Abstract
Background:
The Plasminogen Activator Inhibitor-1 gene 4G/5G (PAI-1 4G/5G) polymorphism has been suggested to be associated with osteonecrosis of the femoral head (ONFH) susceptibility; however, the results are conflicting and inconclusive. We have carried out a comprehensive meta-analysis to derive a more precise estimation of the association.
Methods:
A comprehensive search in PubMed, EMBASE, Google Scholar, and ISI Web of Knowledge databases was conducted to identify all eligible case-control publications investigating the association between PAI-1 4G/5G polymorphism and ONFH risk. Odds ratios (OR) and corresponding 95% confidence intervals (CI) were used to assess the association.
Results:
A total of six studies with 456 cases and 1,019 controls were included in this review. Three studies were from Caucasian descendants and the three others were from East Asian descendants. Overall analysis suggests a significant association between PAI-1 4G/5G polymorphism and ONFH risk under the allele model (4G vs. 5G: OR =1.540, 95% CI =1.055-2.248, P=0.025) and the recessive model (4G4G vs. 4G5G+5G5G: OR=1.931, 95% CI: 1.162-3.207, P=0.011). When stratified by ethnicity, we have found a significant association between PAI-1 4G/5G polymorphism and ONFH risk among the Caucasian (4G5G vs. 5G5G: OR=1.806, 95% CI: 1.064-3.067, P=0.029) and East Asians (4G4G vs. 5G5G: OR=1.619, 95% CI: 1.025-2.556, P=0.039 and 4G4G vs. 4G5G+5G5G: OR=1.665, 95% CI: 1.207-2.297, P=0.002).
Conclusion:
The present meta-analysis suggested that PAI-1 4G/5G (rs1799889) polymorphism is a potential risk factor for development of ONFH. However, large-scale and well-designed case-control studies in different ethnicities are required to validate these results.
Key Words: Meta-analysis, Osteonecrosis of femoral head, Plasminogen activator inhibitor 1, Polymorphism
Introduction
Osteonecrosis (ON) or avascular necrosis (AVN) is a worldwide challenging clinical problem (1). According to the reports, ON is an increasingly common cause of musculoskeletal disability which mostly affects middle-aged active people (2). The femoral head is the most vulnerable site for the development of osteonecrosis (3). ONFH is terminally manifested by death of bone cells that results in impairment of normal reparative processes along micro-fractures in the femoral head (4). It is estimated that 10,000 to 20,000 new cases of ONFH are diagnosed in the United States every year (2, 3, 5). ONFH has traditionally been classified as idiopathic or secondary, depending on the absence or presence of known causes (5). Clinically, ONFH is closely associated with several factors such as chronic alcoholism, smoking, sickle cell disease, decompression sickness, trauma, inheritance, and corticosteroid therapy for either collagen disease or renal and cardiac transplantation (5, 6).
ONFH is a kind of ischemic damage of femoral head, which causes the necrosis of the hip joint cartilage and collapse and degeneration of the femoral head (6–8). The etiology of ONFH has not been fully elucidated (5, 6). It is suggested that thrombophilia genes polymorphisms including factor V Leiden (FVL), Prothrombin, Methylenetetrahydrofolate reductase (MTHFR), and plasminogen activator inhibitor-1 (PAI-1) causing intravascular coagulation conditions may be linked to ONFH (9–11).
To date, several polymorphisms have been identified in the PAI-1 gene such as PAI-1 4G/5G polymorphism (rs1799889). PAI-1 4G/5G polymorphism is an insertion/deletion polymorphism in the promoter region of the SERPINE1 gene, which is also known as plasminogen activator inhibitor type 1 (12, 13). The most common allele is a 5G’s in this SNP, which is commonly called 5G allele (14). Deletion of one nucleotide causes the “4G” allele. It has been reported that 4G allele increases the risk for different conditions such as atherosclerosis and coronary artery disease; while, 5G allele may increase the risk of abdominal aortic aneurysm (AAA) (15, 16). Some studies have reported that PAI-1 4G/5G polymorphism is associated with ONFH, while other publications report no association (17–22). Therefore, we have conducted this meta-analysis to determine whether PAI-1 4G/5G polymorphism is associated with susceptibility to ONFH.
Materials and Methods
Literature Search Strategy
A comprehensive literature search was performed using PubMed, EMBASE, Google Scholar, ISI Web of Knowledge, and China National Knowledge Infrastructure database to identify the studies that have evaluated the association between PAI-1 4G/5G (rs1799889) polymorphism and ONFH up to July 10, 2018. Various combinations of terms and keywords as described by Naderi Ghale-Noie et al were used to screen for potentially relevant studies, including: (“Osteonecrosis” OR “Osteonecrosis of the femoral head” OR “ONFH” “Steroid-induced ONFH” OR “Avascular necrosis of the femoral head” OR “ANFH” OR “Ischemic necrosis of the femoral head” OR “Aseptic necrosis of bone” OR “Osteochondritis dissecans” OR “Perthes disease”) AND (“plasminogen activator inhibitor-1” OR “SERPINE1” OR “PAI-1” OR “PAI-1 4G/5G” OR “rs1799889” OR “c.-816A>G”) AND (“single nucleotide polymorphism” OR “polymorphism” OR “SNP” OR “mutation” OR “variation”) (23). The search was limited to published human studies in all languages with available full-text articles. All eligible studies were retrieved. We also manually searched the references of the retrieved articles, reviews, and previous meta-analyses to identify more potentially relevant articles. When overlapping data on the same subjects were included in more than one publication, only the one with the larger sample size was included in the meta-analysis.
