Summary
H7N9 low pathogenic influenza viruses emerged in China in 2013 and mutated to highly pathogenic strains in 2017, resulting in human infections and disease in chickens. To control spread, a bivalent H5/H7 inactivated vaccine was introduced in poultry in September 2017. To monitor virus evolution and vaccine efficacy, we collected 53,884 poultry samples across China from February 2017 to January 2018. We isolated 252 H7N9 low pathogenic viruses, 69 H7N9 highly pathogenic viruses, and one H7N2 highly pathogenic virus, of which two low pathogenic and 16 highly pathogenic strains were collected after vaccine introduction. Genetic analysis of highly pathogenic strains revealed nine genotypes, one of which is predominant and widespread and contains strains exhibiting high virulence in mice. Additionally, some H7N9 and H7N2 viruses carrying duck virus genes are lethal in ducks. Thus, although vaccination reduced H7N9 infections, the increased virulence and expanded host range to ducks pose new challenges.
In Brief
H7N9 highly pathogenic avian influenza viruses emerged in China in 2017, prompting vaccination in poultry. Shi et al. examine H7N9 viruses across China before and after vaccination, revealing rapid evolution into subtypes and genotypes. Although vaccination reduced infections, some H7N9 and H7N2 viruses exhibit heightened virulence and expansion to ducks.
Graphical Abstract
Introduction
Influenza A viruses are RNA viruses; their genome consists of eight single-strand negative-sense RNA fragments that encode the basic polymerase 2 (PB2), basic polymerase 1 (PB1), acidic polymerase (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix (M), and nonstructural (NS) proteins. The viruses are divided into different subtypes on the basis of the antigenicity of their two surface glycoproteins, HA and NA. Currently, sixteen different HA (H1–H16) and nine different NA (N1–N9) subtypes have been identified from avian species, and two additional HA (H17–18) and NA (N10–11) subtypes have been detected in bats (Tong et al., 2012; Tong et al., 2013).
Influenza A viruses continuously challenge the poultry industry and human health. H1N1, H2N2, and H3N2 viruses have caused human influenza pandemics, and H1N1 and H3N2 viruses still circulate widely in humans around the world. Several subtypes of highly pathogenic H5 and H7 viruses have caused avian influenza outbreaks in poultry and wild birds in many countries since 1959 (Alexander and Brown, 2009; Chen, 2009; Fouchier et al., 2004; Li et al., 2010; Shi et al., 2017), leading to disastrous consequences for the poultry industry. H5N1 and H7N9 viruses have attracted wide attention over the past two decades because they have caused not only problems for the poultry industry, but also severe human infections and deaths (WHO/GIP, 2018a; Wong and Yuen, 2006; Zhou et al., 2013).
Since they emerged in February 2013, the H7N9 influenza viruses have caused 1, 567 human infections in mainland China, Hong Kong, Macau and Taiwan, 615 of which were fatal (WHO/GIP, 2018b), as of June 24, 2018. The viruses were initially found in live poultry markets in several provinces(Chen et al., 2013; Shi et al., 2013; Zhang et al., 2013), and studies indicated that these 2013 H7N9 viruses isolated from birds were nonpathogenic for chickens and mice, and barely replicated in ducks (Pantin-Jackwood et al., 2014; Zhang et al., 2013). Avian and human H7N9 viruses were able to bind to both avian-type and human-type receptors (Belser et al., 2013; Richard et al., 2013; Watanabe et al., 2013; Zhou et al., 2013), but the human isolates were more lethal in mice and more transmissible in ferrets than the avian isolates (Zhang et al., 2013). In early 2017, a few H7N9 HA mutants were detected from samples collected in the live poultry markets in Guangdong province, and animal studies indicated that these mutants were highly pathogenic for chickens (Qi et al., 2018; Shi et al., 2017). Although the index H7N9 HA mutant was not lethal in mice or ferrets, our previous study (Shi et al., 2017) and a study performed by Imai et al. (Imai et al., 2017) revealed that the H7N9 HA mutants could acquire additional mutations during their replication in ferrets or humans, and then become highly lethal in mammals and transmissible in ferrets by respiratory droplet. Yang et al. reported that 50% of human cases of infection with the H7N9 highly pathogenic influenza viruses were fatal (Yang et al., 2017) [since we did not test the virulence in chickens of every strain we obtained in this study, our use of “highly pathogenic” throughout this text simply means that the HA cleavage motif of the virus met the criteria for a highly pathogenic avian influenza virus (Neumann and Kawaoka, 2006; Senne et al., 1996)]. These findings strongly suggest that the H7N9 highly pathogenic influenza viruses pose an increased threat to humans.
Soon after they were detected in the live poultry markets, the H7N9 highly pathogenic viruses were detected in chicken farms, where they had caused severe disease outbreaks (OIE, 2018). Given the damage caused by the H7N9 viruses to poultry and the high risk they pose to human health, control and eradication of both the H7N9 low and highly pathogenic viruses have been of the highest priority for animal disease control authorities in China. In addition to the slaughter of millions of H7N9 highly pathogenic virus-infected poultry, a bivalent H5/H7 inactivated vaccine began to be used in poultry in September 2017 (MoA, 2017).
Despite efforts to control H7N9 influenza, these viruses have not been eradicated in poultry. To reveal the distribution of H7N9 viruses in China, we performed two rounds of large scale surveillance in poultry markets and poultry farms before and after the implementation of the H7N9 poultry vaccination program. A series of H7N9 low pathogenic and highly pathogenic viruses were isolated. We then characterized the genetic evolution and variation in virulence of these viruses in different animals. Our study provides a picture of the evolution of H7N9 highly pathogenic influenza viruses and sheds light on the effectiveness of vaccination in the control of H7N9 influenza in China.
Results
H7N9 influenza viruses were prevalent in China in 2017
To investigate the distribution of the H7N9 viruses, we performed two rounds of active influenza surveillance from February 2017 to January 2018. In the first round (from February 2017 to May 2017), we collected 12,504 samples from 379 live poultry markets, and 17,697 samples from 665 poultry farms; these numbers include the 2,950 samples collected in Guangdong province in February 2017 that were documented in our previous report (Shi et al., 2017). In the second round (from October 2017 to January 2018), we collected 12,967 samples from 204 live poultry markets and 10,716 samples from 374 poultry farms (Table 1). All samples were inoculated individually into 10-day-old embryonated chicken eggs for virus isolation. The HA subtype of the viruses was confirmed by using the hemagglutinin inhibition (HI) test. The NA subtypes of the viruses were determined by direct sequencing as reported by Alvarez et al. (Alvarez et al., 2010). The portion of the HA gene that includes the cleavage site was sequenced to differentiate the virulence of the H7 strains.
Table 1.
H7N9 and H7N2 influenza viruses isolated during surveillance between February 2017 and January 2018d,e.
| Time period | Location | Total number of markets or farms investigated | Number of samples collected | H7N9 low pathogenic virus | H7N9 highly pathogenic virus | |||
|---|---|---|---|---|---|---|---|---|
| Total strains | Province | Total strains | Province | Genotype | ||||
| Feb. 2017–May. 2017 | Market | 379 | 12,504 | 226 | 23 provincesa | 52 | Fujian, Guangdong, Guangxi, and Hunan | G1, G2, G3, G4, G8 |
| Farm | 665 | 17,697 | 24 | 12 provincesb | 4 | Guangxi, Hunan | G2, G4 | |
| Oct. 2017–Jan. 2018 | Market | 204 | 12,967 | 1 | Hunan | 12 | Anhui, Fujian, and Tibet | G2, G8 |
| Farm | 374 | 10,716 | 1 | Liaoning | 2c | Fijian, Yunnan | G2, G9 | |
Anhui, Chongqing, Fujian, Gansu, Guangdong, Guizhou, Hebei, Henan, Hubei, Hunan, Jiangsu, Jiangxi, Liaoning, Inner Mongolia, Ningxia, Shandong, Shanxi, Shanghai, Sichuan, Tibet, Xinjiang, Yunnan, and Zhejiang.
