engineering in Life Sciences, 15(5), 459–468. 10.1002/elsc.201500006 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.