Inclusion and exclusion criteria
Studies included in the meta-analysis had to be consistent with the following criteria: (1) evaluating the association between PAI-1 4G/5G (rs1799889) polymorphism and ONFH risk; (2) using a case-control design; (3) sufficient data of genotypes were presented with estimated odds ratios (ORs); and 95% confidence intervals (CIs). Major reasons for exclusion of studies were as follows: (1) not relevant to PAI-1 4G/5G (rs1799889) polymorphism or ONFH; (2) the design was based on family or lack of the control group; (3) not on human; (4) genotype frequencies not reported; (5) reviews, abstracts, seminars posters, case reports, meta-analyses and letters, (6) and duplicates of previous studies.
Data Extraction
Two authors carefully and independently extracted the data from all eligible publications according to the inclusion criteria using a structured table. The following items were considered: first author’s name, year of publication, country, ethnicity, source of controls (population-based or hospital-based controls), genotyping method, numbers of cases and controls, genotype frequency of cases and controls, minor allele frequencies (MAFs) in control subjects, and the results of Hardy-Weinberg equilibrium (HWE) test. The subjects’ ethnicities were categorized as Caucasian, Asian, or African. Disagreements were resolved in consultation with the third reviewer. An agreement was reached by discussion between the two reviewers whenever there was a conflict, or resolved in consultation with the third reviewer.
Statistical Analysis
The strength of association between PAI-1 4G/5G polymorphism and ONFH risk was tested by odds ratios (ORs) with 95% confidence intervals (CIs). The significance of the pooled OR was determined using the Z-test and a P<0.05 was considered as statistically significant. The pooled ORs were performed in five genetic models: the allele model (4G vs. 5G), the heterozygote model (4G5G vs. 5G5G), the dominant model (4G4G+4G5G vs. 5G5G), and the recessive model (4G4G vs. 4G5G+5G5G). Chi-squared Q-test and I2 statistics were used to identify the heterogeneity among the included publications (24–26). The P-value of <0.05 for the Q-test indicated a lack of heterogeneity among studies. The I2 statistic measures the degree of inconsistency in the studies by calculating what percentage of the total variation across studies is due to heterogeneity rather than by chance (I2<25%, low heterogeneity; 25%≤I2≤75%, moderate heterogeneity; I2>75%, high heterogeneity). If there was significant heterogeneity, a random-effects model (the Der Simonian and Laird method) was selected to pool the data; otherwise, a fixed-effects model (the Mantel–Haenszel method) was selected to pool the data (27, 28). Subgroup analyses by ethnicity were performed to identify the between-study heterogeneity. Additionally, the ef¬fect of each single study on the overall estimate was deter¬mined by application of one-way sensitivity analysis. Hardy-Weinberg Equilibrium (HWE) in the controls was tested by the χ2 test for goodness of fit and P<0.05 was considered to indicate statistically significant HWE (29). Sensitivity analysis was performed by omitting one study at a time. To examine the potential publication bias in the meta-analysis, Begg’s funnel plot and Egger’s test were used; P<0.05 indicated that the result was statistically significant (30, 31). All the statistical analyses were performed by Comprehensive Meta-Analysis (CMA) software version 2.0 (Biostat, USA). Two-sided P values<0.05 were considered as statistically significant.
Results
Study selection and characteristics
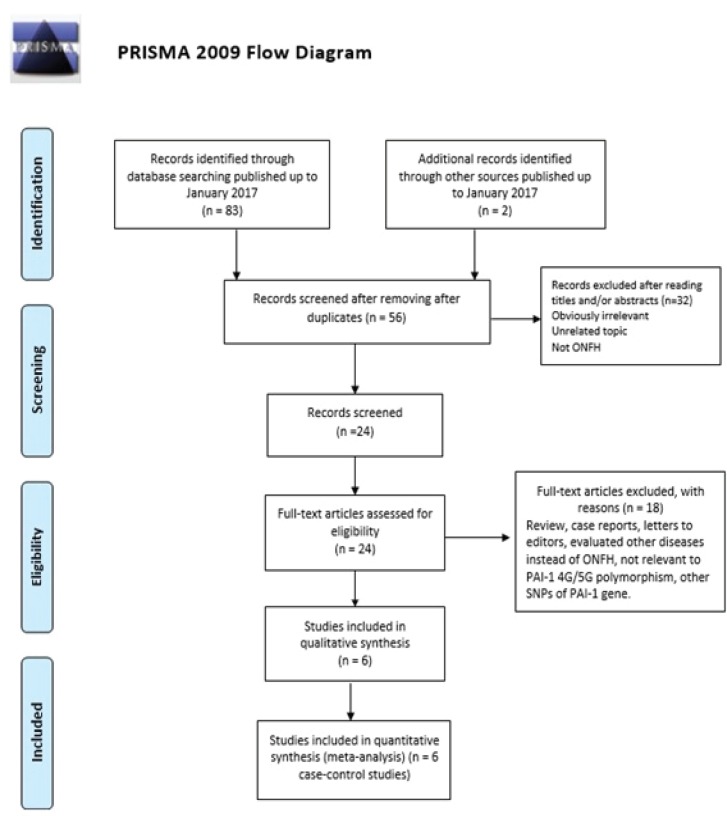
The process of selection of studies for inclusion in the meta-analysis is shown in Figure 1. Based on the search criteria, a total of 131 literatures were identified, among which 37 literatures were excluded as duplicates or not relevant, leaving 94 studies for further selection. Among the remaining studies, 88 articles were excluded because they were irrelevant, review articles, letters to editors, previous meta-analyses, not relevant to PAI-1 4G/5G (rs1799889) polymorphism, not case-control studies, evaluated other diseases instead of ONFH, case reports, and other polymorphisms of PAI-1 gene. Finally, a total of six case-control studies (17–22) were selected in the final meta-analysis concerning the PAI-1 4G/5G (rs1799889) polymorphism, with a total of 456 cases and 1,019 controls [Table 1]. The year of publication ranged between 2001 and 2015. There were 3 studies of Caucasian descendants (USA, Switzerland, and Serbia) and 3 studies of East Asian descendants communities (Japan, China and Korea). All studies showed that the distribution of genotypes in the control group was in agreement with the Hardy-Weinberg equilibrium. Table 1 lists the main characteristics of the six case-control studies about PAI-1 4G/5G (rs1799889) polymorphism.