Beijing, Chongqing, Fujian, Gansu, Guangdong, Guizhou, Hebei, Henan, Hubei, Hunan, Jiangsu, and Liaoning.
One virus was H7N2.
Viruses of other subtypes are shown in Table S1.
Geographic distribution of the H7N9 viruses is shown in Figure S1
From the first round of surveillance, we isolated a total of 843 avian influenza viruses of 10 different HA subtypes, including H1 (22 strains), H3 (73 strains), H4 (12 strains), H5 (57 strains), H6 (125 strains), H7N9 (306 strains), H9N2 (243 strains), H10N8 (1 strain), H11 (3 strains), and H12N5 (1 strain) (Table 1, Table S1). The 306 H7N9 viruses included 250 low pathogenic strains and 56 highly pathogenic strains. Of note, 16 of the samples we collected in the poultry markets contained both H7N9 low pathogenic and H9N2 viruses. Among the 250 low pathogenic strains, 226 strains were isolated from live poultry markets in 23 provinces, and 24 strains were isolated from poultry farms in 12 provinces (Table 1, Figure S1A); among the 56 highly pathogenic H7N9 strains, 52 strains were isolated from the live poultry markets in four provinces, and four strains came from poultry farms in Guangxi and Hunan provinces (Table 1, Figure S1A).
In the second round of surveillance, we isolated a total of 932 avian influenza viruses of 9 different HA subtypes, including H1 (21 strains), H3 (107 strains), H4 (16 strains), H5 (225 strains), H6 (111 strains), H7 (16 strains), H9N2 (424 strains), H10 (10 strains), and H11 (2 strains) (Table 1, Table S1). The 16 H7 viruses included two H7N9 low pathogenic strains, as well as 13 H7N9 and one H7N2 highly pathogenic strains (Table 1). One of the low pathogenic viruses was isolated from a poultry market in Hunan province, and the other was isolated from a farm in Liaoning province. Twelve of the H7N9 highly pathogenic strains were isolated from poultry markets in the provinces of Anhui, Fujian, and Tibet (Table 1, Figure S1B) and the other H7N9 highly pathogenic virus was isolated from a chicken farm in Yunnan province. The H7N2 highly pathogenic virus was isolated from a duck farm in Fujian province (Table 1, Figure S1B).
The H7N9 highly pathogenic viruses spread to poultry farms and caused disease outbreaks in chickens in eight provinces from March to August in 2017 (Figure S1A); nine strains were isolated from the samples that were presented to our laboratory for disease diagnosis (Figure 1, virus names labeled with double asterisks), and all of these strains were highly virulent in chickens with an intravenous pathogenicity index of 3, as tested by the method described in the manual of the World Organization of Animal Health (OIE) (OIE, 2011).
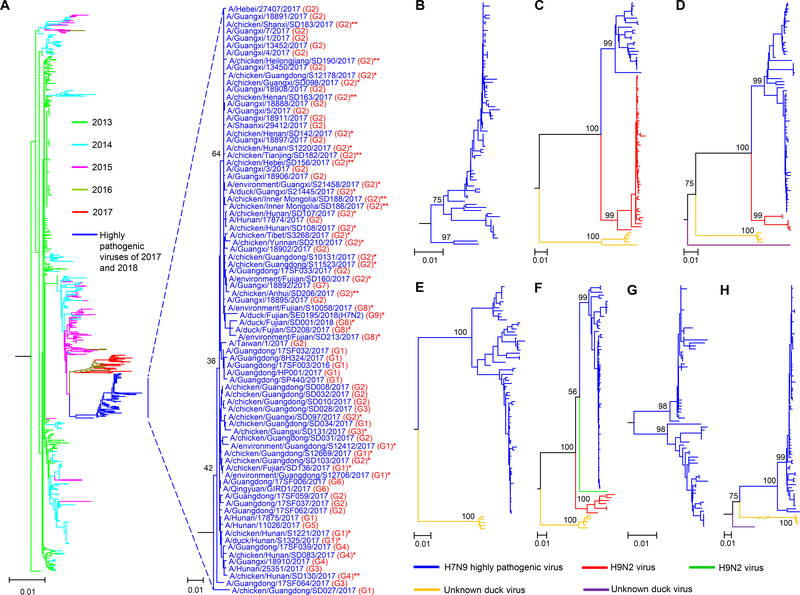
Figure 1. Phylogenetic analyses of H7N9 influenza viruses isolated in China.
(A) The big tree shows the phylogenetic relationship of the HA genes of 1002 viruses, including 921 H7N9 low pathogenic viruses [24 of which were sequenced in this study (Table S2)] that were isolated in China from 2013 to 2017, 80 H7N9 highly pathogenic viruses, and an H7N2 virus; the viral names of the 81 H7 highly pathogenic viruses are shown in the small tree. Highly pathogenic viruses sequenced in this study are labeled with single asterisk or double asterisks; double asterisks denote the viruses that have caused disease outbreaks in poultry farms. (B) The phylogenetic tree of the NA genes of the 80 H7N9 highly pathogenic viruses. The phylogenetic trees of the PB2 (C), PB1 (D), PA (E), NP (F), M (G), and NS (H) genes were generated from the relative genes of the 80 H7N9 highly pathogenic viruses and the H7N2 virus. The phylogenetic trees of NA, PB2, and PB1 with the virus names are shown in Figure S2. The phylogenetic trees of PA, NP, M, and NS with the virus names are shown in Figure S3.
These data demonstrate that several different subtypes of avian influenza viruses were present in poultry in China and that the H3, H5, H6, and H9 viruses were widespread during the two rounds of surveillance. However, although the H7N9 viruses were widespread in poultry in China during our first round of surveillance, the prevalence of these viruses dramatically decreased during our second round of surveillance.
The H7N9 highly pathogenic influenza viruses evolved rapidly and formed multiple genotypes
Our previous study suggested that the seven H7N9 highly pathogenic viruses detected in Guangdong in early 2017 may have arisen from two different H7N9 low pathogenic viruses (Shi et al., 2017). As indicated by our surveillance data above, both low and highly pathogenic H7N9 viruses were detected in several provinces in addition to Guangdong province (Figure S1A); however, it was not clear whether these highly pathogenic viruses were derived from local H7N9 low pathogenic viruses or were introduced from Guangdong province. To investigate the origins and genetic relationships of the H7N9 and H7N2 highly pathogenic viruses, we sequenced the full genomes of 37 viruses, including the H7N2 duck virus, the nine viruses that caused disease outbreaks, and 27 representative H7N9 highly pathogenic viruses from different sampling times, locations, and species. We then assessed their phylogenetic relationships with 44 H7N9 highly pathogenic viruses isolated from humans and chickens that were previously reported by us and others (Shi et al., 2017; Yang et al., 2017).
The HA gene of the 81 H7 highly pathogenic viruses shared 97.4%–100% identity at the nucleotide level and formed a unique cluster separate from the HA gene of the 921 H7N9 low pathogenic viruses [24 of which were sequenced in this study (Table S2)] detected from 2013–2017 in the phylogenetic tree (Figure 1 A). Five different motifs were detected in the HA cleavage sites of these strains: -KGKRTAR/G-, -KRKRAAR/G-, -KRKRTAR/G-, -KGKRIAR/G-, and -KRRRTAR/G- (the four amino acids underlined were insertions, whereas the amino acid R shown in italics was a mutation of the amino acid G at that position). The motif -KRKRAAR/G- was detected only in a human H7N9 virus, whereas the motifs -KGKRIAR/G- and -KRRRTAR/G- were detected in an environmental isolate and a chicken isolate, respectively. The motif -KGKRTAR/G- was detected in 15 viruses isolated from poultry, and the motif -KRKRTAR/G- was detected in 63 viruses that were isolated from birds and humans (Figure 2).