Figure 1.
The flow diagram for the review process and outcomes of inclusion and exclusion.
Table 1.
Characteristics of studies Included in the meta-analysis of PAI-1 4G/5G (rs1799889) polymorphism and ONFH
| First author |
Country
(Ethnicity) |
SOC |
Genotyping
Method |
Case/Control |
Cases
|
Controls
|
MAF | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Genotype
|
Allele
|
Genotype
|
||||||||||||||
| 5G5G | 4G5G | 4G4G | 5G | 4G | 5G5G | 4G5G | 4G4G | 5G | 4G | |||||||
| Glueck 2001 | USA (Caucasian) | HB | PCR | 95/234 | 15 | 44 | 36 | 74 | 116 | 84 | 103 | 47 | 271 | 197 | 0.420 | 0.137 |
| Ferrari 2002 | Switzerland (Caucasian) | PB | PCR | 26/326 | 2 | 8 | 16 | 12 | 40 | 93 | 166 | 67 | 352 | 300 | 0.460 | 0.652 |
| Asano 2004 | Japan (Asian) | HB | AS-PCR | 31/106 | 4 | 11 | 16 | 19 | 43 | 14 | 56 | 36 | 84 | 128 | 0.603 | 0.283 |
| Sun 2008 | China (Asian) | HB | PCR-SPOLA | 61/52 | 16 | 22 | 23 | 54 | 68 | 12 | 27 | 13 | 51 | 53 | 0.509 | 0.779 |
| Kim 2011 | Korea (Asian) | HB | TaqMan | 206/251 | 29 | 95 | 82 | 140 | 259 | 46 | 130 | 75 | 222 | 280 | 0.557 | 0.429 |
| Srzentić 2015 | Serbia (Caucasian) | NS | Sequencing | 37/50 | 9 | 22 | 6 | 40 | 34 | 8 | 27 | 15 | 43 | 57 | 0.570 | 0.472 |
SOC: Source of controls; HB: hospital-based; PB: Population-based; PCR- SPOLA: PCR-solid phase oligonucleotide assay; MAF: minor allele frequency; HWE: Hardy–Weinberg equilibrium.
Main Results of Meta-Analysis
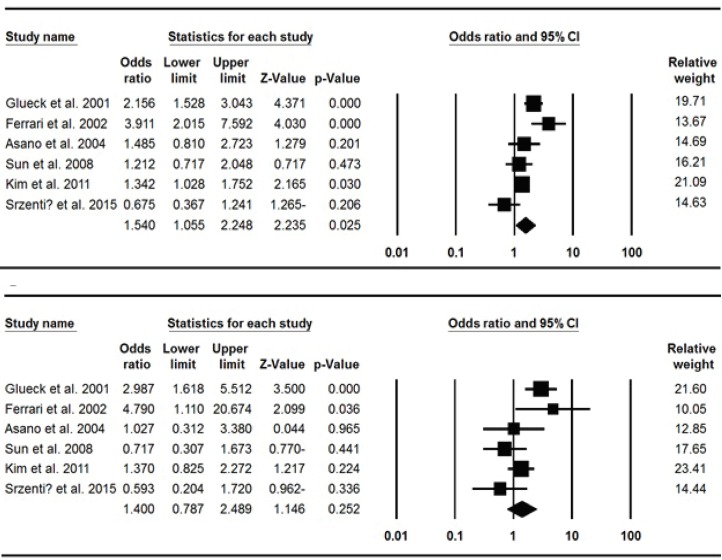
Table 2 shows the main results of the meta-analysis of PAI-1 4G/5G (rs1799889) polymorphism and risk of ONFH. When all the eligible studies were pooled into the meta-analysis of PAI-1 4G/5G polymorphism, significantly increased risk of ONFH was observed in allele model (4G vs. 5G: OR=1.540, 95% CI: 1.055-2.248, P=0.025) and recessive model (4G4G vs. 4G5G+5G5G: OR=1.931, 95% CI: 1.162-3.207, P=0.011) [Figure 2]. In the subgroup analysis by ethnicity, significantly increased ONFH risk was observed in Caucasians under heterozygote model (4G5G vs. 5G5G: OR=1.806, 95% CI: 1.064-3.067, P=0.029) by using fixed-effect model and in East Asians under homozygote model (4G4G vs. 5G5G: OR=1.619, 95% CI: 1.025-2.556, P=0.039) and recessive model (4G4G vs. 4G5G+5G5G: OR=1.665, 95% CI: 1.207-2.297, P=0.002).
Table 2.
Meta-analysis of the association of PAI-1 4G/5G (rs1799889) polymorphism and ONFH
| Genetic model | Type of model |
Heterogeneity
|
Odds ratio
|
Publication bias
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| (%) I 2 | P H | OR | 95% CI | P OR | P Beggs | P Eggers | |||
| Overall | |||||||||
| 4G vs. 5G | Random | 75.17 | 0.001 | 1.540 | 1.055-2.248 | 0.025 | 0.707 | 0.959 | |
| 4G4G vs. 5G5G | Random | 70.39 | 0.005 | 1.986 | 0.962-4.101 | 0.064 | 1.000 | 0.839 | |
| 4G5G vs. 5G5G | Fixed | 40.50 | 0.135 | 1.232 | 0.880-1.725 | 0.225 | 1.000 | 0.553 | |
| 4G4G+4G5G vs. 5G5G | Random | 63.27 | 0.018 | 1.400 | 0.787-2.489 | 0.252 | 1.000 | 0.765 | |
| 4G4G vs. 4G5G+5G5G | Random | 69.95 | 0.005 | 1.931 | 1.162-3.207 | 0.011 | 1.000 | 0.967 | |
| Ethnicity | |||||||||
| Caucasian | |||||||||
| 4G vs. 5G | Random | 87.62 | 0.001 | 1.786 | 0.747-4.268 | 0.192 | 1.000 | 0.875 | |
| 4G4G vs. 5G5G | Random | 85.27 | 0.001 | 2.581 | 0.449-14.825 | 0.288 | 1.000 | 0.810 | |
| 4G5G vs. 5G5G | Fixed | 41.25 | 0.182 | 1.806 | 1.064-3.067 | 0.029 | 1.000 | 0.695 | |
| 4G4G+4G5G vs. 5G5G | Random | 74.56 | 0.020 | 1.985 | 0.623-6.327 | 0.246 | 1.000 | 0.836 | |
| 4G4G vs. 4G5G+5G5G | Random | 86.13 | 0.001 | 1.990 | 0.586-6.757 | 0.270 | 1.000 | 0.757 | |
| Asian | |||||||||
| 4G vs. 5G | Fixed | 0.00 | 0.882 | 1.336 | 1.071-1.667 | 0.010 | 1.000 | 0.974 | |
| 4G4G vs. 5G5G | Fixed | 0.00 | 0.900 | 1.619 | 1.025-2.556 | 0.039 | 1.000 | 0.444 | |
| 4G5G vs. 5G5G | Fixed | 0.00 | 0.443 | 0.949 | 0.613-1.469 | 0.816 | 1.000 | 0.308 | |
| 4G4G+4G5G vs. 5G5G | Fixed | 0.00 | 0.431 | 1.139 | 0.757-1.714 | 0.531 | 1.000 | 0.506 | |
| 4G4G vs. 4G5G+5G5G | Fixed | 0.00 | 0.798 | 1.665 | 1.207-2.297 | 0.002 | 1.000 | 0.221 | |
Figure 2.
Forest plots showed significant association between PAI-1 4G/5G polymorphism and ONFH. A: Allele model (4G vs. 5G); B: Dominant model (4G4G+4G5G vs. 5G5G).
Heterogeneity Test
When we pooled the data for PAI-1 4G/5G (rs1799889) polymorphism a significant heterogeneity was observed in four model including Allele model (4G vs. 5G): I2=75.17%, Ph=0.001; Heterozygote model (4G5G vs. 5G5G): I2=70.39%, Ph=0.005; Dominant model (4G4G+4G5G vs. 5G5G): I2= 63.27%, Ph=0.018, and Recessive model (4G4G vs. 4G5G+5G5G): I2=69.95%, Ph=0.005 [Table 2]. After stratification of the subjects by ethnicity, the heterogeneity was obviously disappeared in East Asians under all genetic models. However, heterogeneity was still present among the Caucasians [Table 2].
Sensitivity Analyses
We performed sensitivity analyses to assess the influence of each individual study on the pooled OR by sequential removal of individual studies. However, the results suggested that no individual study significantly affected the pooled OR, suggesting that the results of this meta-analysis are stable sufficiently (data not shown).