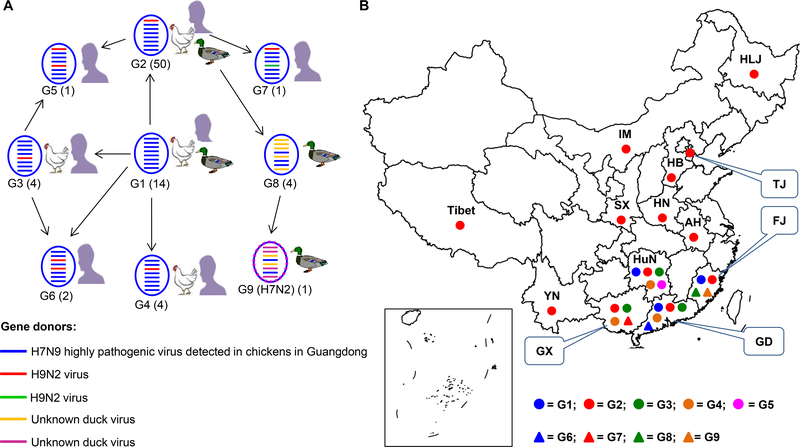
Figure 2. Genotypes and distribution of H7N9 and H7N2 highly pathogenic viruses detected in China.
(A) Genotypes of H7N9 and H7N2 viruses and the hosts from which these genotypes were detected. Numbers of strains of each genotype are indicated in parentheses. (B). Geographic distribution of the H7N2 virus and of the different genotypes of H7N9 viruses.
The NA gene of the 80 H7N9 highly pathogenic viruses shared 96.1%–100% identity at the nucleotide level, and the NA gene of the H7N2 virus shared the highest identity (98%) with the NA gene of the H3N2 duck virus A/duck/Guangxi/135D20/2013 (accession number in GenBank: KT022302.1). The M gene of the 81 viruses shared 95.1%–100% identity at the nucleotide level. The PB2, PB1, PA, NP, and NS genes of these 81 highly pathogenic viruses showed distinct diversity, sharing 85.5%–100%, 86.7%–100%, 90.7%–100%, 88.2%–100%, and 65.2%–100% identity, respectively, at the nucleotide level. They formed 2–4 different clusters shown in different colors in their phylogenetic trees (Figure 1C–F, H, Figure S2, and Figure S3); the gene identity between the clusters was less than 95%.
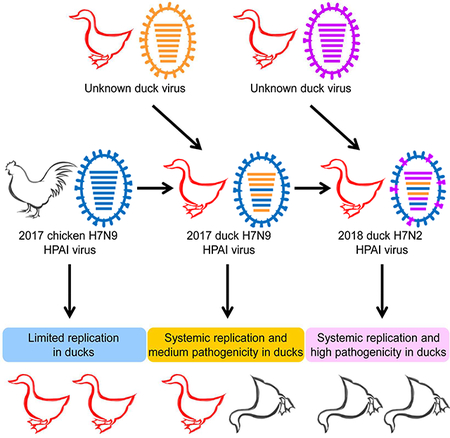
This phylogenetic analysis indicated that the H7N9 and H7N2 highly pathogenic viruses that were detected in provinces other than Guangdong were descendants of the earlier Guangdong H7N9 highly pathogenic viruses, rather than mutated derivatives of the local H7N9 low pathogenic viruses. According to the gene identity from the phylogenetic analysis, the H7N9 highly pathogenic viruses were divided into nine genotypes (Figure 2A), which resulted from frequent reassortment with other chicken and duck viruses. The genotype 2–7 (G2-G7) viruses were reassortants of genotype 1 (G1) H7N9 highly pathogenic viruses (shown in blue) and different local H9N2 chicken viruses (shown in green or red). The genotype 8 (G8) viruses were reassortants of the genotype 2 viruses and unknown duck viruses (shown in dark yellow). The genotype 8 viruses further reassorted with other unknown duck viruses (shown in purple) and generated the H7N2 virus, which was designated as a genotype 9 (G9) virus (Figure 2A). Viruses of G1 and G2 were detected in chickens, humans, and ducks; viruses of G3 and G4 were detected in chickens and humans, viruses of G5, G6, and G7 were detected in only humans, whereas viruses of G8 and G9 were detected in only ducks (Figure 2A). Of note, the G2 viruses were predominant as 50 of the 80 strains analyzed here belonged to G2 and they were detected in 13 provinces (Figure 2A, B).
H7N9 highly pathogenic viruses have become highly virulent in mice
We previously reported that the H7N9 highly pathogenic virus was not lethal in mice (Shi et al., 2017), but other groups reported that some chicken H7N9 highly pathogenic viruses had killed certain mice after inoculation with 106 50% egg infectious dose (EID50) of virus (Liu et al., 2018; Qi et al., 2018). H7N9 viruses with four different motifs in their HA cleavage sites were all highly lethal in chickens (Shi et al., 2017); however, it is not known whether differences in these motifs affect the virulence of the viruses in mammals. To fully understand the pathotypes of H7N9 highly pathogenic viruses in mammals, we tested the replication and virulence in mice of 18 viruses, including the H7N2 duck virus and 17 H7N9 viruses that were selected from genotypes 1, 2, 3, 4, and 8 and bore different motifs in their HA cleavage site (Figure 3).
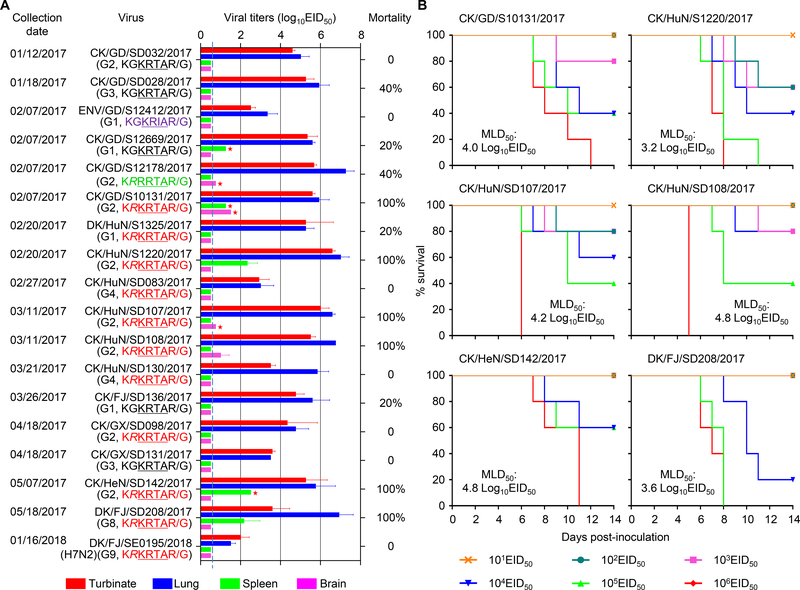
Figure 3. Replication and virulence of the H7N9 and H7N2 viruses in mice.
(A) Viral titers in organs and mortality of mice after inoculation with 106 50% egg infectious dose (EID50) of different viruses. Virus titers were determined in eggs. Color bars show the mean (n = 3), and the error bars represent the standard deviations. The value labeled with a red star indicates that the virus was only detected in the organ(s) of one mouse. The dashed lines in these panels indicate the lower limit of virus detection. The mortality of each virus was determined by inoculating five mice. (B) Mouse lethal dose of different H7N9 viruses.
All of the viruses replicated in the turbinates and lungs of the mice, although the replication titers of the H7N2 virus were notably lower than those of the H7N9 viruses (Figure 3A). Virus was not detected in the kidneys of any mice inoculated with any of the 18 viruses. Low viral titers were detected in the brains and the spleens of mice inoculated with four and five of the H7N9 viruses, respectively (Figure 3A). The H7N2 virus and six of the H7N9 viruses did not kill any mice, whereas three H7N9 viruses killed 20% of the infected mice, two H7N9 viruses killed 40% of the infected mice, and the other six viruses killed all of the infected mice during the observation period (Figure 3A).
We then tested the 50% mouse lethal dose (MLD50) by inoculating groups of five mice intranasally (i.n.) with 101.0–106.0 EID50 of the six viruses that killed all of the mice at the dose of 106EID50 and monitored their survival for 2 weeks. As shown in Figure 3B, the MLD50 values of the six viruses ranged from 3.2 log10EID50 to 4.8 log10EID50 (Figure 3B). The CK/HuN/S1220/2017 virus (MLD50=3.2 log10EID50) was 1,200-fold more lethal than the CK/GX/SD098/2017 virus (MLD50 > 6.5 log10EID50); these two viruses belong to the same genotype and have the same motif in their HA cleavage site. These results indicate that, after circulating in poultry for only a few months, some H7N9 highly pathogenic viruses have become virulent in mice, although the genetic changes that contribute to the increased virulence of these viruses in mammals remain unknown.
H7N9 and H7N2 viruses bearing genes from duck influenza viruses replicate efficiently and are lethal in ducks
Previous studies indicated that the H7N9 low pathogenic viruses isolated in 2013 were not able to replicate efficiently in ducks (Pantin-Jackwood et al., 2014; Zhang et al., 2013). However, during our 2017 surveillance, both H7N9 and H7N2 highly pathogenic viruses were isolated from samples collected from ducks. We therefore tested the replication and virulence of the H7N2 virus and seven H7N9 viruses of genotypes 1, 2, and 8 in ducks.
The eight viruses had distinct replication and lethality in ducks (Table 2). Three H7N9 viruses, EN/GD/S12412/2017, CK/GD/SD032/2017, and CK/GD/S12178/2017, were detected in the pharyngeal swabs of four or five of the eight ducks inoculated, and they were not detected in the cloacal swabs or any organs tested. Moreover, only one or two of the five ducks in the groups that were inoculated with these three viruses seroconverted (Table 2). The CK/HuN/S1220/2017, DK/HuN/S1325/2017, and DK/GX/S21445/2017 viruses were detected in the pharyngeal swabs and cloacal swabs of some ducks, and low virus titers were also detected in some organs of one or two ducks inoculated with these viruses (Table 2). All of the ducks inoculated with these three viruses seroconverted (Table 2). The DK/FJ/SD208/2017 virus was detected in the pharyngeal swabs of all eight ducks inoculated and in the cloacal swabs of five of the eight inoculated ducks. This virus was also detected in the lungs, spleens, kidneys, and brains of all three ducks tested. Two ducks died and the three that survived all seroconverted (Table 2). The H7N2 virus DK/FJ/SE0195/2018 replicated efficiently in ducks. It was detected in the pharyngeal and cloacal swabs of all eight ducks inoculated and in the organs of all three ducks that were euthanized. All five remaining ducks died within seven days of virus inoculation (Table 2).
Table 2.
Replication and virulence of the H7N9 and H7N2 influenza viruses in ducksa.
| Virus (Genotype) | Virus shedding on day 3 p.i. (log10 EID50) (positive/total)b | Virus titer in organ on day 3 p.i. (log10 EID50) (positive/total)b | Death/Total | Serocon -version (Positive /total) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Pharynx | Cloacae | Lung | Liver | Spleen | Kidney | Brain | |||
| EN/GD/S12412/2017 (G1) | 2.8±0.5 (4/8) | < | < | < | < | < | < | 0/5 | 2/5 |
| CK/GD/SD032/2017 (G2) | 2.2±0.4 (5/8) | < | < | < | < | < | < | 0/5 | 2/5 |
| CK/GD/S12178/2017 (G2) | 2.6±0.8 (4/8) | < | < | < | < | < | < | 0/5 | 1/5 |
| CK/HuN/S1220/2017 (G2) | 2.8±0.9 (8/8) | 0.8 (1/8) | 0.8 (1/3) | < | 1.5 (1/3) | 0.8 (1/3) | < | 0/5 | 5/5 |
| DK/HuN/S1325/2017 (G1) | 1.9±0.5 (3/8) | 1.3 (1/8) | < | 0.8±0 (2/3) | < | 0.8 (1/3) | < | 0/5 | 5/5 |
| DK/GX/S21445/2017 (G2) | 2.4±0.7 (4/8) | 1.3+0 (2/8) | 0.8 (1/3) | 1.3 (1/3) | < | 0.8 (1/3) | < | 0/5 | 5/5 |
| DK/FJ/SD208/2017 (G8) | 3.5±1.1 (8/8) | 2.7±0.7 (5/8) | 4.0±1.4 (3/3) | 2.5±0.9 (3/3) | 3.2±1.2 (3/3) | 3.6±1.9 (3/3) | 4.7±0.9 (3/3) | 2/5c | 3/3 |
| DK/FJ/SE0195/2018 (H7N2) (G9) | 4.0±0.8 (8/8) | 2.1±0.9 (8/8) | 5.4±0.6 (3/3) | 5.1±0.6 (3/3) | 3.7±0.1 (3/3) | 5.7±0.1 (3/3) | 4.9±0.7 (3/3) | 5/5d | / |
Groups of eight three-week-old specific-pathogen-free ducks were inoculated intranasally with 106 50% egg infectious dose (EID50) of each virus in a 0.1-ml volume. Pharyngeal and cloacal swabs were collected from all birds on day 3 post inoculation (p.i.), and then three birds in each group were euthanized, and their organs were collected for virus titration in eggs. The remaining five birds in each group were observed for survival for two weeks. <, virus was not detected from the undiluted samples; /, all birds in that group died before the scheduled serum collection at two weeks after virus inoculation.
Data are mean ± standard deviation of the values from the birds that had detectable viruses.
The two birds died on day 4 p.i. and day 6 p.i. respectively.
The birds died within 7 days of infection; mean death time = 5.4 days.
These results indicate that six of the seven H7N9 viruses we tested here replicated poorly and were not lethal in ducks, but that the H7N9 virus DK/FJ/SD208/2017 and the H7N2 virus DK/FJ/SE0195/2018 replicated systemically and were lethal in ducks. Of note, these two duck-lethal viruses are reassortants of the chicken H7N9 viruses and some unknown duck virus(es) (Figure 2A). It is highly likely that the reassortant gene constellations favored the adaptation, replication, and lethality of these two viruses in ducks.
The H5/H7 bivalent inactivated vaccine provides protection against different H7 viruses in chickens and ducks
Because the H7N9 viruses mainly circulated in chickens, and most strains had limited replication in ducks, the H5/H7 bivalent inactivated vaccine was fully evaluated and mainly used in chickens. The protective efficacy of the H5/H7 bivalent inactivated vaccine in chickens against the challenge of three representative H7 viruses, including the H7N9 low pathogenic virus A/chicken/Chongqing/SD057/2017 (CK/CQ/SD057/2017), the H7N9 highly pathogenic virus A/chicken/Guangdong/SD008/2017 (CK/GD/SD008/2017) (the index strain) (Shi et al, 2017), and the H7N2 highly pathogenic virus DK/FJ/SE0195/2018 is shown in Figure 4. The HA genes of CK/CQ/SD057/2017, CK/GD/SD008/2017, and DK/FJ/SE0195/2018 share 97.8%, 98.3%, and 97.1% identity, respectively, with that of the H7N9 vaccine seed virus.
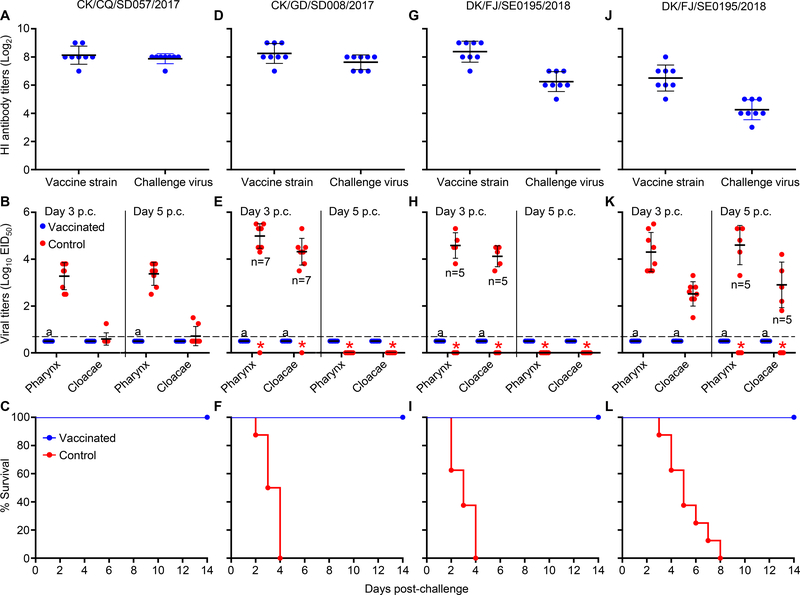
Figure 4. Protective efficacy of the H5/H7 bivalent inactivated vaccine against challenge with different H7 viruses in chickens and ducks.