Publication Bias
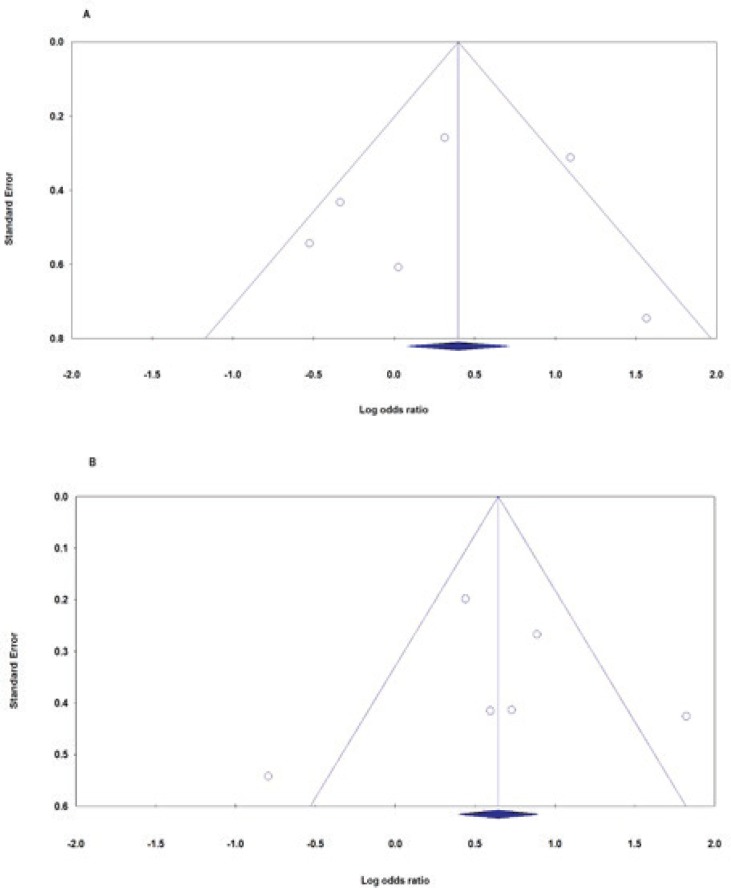
Begg’s funnel plot and Egger’s test were performed to access the small study effects of the studies in the meta-analysis. The funnel plot identified no obvious publication bias for PAI-1 4G/5G (rs1799889) polymorphism, and this was confirmed by Begg’s test and Egger’s test. As followed for overall tests: allele model (4G vs. 5G): Begg’s test p=0.707, Egger’s test p=0.959, homozygote model (4G4G vs. 5G5G): Begg’s test p=1.000, Egger’s test p=0.839, heterozygote model (4G5G vs. 5G5G): Begg’s test p=1.000, Egger’s test p=0.553, dominant model (4G4G+4G5G vs. 5G5G): Begg’s test p=1.000, Egger’s test p=0.765 and recessive model (4G4G vs. 4G5G+5G5G): Begg’s test p=1.000, Egger’s test p=0.967 [Figure 3].
Figure 3.
Begg’s funnel plots of PAI-1 4G/5G polymorphism and ONFH risk for publication bias test. Each point represents a separate study for the indicated association. A: Allele model (4G vs. 5G); B: Dominant model (4G4G+4G5G vs. 5G5G).
Minor Allele Frequency (MAFs)
The minor allele frequencies of PAI-1 4G/5G (rs1799889) polymorphism in healthy controls are shown in Table 1. The genetic distributions of PAI-1 4G/5G polymorphism in all studies followed the law of HWE (P>0.05). The minor allele frequency of PAI-1 4G/5G was 50.15% (42.0%-60.3%) for controls.
Discussion
PAI-1 gene, also known as serpin E1, is located in human chromosome 7q21.3-q22, spans 12.3 kb and contains 9 exons (32). PAI-1 gene is genetically linked with different conditions including thrombosis and atherosclerosis (33). PAI-1 is a fast-acting inhibitor of fibrinolysis, and increased plasma levels is associated with increased incidence of thrombophilia and osteonecrosis (11). High levels of PAI-1, induced by PAI-1 4G/5G polymorphism causes suppression of fibrinolysis by inhibition of plasminogen activator and promotion of thrombosis, and consequently restricts flow to the femoral head and osteonecrosis (34, 35). Three major polymorphisms of the PAI-1 gene have been identified, including PAI-1 4G/5G insertion/deletion (rs1799889) polymorphism at -675 in the promoter region, G-A substitution at position -844 (rs2227631), and c.43G<A (p.A15T, rs6092) (36). To date, a few studies have analyzed the association between PAI-1 4G/5G (rs1799889) polymorphism and ONFH (17–22). In the other hand, the results of the studies about the role of the PAI-1 4G/5G polymorphism in relation to ONFH susceptibility are conflicting. For example, studies by Glueck et al., Ferrari et al., Kim et al., and Sun et al. found an increased risk for ONFH associated with the PAI-1 4G/5G polymorphism and the other studies by Asano et al. did not detect the association (17-21). It seems that the conflicting findings among those studies might be due to different issues including the definition of the osteonecrosis, criteria of subjects, sample size, source of controls, genotyping technique and so on. In addition, it is obvious that ONFH is a complex condition and potential gene–gene and gene–environment interactions may also play vital roles in its formation. Hence, we have performed a more precise meta-analysis. After pooling the data from the eligible case-control studies, our meta-analysis indicated that PAI-1 4G/5G (rs1799889) polymorphism is associated with increased ONFH risk in overall population and by ethnicity.