Hemagglutinin inhibition (HI) antibody titers, virus shedding titers, and survival patterns of chickens challenged with the H7N9 low pathogenic virus CK/CQ /SD057/2017 are shown in panels A, B, and C, respectively, those of chickens challenged with the H7N9 highly pathogenic virus CK/GD/SD008/2017 are shown in panels D, E, and F, respectively, and those of chickens challenged with the H7N2 highly pathogenic virus DK/FJ/SE0195/2018 are shown in panels G, H, and I, respectively. The HI antibody titers, virus shedding titers, survival patterns of ducks challenged with the H7N2 highly pathogenic virus DK/FJ/SE0195/2018 are shown in panels J, K, and L, respectively. Virus titers shown in panels B, E, H, and K are the means ± standard deviations from the birds that survived. A value of 0.5 was assigned to virus shedding-negative birds for statistical purposes. The red asterisk indicates that the bird(s) died before that day, and therefore virus shedding data were not available for the statistical analysis. The number of birds that survived is shown in the panel when fewer than eight survived. The dashed lines in these panels indicate the lower limit of virus detection. a, p<0.001 compared with the corresponding titers of the control birds.
The mean HI antibody titers in the three groups of vaccinated chickens ranged from 8.1 log2 to 8.4 log2 against the H7N9 vaccine strain, and were 7.9 log2, 7.6 log2, and 6.3 log2 against CK/CQ/SD057/2017, CK/GD/SD008/2017, and DK/FJ/SE0195/2018, respectively (Figure 4A, D, G). The chickens in the control groups did not have detectable antibodies against these viruses (data not shown).
In the CK/CQ/SD057/2017 strain challenged groups, all eight control chickens shed virus through the pharynx with mean titers of 3.3 log10EID50 and 3.4 log10EID50 on days 3 and 5 post challenge (p.c.)., respectively, but only one and two chickens shed detectable virus through the cloacae on days 3 and 5 p.c., respectively. However, virus shedding was not detected from any of the vaccinated chickens (Figure 4B). The chickens in both the vaccinated and control groups remained healthy and survived for the duration of the observation period (Figure 4C).
In the CK/GD/SD008/2017 challenged groups, seven control chickens survived on day 3 p.c. and shed virus with mean titers of 5.0 log10EID50 and 4.3 log10EID50 in their pharyngeal and cloacal swabs, respectively (Figure 4E). In the DK/FJ/SE0195/2018 challenged groups, six control chickens survived on day 3 p.c. and shed virus with mean titers of 4.6 log10EID50 and 4.1 log10EID50 in their pharyngeal and cloacal swabs, respectively (Figure 4H). All of the control chickens challenged with these two viruses died within 4 days of challenge (Figure 4F, I). Virus shedding was not detected from any of the vaccinated chickens challenged with these two viruses, and all of the chickens remained healthy and survived (Figure 4F, I). These results indicate that the H5/H7 bivalent vaccine is immunogenic in chickens and provides sound protection in chickens against challenge with H7 low and highly pathogenic viruses.
Post-vaccination serological surveillance was performed from October 2017 until January 2018. Sera were collected from chickens in 251 farms, including 21 farms of fast-growing broilers (meat chickens that are usually slaughtered at 40 days of age) and 230 farms of layer chickens, breeders, and slow-growing meat chickens (locally bred meat chickens that are usually taken to the live poultry markets at 90–120 days of age). Our results showed that one of the 21 broiler farms had vaccinated with the H5/H7 bivalent vaccine and two of these farms had vaccinated with the H5 single vaccine. Among the 230 farms of other bird species, 168 farms (73.1%) had vaccinated with the H5/H7 bivalent vaccine, 47 farms had vaccinated with the H5 vaccine, and 15 farms had not effectively vaccinated with either the H5 or H5/H7 vaccine.
As shown in Table 2, some strains have acquired the ability to replicate efficiently and kill ducks; we therefore evaluated the protective efficacy of the H5/H7 bivalent inactivated vaccine in ducks. The mean HI antibody titers in the vaccinated ducks were 6.5 log2 and 4.3 log2 against the H7N9 vaccine strain and the H7N2 challenge virus, respectively (Figure 4J). The control ducks shed virus on both day 3 p.c. and day 5 p.c. (Figure 4K). The mean viral titers on day 3 p.c. were 4.3 log10EID50 and 2.5 log10EID50 in the pharyngeal swabs and cloacal swabs, respectively, and the mean titers on day 5 p.c. were 4.6 log10EID50 and 2.9 log10EID50 in the pharyngeal swabs and cloacal swabs, respectively. However, virus shedding was not detected from any of the vaccinated ducks. The ducks in the control group died within eight days of virus challenge, whereas all of the vaccinated ducks remained healthy and survived (Figure 4L). These results indicate that the H5/H7 bivalent inactivated vaccine is immunogenic in ducks and could provide complete protection of ducks from lethal challenge with H7N2 virus.
Discussion
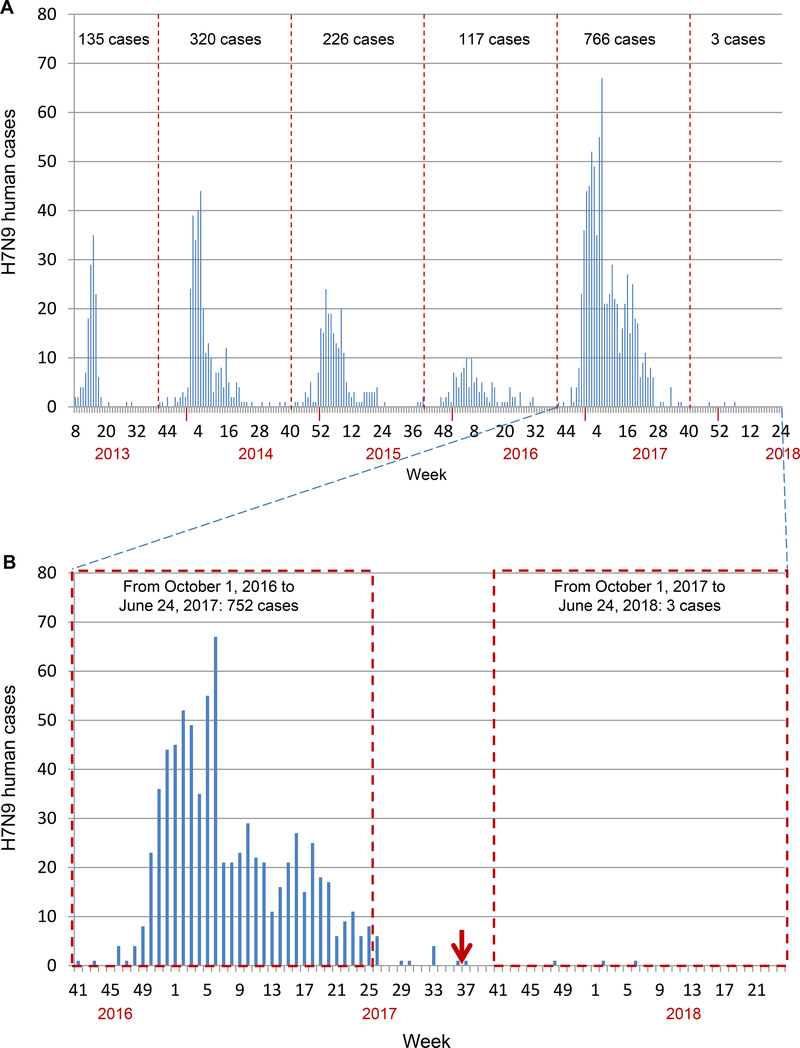
Our study here showed that the H7N9 viruses were widely prevalent in poultry markets and poultry farms in early 2017, but their prevalence dramatically decreased during our second round of surveillance. As previously reported, the H7N9 highly pathogenic influenza viruses are not only disastrous to poultry, but also pose an increased threat to humans (Imai et al., 2017; Shi et al., 2017); therefore, a series of actions were taken to control and eradicate the H7N9 viruses in China. In addition to the slaughter of poultry infected with H7N9 lethal viruses, an H5/H7 bivalent inactivated vaccine was fully evaluated in laboratories and farmed poultry in Guangdong, Guangxi, and Heilongjiang provinces, before being administered to poultry (mainly chickens) throughout China in September 2017. Serological surveillance performed from October 2017 to January 2018 in chicken farms indicated that, although the vaccination coverage in fast-growing meat chickens was very low and some chicken farms only vaccinated with the previously purchased H5 single vaccine, 73.1% of the 230 farms of layer chickens and slow-growing meat chickens were vaccinated with the H5/H7 bivalent vaccine. The decreased prevalence of the H7N9 viruses in poultry is direct evidence that the vaccine has played an important role in preventing H7N9 virus infection in poultry. Moreover, as shown in Figure 5, the H7N9 viruses have caused five waves of human infection since 2013, amongst which the second to fifth waves started around the beginning of October of each year (Figure 5A). There were 766 human cases detected between October 1, 2016 and September 30, 2017; in comparison, only three H7N9 human cases have been reported since October 1, 2017 (Figure 5B), indicating that the vaccination of poultry has prevented and eliminated the “sixth wave” of human infection with H7N9 virus in China.