As seen in Table 1, most of the included studies in the current meta-analysis had small sample sizes. Single studies, especially the one with relatively small sample size, may do not have enough statistical power to identify a genetic association. Meta-analysis has the capability of quantitative combining and synthetic evaluation in terms of the studies with the same objective and multiple independent results, so as to improve the inspection efficiency. The current meta-analysis reviewed the case–control studies, which provided evidence for ONFH risk assessment comprehensively upon PAI-1 4G/5G (rs1799889) polymorphism and ONFH risk. Overall analysis suggests a significant association between PAI-1 4G/5G polymorphism and ONFH under the allele model (4G vs. 5G: OR=1.540, 95% CI=1.055-2.248, p=0.025) and the recessive model (4G4G vs. 4G5G+5G5G: OR=1.931, 95% CI=1.162-3.207, p=0.011). When stratified by ethnicity, we found an association among the Caucasian population under heterozygote model (4G5G vs. 5G5G: OR=1.806, 95% CI=1.064-3.067, P=0.029) and East Asians under homozygote model (4G4G vs. 5G5G: OR=1.619, 95% CI: 1.025-2.556, P=0.039) and recessive model (4G4G vs. 4G5G+5G5G: OR=1.665, 95% CI: 1.207-2.297, P=0.002). Similarly, Zeng et al in a meta-analysis reported an association between PAI-1 4G/5G polymorphism and the increasing risk of ONFH under the allele model (RR=1.24, 95% CI: 1.16-1.33) and the dominant model (RR=1.12, 95% CI: 1.05-1.18) (10). In addition, in another meta-analysis, Liang et al found a significant association between PAI-1 4G/5G polymorphism and ONFH susceptibility under four allele, dominant, recessive, and homozygote genetic models (11). Moreover, Kim et al., in a case-control study with 206 ONFH cases and 251 controls, found that in addition to PAI-1 4G/5G (rs1799889) polymorphism, two other polymorphisms including rs2227631 and rs11178 of the PAI-1 gene were associated with ONFH risk (21).
Between-study heterogeneity is a potential and multifactorial problem when interpreting the results of a meta-analysis, and identifying the sources of heterogeneity is one of the most important goals of meta-analysis (37–40). It seems that factors such as age, gender distribution, personal history, diversity in study design, difference of ethnicity, sample sizes, source sampling, selection of controls, measurement errors, and so on might also be responsible for the between-study heterogeneity (40, 41). Obviously, there were moderate to high level heterogeneity in this meta-analysis, thus we used random-effect model for those genetic models. We performed subgroup analysis by genotyping method to find the source of between-study heterogeneity. After subject stratification by ethnicity, the heterogeneity was obviously disappeared in East Asians under all genetic models, but not in the Caucasians. Therefore, it was deducted that ethnicity was the main source of heterogeneity in the meta-analysis.
Our meta-analysis also had some limitations. Firstly, only six case-control studies were finally included into the meta-analysis. The limited number of studies may increase the risk of bias in the meta-analysis, especially in the subgroup analysis by ethnicity. Therefore, more case-control design studies with large samples are required for a more comprehensive meta-analysis. Secondly, although all the eligible studies were collected, the cumulative sample size was not large enough. This could increase the chance of type I and type II errors. Third, due to the unavailability of potential correlative factors data such as age, gender, environmental factors and lifestyle habits, our meta-analysis was based on single-factor estimates unadjusted ORs with 95% CIs, which may influence the power and reliability of results. Fourth, as other malignancies, ONFH is a multifactorial condition that results from complex interactions between various genetic and environmental factors. Further evaluation of melanoma risk would pay more attention to the potential interactions among gene–gene, gene–environment. Finally, in the current meta-analysis we have focused only on the PAI-1 4G/5G single polymorphism on ONFH risk rather than combined effects of thrombophilia genes well-known polymorphisms such as Prothrombin G20210A, Factor V Leiden G1691A, MTHFR C677T and A1298C, which could help to improve a precise estimation of the roles of PAI-1 4G/5G polymorphism in the development of ONFH. Therefore, more well-designed studies are needed in the future, and ORs adjusted for other confounding factors need reporting. Despite these limitations, our meta-analysis has some clear advantages such as, lack of publication bias.
In summary, the results of this meta-analysis suggested that the PAI-1 4G/5G (rs1799889) polymorphism may be associated with risk of ONFH. Large-scale well-designed studies with more information about the potential correlative factors in different populations are needed for better estimation of the possible gene-gene or gene-environment interactions.
The authors declare no competing financial interests.