Figure 5. Human infections with H7N9 viruses.
(A) The total H7N9 human case counts as of June 24, 2018. The dashed lines indicate the date of October 1st of each year. (B) The H7N9 human case counts from October 1, 2016 to June 24, 2018. The red arrow indicates when H5/H7 vaccine administration to poultry was initiated in China. The two dashed line boxes show the H7N9 human case counts for the two similar time periods before and after the poultry vaccine was used.
The virulence of influenza virus is a polygenic trait, and several important genetic markers in different genes have been reported to contribute to the virulence of influenza viruses in different hosts (Bussey et al., 2010; Fan et al., 2009; Feng et al., 2016; Hatta et al., 2001; Hu et al., 2013; Jiao et al., 2008; Li et al., 2005; Li et al., 2006; Mehle and Doudna, 2009; Song et al., 2011; Subbarao et al., 1993; Zhao et al., 2017; Zhu et al., 2008). Studies indicate that after replication in mammalian hosts, the H7N9 influenza viruses could easily obtain more mutations, primarily the PB2 627K or PB2 701N mutation, and then become more virulent in mammals (Mok et al., 2014; Shi et al., 2017; Zhang et al., 2014; Zhang et al., 2013). In this study, we found that, even without replication in any mammal, the H7N9 highly pathogenic viruses have become more virulent in mice. Given that previous studies indicated that the virulence of H5N1 virus in mice correlates with its virulence in humans (Gao et al., 1999; Gubareva et al., 1998; Lu et al., 1999), it is reasonable to speculate that the mouse-lethal H7N9 strains may also be more lethal in humans. The virulence difference in mice between the CK/GX/SD098/2017 and CK/HuN/S1220/2017 viruses was greater than 1000-fold (MLD50: >6.5 log10EID50 versus 3.2 log10EID50). These two viruses belong to the same genotype, bear the same motif in their HA cleavage site, and have the same amino acids at positions that have previously been reported to affect the virulence of influenza viruses in mice (Bussey et al., 2010; Fan et al., 2009; Feng et al., 2016; Hatta et al., 2001; Hu et al., 2013; Jiao et al., 2008; Li et al., 2005; Mehle and Doudna, 2009; Zhao et al., 2017). Therefore, these two viruses may be used as models to explore the unidentified molecular markers that contribute to the virulence of influenza viruses.
Gene fragment reassortment is a major mechanism for influenza virus evolution. Eight different genotypes of H7N9 highly pathogenic viruses and an H7N2 highly pathogenic virus have been generated in nature in a relatively short time period. Three genotypes (G5, G6, and G7) of H7N9 viruses were detected in humans only. It is highly likely that other similar reassortants have occurred in poultry, but were not detected by us; although we cannot rule out the possibility that these reassortants may have originated in humans, given that multiple H9N2 humans cases have been reported (WHO/GIP, 2018b), and that H9N2 influenza viruses readily infect humans as we have previously reported (Li et al., 2014). The eight genotypes of the H7N9 viruses were generated in Guangdong, Guangxi, Hunan, and Fujian provinces from January to May of 2017, indicating that the poultry were actively co-infected by H7N9 highly pathogenic viruses and other viruses during that period.
The H7N9 low pathogenic viruses have been circulating in chickens for several years, but they were unable to replicate in and adapt to ducks. However, our study here indicates that the H7N9 highly pathogenic virus has extended its host range by acquiring genes from duck influenza viruses and has now adapted to ducks. Although our data thus far indicate that these adapted viruses are only found in ducks in Fujian province (Figure 2B), it is a concern that these viruses will spread widely sooner or later, because many ducks are reared in open fields and traded through the live poultry market system in China. These ducks will, therefore, have the opportunity to spread the viruses to different locations and to other avian species, including the wild birds that they come into contact with. Therefore, the adaptation and circulation of H7N9 and H7N2 lethal viruses in ducks will pose more challenges to the control of avian influenza in China.
Vaccines have been used in poultry to prevent H5 influenza virus infection in many countries, including China (Chen and Bu, 2009; Li et al., 2014; Swayne, 2012; Swayne et al., 2011). This strategy will only be successful if the vaccine is antigenically well matched with the target viruses and the vaccination coverage reaches at least 70% (van der Goot et al., 2005). Our serological surveillance over the past few years has revealed that H5 vaccination coverage in ducks is very low (about 30%), but it could easily reach 70% in chickens. Therefore, the viruses can be controlled or even eradicated when their replication is confined to chickens. One successful example is the eradication of the clade 7.2 H5 viruses in China. The clade 7.2 viruses only replicated and circulated in chickens (Li et al., 2010; Liu et al., 2016), and were completely eradicated shortly after the H5N1 Re-7 vaccine (Liu et al., 2016) was widely used in China. A second example is the dramatic decrease in H7N9 virus prevalence after widespread application of the H5/H7 bivalent vaccine. Our study showed that H7N9 and H7N2 highly pathogenic viruses have been detected in ducks and that the H5/H7 bivalent vaccine is immunogenic and could provide solid protection against a lethal challenge by H7 virus in ducks. Immediate application of this vaccine to ducks would be required to speed the process of H7N9 and H7N2 virus control and eradication.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for laboratory resources and reagents should be directed to and will be fulfilled by the corresponding author, Hualan Chen (chenhualan@caas.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Eggs
Embryonated chicken eggs obtained from Harbin Weike Biotechnology Development Company were incubated at 37°C and 80% humidity for 10 days before being used for virus isolation or titration.
Animals
All experiments with animals were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. The protocols were approved by the Committee on the Ethics of Animal Experiments of the HVRI of the CAAS.
Mice
Five-week-old female BALB/c mice were purchased from Vital River Laboratories, Beijing, China and housed in ventilated cages (maxima eight mice per cage) in the enhanced animal biosafety level 3 (ABSL3+) facility at the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS). The mice were maintained on a 12/12-hour light/dark cycle, 22–26°C, and 40%–50% relative humidity. They were six weeks old at the time of infection and were checked twice a day for disease and death during the infection studies.
Chickens
Two-week-old female specific-pathogen-free (SPF) chickens (White Leghorn) were obtained from the Experimental Animal Division of HVRI and housed in ventilated isolators (maxima eight chickens per isolator) in the enhanced animal biosafety level 3 (ABSL3+) facility at the HVRI of the CAAS. The chickens were maintained on a 14/10-hour light/dark cycle, 22–26°C, and 40%–50% relative humidity. They were three weeks old at the time of vaccination and were checked twice a day for disease and death after challenge.