References
- 1.Schmitt-Sody M, Kirchhoff C, Mayer W, Goebel M, Jansson V. Avascular necrosis of the femoral head: inter- and intraobserver variations of Ficat and ARCO classifications. Int Orthop. 2008;32(3):283–7. doi: 10.1007/s00264-007-0320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heidari B. Knee osteoarthritis prevalence, risk factors, pathogenesis and features: part I. Caspian J Intern Med. 2011;2(2):205–12. [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan HF, Zhang J, Guo CA, Yan ZQ. Clinical outcomes of osteonecrosis of the femoral head after autologous bone marrow stem cell implantation: a meta-analysis of seven case-control studies. Clinics. 2016;71(2):110–3. doi: 10.6061/clinics/2016(02)10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pouya F, Kerachian MA. Avascular necrosis of the femoral head: are any genes involved? Arch Bone Jt Surg. 2015;3(3):149–55. [PMC free article] [PubMed] [Google Scholar]
- 5.Gagala J, Buraczynska M, Mazurkiewicz T, Ksiazek A. Prevalence of genetic risk factors related with thrombophilia and hypofibrinolysis in patients with osteonecrosis of the femoral head in Poland. BMC Musculoskelet Disord. 2013;14(1):264. doi: 10.1186/1471-2474-14-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi HR, Steinberg ME, Cheng E. Osteonecrosis of the femoral head: diagnosis and classification systems. Curr Rev Musculoskelet Med. 2015;8(3):210–20. doi: 10.1007/s12178-015-9278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen RK. Management of avascular necrosis of femoral head at pre-collapse stage. Indian J Orthop. 2009;43(1):6–16. doi: 10.4103/0019-5413.45318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, Dong Z, Li S, Song H. Steroid-induced ischemic bone necrosis of femoral head: treatment strategies. Pak J Med Sci. 2015;31(2):471–6. doi: 10.12669/pjms.312.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang JD, Hur M, Lee SS, Yoo JH, Lee KM. Genetic background of nontraumatic osteonecrosis of the femoral head in the Korean population. Clin Orthop Relat Res. 2008;466(5):1041–6. doi: 10.1007/s11999-008-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Z, Wang B, Pan H. Relation between osteonecrosis of the femoral head and PAI-1 4G/5G gene polymorphism: a meta-analysis. Int J Clin Exp Med. 2015;8(11):20337–42. [PMC free article] [PubMed] [Google Scholar]
- 11.Liang XN, Xie L, Cheng JW, Tan Z, Yao J, Liu Q, et al. Association between PAI-1 4G/5G Polymorphisms and osteonecrosis of femoral head: a meta-analysis. Thromb Res. 2013;132(2):158–63. doi: 10.1016/j.thromres.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 12.Kamali M, Hantoushzadeh S, Borna S, Neamatzadeh H, Mazaheri M, Noori-Shadkam M, et al. Association between thrombophilic genes polymorphisms and recurrent pregnancy loss susceptibility in the Iranian population: a systematic review and meta-analysis. Iran Biomed J. 2018;22(2):78–89. doi: 10.22034/ibj.22.2.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsantes AE, Nikolopoulos GK, Bagos PG, Bonovas S, Kopterides P, Vaiopoulos G. The effect of the plasminogen activator inhibitor-1 4G/5G polymorphism on the thrombotic risk. Thromb Res. 2008;122(6):736–42. doi: 10.1016/j.thromres.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Bouchard L, Vohl MC, Lebel S, Hould FS, Marceau P, Bergeron J, et al. Contribution of genetic and metabolic syndrome to omental adipose tissue PAI-1 gene mRNA and plasma levels in obesity. Obes Surg. 2010;20(4):492–9. doi: 10.1007/s11695-010-0079-1. [DOI] [PubMed] [Google Scholar]
- 15.Onalan O, Balta G, Oto A, Kabakci G, Tokgozoglu L, Aytemir K, et al. Plasminogen activator inhibitor-1 4G4G genotype is associated with myocardial infarction but not with stable coronary artery disease. J Thromb Thrombolysis. 2008;26(3):211–7. doi: 10.1007/s11239-007-0083-z. [DOI] [PubMed] [Google Scholar]
- 16.French D, Hamilton LH, Mattano LA Jr, Sather HN, Devidas M, Nachman JB, et al. A PAI-1 (SERPINE1) polymorphism predicts osteonecrosis in children with acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2008;111(9):4496–9. doi: 10.1182/blood-2007-11-123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrari P, Schroeder V, Anderson S, Kocovic L, Vogt B, Schiesser D, et al. Association of plasminogen activator inhibitor-1 genotype with avascular osteonecrosis in steroid-treated renal allograft recipients. Transplantation. 2002;74(8):1147–52. doi: 10.1097/00007890-200210270-00016. [DOI] [PubMed] [Google Scholar]
- 18.Asano T, Takahashi KA, Fujioka M, Inoue S, Ueshima K, Hirata T, et al. Relationship between postrenal transplant osteonecrosis of the femoral head and gene polymorphisms related to the coagulation and fibrinolytic systems in Japanese subjects. Transplantation. 2004;77(2):220–5. doi: 10.1097/01.TP.0000101433.99651.96. [DOI] [PubMed] [Google Scholar]
- 19.Glueck CJ, Fontaine RN, Gruppo R, Stroop D, Sieve-Smith L, Tracy T, et al. The plasminogen activator inhibitor-1 gene, hypofibrinolysis, and osteonecrosis. Clin Orthop Relat Res. 1999;366(1):133–46. doi: 10.1097/00003086-199909000-00017. [DOI] [PubMed] [Google Scholar]