Ducks
Two-week-old female SPF ducks (Shaoxin shelduck, a local bred) were obtained from the Experimental Animal Division of HVRI and housed in ventilated isolators (maxima eight ducks per isolator) in the enhanced animal biosafety level 3 (ABSL3+) facility at the HVRI of the CAAS. The ducks were maintained on a 14/10-hour light/dark cycle, 22–26°C, and 40%–50% relative humidity. They were three weeks old at the time of infection or vaccination and were checked twice a day for disease and death after live virus inoculation.
METHOD DETAILS
Sample collection and virus isolation
We performed two rounds of large scale active influenza virus surveillance, from February 2017 to January 2018, during which 53,884 samples from live poultry markets and poultry farms were collected in 26 provinces in China (Table 1). Each sample was placed in 2 mL of minimal essential medium supplemented with penicillin (2000 U/mL) and streptomycin (2000 U/mL). The samples were collected and maintained in 2–8°C, shipped to the laboratory in sealed containers, and inoculated into eggs within 72 hours of collection. We also received organs of dead chickens from nine different chicken farms for disease diagnosis. All of the individual samples were inoculated into 10-day-old embryonated chicken eggs for 48 h at 37 °C. The allantoic fluid was collected and tested for HA activity with 0.5% chicken red blood cells. When the HA assay was positive, hemagglutinin inhibition (HI) assays were performed by using antisera against the antigens of 16 HA subtypes of avian influenza viruses and Newcastle disease virus, another avian virus frequently isolated from avian species. The NA subtypes of the viruses were determined by direct sequencing as reported by Alvarez et al. (Alvarez et al., 2010). A portion of the HA gene that included the cleavage site was sequenced to differentiate the virulence of the H7 strains.
Serum samples were also collected from 251 chicken farms (10 chickens per farm) when we collected the samples for virus isolation between October 2017 and January 2018. Antibody titers of these serum samples were tested by means of the HI assay using the H5N1 antigen and H7N9 antigen supplied by the Harbin Weike Biotechnology Development Company (Harbin, China). Chicken farms were deemed to be vaccinated when at least seven chickens had HI antibody titers and the mean titer of the ten chickens was ≥4 log2.
Sequence and phylogenetic analysis
The HA gene of 24 low pathogenic H7N9 viruses that were isolated from different locations in 2017 and the full genome of 36 H7N9 viruses and one H7N2 highly pathogenic virus isolated between 2017 and 2018 were sequenced in this study. The viral RNA of the H7N9 viruses was extracted from virus-infected allantoic fluid by using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany). RT-PCR was performed with a set of gene-specific primers, and the products were sequenced on an Applied Biosystems DNA analyzer. The nucleotide sequences were edited using the Seqman module of the DNAStar package.
We performed the phylogenetic analysis using the Mega 6.0.6 ClustalW software package, implementing the neighbor-joining method. For this analysis, in addition to the HA gene of the 24 low pathogenic H7N9 viruses isolated in 2017 and the full genome of the 36 H7N9 and one H7N2 highly pathogenic viruses we sequenced in this study, we also downloaded the HA gene of 897 H7N9 low pathogenic viruses and the full genome sequences of 44 H7N9 highly pathogenic viruses from the public database that were previously submitted by us and others (Shi et al., 2017; Yang et al., 2017). The trees of the other genes were developed from the genes of the 80 H7N9 highly pathogenic viruses and the H7N2 virus. The tree topology was evaluated by 1,000 bootstrap analyses; 95% sequence identity cut-offs were used to categorize the clusters of each gene segment in the phylogenetic trees.
Animal studies
Randomization and blinding were not used for the allocation of animals to experimental groups.
Mouse study
Six-week-old female BALB/c mice (Vital River Laboratories, Beijing, China) were used in this study. To assess virus replication and virulence, groups of mice (n=8) were lightly anesthetized with CO2 and inoculated intranasally (i.n.) with 106 50% egg infectious dose (EID50) of the test virus in a volume of 50 μl. Three mice in each group were euthanized on day 3 post inoculation (p.i.) and their nasal turbinates, lungs, spleens, kidneys, and brains were collected and titrated for virus infectivity in eggs. The other five mice in each group were monitored for up to 14 days for mortality. The 50% mouse lethal dose values of the six viruses that killed all of the mice at the dose of 106EID50 were determined by inoculating groups of five mice with 10-fold serial dilutions containing 101 to 106 EID50 of each virus in a volume of 50 μl. The mice were monitored for 14 days for mortality.
Duck study
To determine the replication and pathogenicity of the viruses in ducks, groups of three-week-old female SPF ducks (Shaoxin shelduck, a local bred) (n=8) were inoculated i.n. with 106 EID50 of each virus in a 0.1-ml volume. Pharyngeal and cloacal swabs were collected from all birds on day 3 p.i., and then three birds in each group were euthanized, and their organs, including lungs, livers, spleens, kidneys, and brains, were collected for virus titration in eggs. The other five birds in each group were observed for survival for two weeks. Sera were collected from the surviving birds at the end of the observation period to test for seroconversion by using the HI assay.
Vaccine tests in chickens and ducks
The H5/H7 bivalent vaccine (lot#: 2017002) was supplied by the Harbin Weike Biotechnology Development Company (Harbin, China). It is a formalin-inactivated oil-emulsion vaccine, with three parts inactivated allantonic fluid emulsified in two parts paraffin oil (volume/volume). The H5 seed virus (Re-8) contains the HA and NA genes from the clade 2.3.4.4 virus A/chicken/Guizhou/4/2013 (H5N1) and its six internal genes from the high-growth A/Puerto Rico/8/1934(H1N1) (PR8) virus and has been used in China since 2015. The H7 seed virus (H7-Re1) is a reassortant bearing the HA and NA genes of the H7N9 low pathogenic virus A/pigeon/Shanghai/1069/2013 and the six internal genes of PR8.
To evaluate the protective efficacy of the H5/H7 vaccine in chickens and ducks, groups of three-week-old female SPF chickens (White Leghorn) (n=8) or female SPF ducks (n=8) were inoculated intramuscularly with 0.3 ml (chickens) or 0.5 ml (ducks) of the vaccine or with equal volume of PBS as a control. Three weeks post-vaccination, serum was collected from the birds for HI antibody testing. The chickens in each group were then challenged i.n. with 106EID50 of the H7N9 low pathogenic virus CK/CQ/SD057/2017, the H7N9 highly pathogenic virus CK/GD/SD008/2017 (Shi et al., 2017), or the H7N2 highly pathogenic virus DK/FJ/SE0195/2018. The ducks were challenged with the H7N2 virus DK/FJ/SE0195/2018. Pharyngeal and cloacal swabs were collected from all of the surviving birds on days 3 and 5 post challenge and titrated in eggs. The birds were observed for signs of disease and death for two weeks.
QUANTIFICATION AND STATISTICAL ANALYSIS
Virus titers of control and vaccinated birds were statistically analyzed by using the one-tailed unpaired t-test. A value of 0.5 was assigned to the virus shedding-negative birds for statistical purposes. The mean virus titers and standard deviations were calculated from the samples of birds that survived at each time points (day 3 and day 5 p.c.). The data from each time point were analyzed separately, as were those of the different challenge strains. The number of birds that survived in each of the four vaccinated groups at the two time points was eight. The number of chickens that survived at the two time points in the control group that was challenged with CK/CQ/SD057/2017 was eight; the numbers of chickens that survived in the control group that was challenged with CK/GD/SD008/2017 was seven and zero on day 3 p.c. and day 5 p.c., respectively; the numbers of chickens that survived in the control group that was challenged with DK/FJ/SE0195/2018 was five and zero on day 3 p.c. and day 5 p.c., respectively; the numbers of ducks that survived in the control group that was challenged with DK/FJ/SE0195/2018 was eight and five on day 3 p.c. and day 5 p.c., respectively. P values of < 0.05 were considered significant.
We did not use any methods to determine whether the data met the assumptions of the statistical approach.