- 20.Sun W, Li Z, Shi Z, Wang B, Gao F, Yang Y, et al. Relationship between post-SARS osteonecrosis and PAI-1 4G/5G gene polymorphisms. Eur J Orthop Surg Traumatol. 2014;24(4):525–9. doi: 10.1007/s00590-013-1223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Cho C, Cho Y, Cho S, Yoon K, Kim K. Significant associations of PAI-1 genetic polymorphisms with osteonecrosis of the femoral head. BMC Musculoskelet Disord. 2011;12(1):160. doi: 10.1186/1471-2474-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srzentić S, Nikčević G, Spasovski D, Baščarević Z, Živković Z, Terzic-Šupić Z, et al. Predictive genetic markers of coagulation, inflammation and apoptosis in Perthes disease—Serbian experience. Eur J Pediatr. 2015;174(8):1085–92. doi: 10.1007/s00431-015-2510-z. [DOI] [PubMed] [Google Scholar]
- 23.Ghale-Noie ZN, Hassani M, Kachooei AR, Kerachian MA. High serum alpha-2-macroglobulin level in patients with osteonecrosis of the femoral head. Arch Bone Jt Surg. 2018;6(3):219–24. [PMC free article] [PubMed] [Google Scholar]
- 24.Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21(18):3672–3. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 25.Khoram-Abadi KM, Forat-Yazdi M, Kheirandish S, Saeidi N, Zarezade Z, Mehrabi N, et al. DNMT3B -149 C>T and -579 G>T polymorphisms and risk of gastric and colorectal cancer: a meta-analysis. Asian Pac J Cancer Prev. 2016;17(6):3015–20. [PubMed] [Google Scholar]
- 26.Jafari Nedooshan J, Forat Yazdi M, Neamatzadeh H, Zare Shehneh M, Kargar S, Seddighi N. Genetic association of XRCC1 gene rs1799782, rs25487 and rs25489 polymorphisms with risk of thyroid cancer: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18(1):263–70. doi: 10.22034/APJCP.2017.18.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48. [PubMed] [Google Scholar]
- 28.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 29.Namazi A, Forat-Yazdi M, Jafari M, Farahnak S, Nasiri R, Foroughi E, et al. Association of interleukin-10 -1082 A/G (RS1800896) polymorphism with susceptibility to gastric cancer: meta-analysis of 6,101 cases and 8,557 controls. Arq Gastroenterol. 2018;55(1):33–40. doi: 10.1590/S0004-2803.201800000-18. [DOI] [PubMed] [Google Scholar]
- 30.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101. [PubMed] [Google Scholar]
- 31.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Huang P. Plasminogen activator inhibitor-1 4G/5G polymorphism is associated with type 2 diabetes risk. Int J Clin Exp Med. 2013;6(8):632–40. [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer K, Müller K, Hecke A, Mounier E, Goebel J, Loskutoff DJ, et al. Enhanced thrombosis in atherosclerosis-prone mice is associated with increased arterial expression of plasminogen activator inhibitor-1. Arterioscler Thromb Vasc Biol. 2003;23(11):2097–103. doi: 10.1161/01.ATV.0000097766.36623.DF. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, Shi X, Xun X, Rao L. Endothelial nitric oxide synthase gene single nucleotide polymorphisms and the risk of hypertension: a meta-analysis involving 63,258 subjects. Clin Exp Hypertens. 2017;39(2):175–82. doi: 10.1080/10641963.2016.1235177. [DOI] [PubMed] [Google Scholar]
- 35.Cesari M, Pahor M, Incalzi RA. Plasminogen activator inhibitor-1 (PAI-1): a key factor linking fibrinolysis and age-related subclinical and clinical conditions. Cardiovasc Ther. 2010;28(5):e72–91. doi: 10.1111/j.1755-5922.2010.00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meigs JB, Dupuis J, Liu C, O’Donnell CJ, Fox CS, Kathiresan S, et al. PAI-1 gene 4G/5G polymorphism and risk of type 2 diabetes in a population-based sample. Obesity. 2006;14(5):753–8. doi: 10.1038/oby.2006.85. [DOI] [PubMed] [Google Scholar]
- 37.Yazdi MM, Jamalaldini MH, Sobhan MR, Jafari M, Mazaheri M, Zare-Shehneh M, et al. Association of ESRα gene Pvu II T>C, XbaI A>G and BtgI G>A polymorphisms with knee osteoarthritis susceptibility: a systematic review and meta-analysis based on 22 case-control studies. Arch Bone Jt Surgery. 2017;5(6):351–62. [PMC free article] [PubMed] [Google Scholar]
- 38.Sobhan MR, Mahdinezhad-Yazdi M, Jafari M, Mazaheri M, Neamatzadeh H, Daliri K. Association of ESRα XbaI A>G, PvuII T>C and ESRβ AlwNI T>C polymorphisms with the risk of adolescent idiopathic scoliosis: a systematic review and genetic meta-analysis. Rev Bras Ortop. 2018 doi: 10.1016/j.rboe.2018.03.001. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobhan MR, Forat Yazdi M, Mazaheri M, Zare Shehneh M, Neamatzadeh H. Association between the DNA repair gene XRCC3 rs861539 polymorphism and risk of osteosarcoma: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18(2):549–55. doi: 10.22034/APJCP.2017.18.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehdinejad M, Sobhan MR, Mazaheri M, Shehneh MZ, Neamatzadeh H, Kalantar SM. Genetic association between ERCC2, NBN, RAD51 gene variants and osteosarcoma risk: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18(5):1315–21. doi: 10.22034/APJCP.2017.18.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobhan MR, Mehdinejad M, Jamaladini MH, Mazaheri M, Zare-Shehneh M, Neamatzadeh H. Association between aspartic acid repeat polymorphism of the asporin gene and risk of knee osteoarthritis: a systematic review and meta-analysis. Acta Orthop Traumatol Turc. 2017;51(5):409–15. doi: 10.1016/j.aott.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]