Biosafety statement and facility
Routine surveillance samples were processed in the enhanced biosafety level 2 (BSL2+) facility in the HVRI of the CAAS. Our staff wear gloves, N95 masks, and disposable coveralls when working in the facility, and all waste is autoclaved before being removed from the facility. The diagnosis of H7N9 and all experiments with live H7N9 viruses were conducted in the enhanced animal biosafety level 3 (ABSL3+) facility in the HVRI of the CAAS, which is approved for such use by the Ministry of Agriculture and Rural Affairs of China. All animal studies were approved by the Review Board of the HVRI, CAAS.
DATA AND SOFTWARE AVAILABILITY
Genome sequences generated in this study are publicly available in the GenBank database under the accession numbers: MH209256-MH209575.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| The swabs and environmental samples for virus surveillance collected from different live poultry markets and poultry farms in China (see Table 1). | This study | N/A |
| The serum samples from chickens for antibody detection collected from different chicken farms in China. | This study | N/A |
| Antiserum | ||
| Antiserum of H1–H16 subtype avian influenza viruses | National Avian Influenza Reference Laboratory, Harbin, China | N/A |
| Bacterial and Virus Strains | ||
| A/chicken/Guangdong/SD008/2017(H7N9) | Shi et al., 2017 | N/A |
| The 1,775 strains of the different subtypes of avian influenza viruses (see Table 1 and Table S1). | This study | N/A |
| Deposited Data | ||
| The HA gene of 24 low pathogenic H7N9 viruses that were isolated from different locations in 2017 and the full genome of 36 H7N9 viruses and one H7N2 highly pathogenic virus isolated between 2017 and 2018 | This study | National Center for Biotechnology Information (NCBI) Genbank: MH209256 to MH209575. |
| The full genome sequences of 44 H7N9 highly pathogenic viruses | Shi et al., 2017 | NCBI Genbank: MF630034-MF630049, MF630106-MF630121, MF630130-MF630153. |
| Database of GISAID | Database of GISAID: EPI917062-EPI917069, EPI918730, EPI918732, EPI918734, EPI918736–918740, EPI919592-EPI919607, EPI960354-EPI960369, EPI973363-EPI973370, EPI1018236-EPI1018251, EPI1018193, EPI1018196–1018197, EPI1018199–1018203, EPI1018260-EPI1018267, EPI997214-EPI997221, EPI1013218-EPI1013273, EPI1018081-EPI1018176, EPI1022610-EPI1022633, EPI1022650-EPI1022673. |
|
| The HA gene of 897 H7N9 low | Shi et al., 2013 | Database of Global Initiative on Sharing All |
| pathogenic viruses | Influenza Data (GISAID): EPI440685, EPI440693, EPI440701. | |
| Zhang et al., 2013 | NCBI Genbank: CY146908, CY146916, CY146924, CY146932, CY146940, CY146948, CY146964, CY146972, CY146980, CY146988, CY146996, CY147004, CY147012, CY147020, CY147028, CY147036, CY147044, CY147052, CY147060, CY147068, CY147076, CY147084, CY147092, CY147100, CY147108, CY147116, CY147132, CY147140, CY147148, CY147156, CY147164, CY147180, CY147188, CY147196. |
|
| Shi et al., 2017 | NCBI Genbank: MF629925, MF629933, MF629941, MF629949, MF629957, MF629965, MF629973, MF629981, MF629989, MF629997, MF630005, MF630013, MF630021, MF630029, MF630053, MF630061, MF630069, MF630077, MF630085, MF630093, MF630101, MF630125, MF630157, MF630165, MF630173, MF630181, MF630189, MF630197, MF630205, MF630213, MF630221, MF630229, MF630237, MF630245, MF630253, MF630261, MF630269, MF630277, MF630285, MF630293, MF630301, MF630309, MF630317, MF630325, MF630333, MF630341, MF630349, MF630357, MF630365, MF630373, MF630381, MF630389, MF630397, MF630405, MF630413, MF630421, MF630429, MF630461, MF630469, MF630477, MF630485, MF630493, MF630517, MF630541, MF630549, MF630557, MF630573. |
|
| Database of GISAID: https://platform.gisid.org/ |
Database of GISAID: EPI453604, EPI453609, EPI560398, EPI621120, EPI621127, EPI682919, EPI682920, EPI682921, EPI682922, EPI682923, EPI682924, EPI460751, EPI460767, EPI460775, EPI450526, EPI636673, EPI636680, EPI636687, EPI636694, EPI636701, EPI636708, EPI636715, EPI636722, EPI636730, EPI636737, EPI636745, EPI636752, EPI636759, EPI636766, EPI636773, EPI636780, EPI636787, EPI636794, EPI636804, EPI636812, EPI636819, EPI636826, EPI636833, EPI636840, EPI636849, EPI636856, EPI636863, EPI636870, EPI636877, EPI636884, EPI636892, EPI636900, EPI636907, EPI636914, EPI636921, EPI636930, EPI636937, EPI636944, EPI636951, EPI636958, EPI636965, EPI636972, EPI636979, EPI636986, EPI636993, EPI637000, EPI637007, EPI637014, EPI637021, EPI637028, EPI637035, EPI637042, EPI637049, EPI637056, EPI637063, EPI637070, EPI637077, EPI637084, EPI637091, EPI637098, EPI637105, EPI637112, EPI637119, EPI637126, EPI637133, EPI637140, EPI637147, EPI637154, EPI637161, EPI637169, EPI637177, EPI637184, EPI637191, EPI637198, EPI637217, EPI637224, EPI637231, EPI637238, EPI637245, EPI637252, EPI637259, EPI637268, EPI637275, EPI637282, EPI637290, EPI637297, EPI637305, EPI637312, EPI637319, EPI637326, EPI637333, EPI637340, EPI637347, EPI637355, EPI637362, EPI637369, EPI637376, EPI637383, EPI637390, EPI637397, EPI637404, EPI637411, EPI637418, EPI637425, EPI637432, EPI637439, EPI637446, EPI637453, EPI637460, EPI637467, EPI637474, EPI637481, EPI637493, EPI637500, EPI637507, EPI637514, EPI637521, EPI637528, EPI637535, EPI637542, EPI637549, EPI637556, EPI637563, EPI637570, EPI637577, EPI637584, EPI637591, EPI637598, EPI637605, EPI637612, EPI637619, EPI637626, EPI637633, EPI637640, EPI637647, EPI637654, EPI637661, EPI637668, EPI637675, EPI637682, EPI637689, EPI637696, EPI637703, EPI637710, EPI637717, EPI637724, EPI637731, EPI637738, EPI637745, EPI637752, EPI637759, EPI637766, EPI637773, EPI637780, EPI637787, EPI637794, EPI637801, EPI637808, EPI637815, EPI637822, EPI637829, EPI637836, EPI637843, EPI637850, EPI637857, EPI637864, EPI637871, EPI637878, EPI637885, EPI637892, EPI637899, EPI637906, EPI637913, EPI637920, EPI637927, EPI637934, EPI637941, EPI637948, EPI637955, EPI637962, EPI637969, EPI637976, EPI637983, EPI637990, EPI637997, EPI638004, EPI638011, EPI638018, EPI638025, EPI638032, EPI638039, EPI638046, EPI638053, EPI638060, EPI638067, EPI638074, EPI638081, EPI638088, EPI638095, EPI638102, EPI515796, EPI531769, EPI531777, EPI531785, EPI531793, EPI505122, EPI503499, EPI476697, EPI497814, EPI497829, EPI497836, EPI497849, EPI497857, EPI497865, EPI497873, EPI497894, EPI497902, EPI497910, EPI497918, EPI498087, EPI498098, EPI504725, EPI515470, EPI515869, EPI515877, EPI515885, EPI469658, EPI469660, EPI577932, EPI577938, EPI578145, EPI581738, EPI581750, EPI581756, EPI581762, EPI581832, EPI581838, EPI581865, EPI581873, EPI581916, EPI581922, EPI581972, EPI581991, EPI582340, EPI582347, EPI582354, EPI582361, EPI582937, EPI582944, EPI582959, EPI582966, EPI582983, EPI583007, EPI583014, EPI583021, EPI583072, EPI583088, EPI583095, EPI583102, EPI583108, EPI583115, EPI583122, EPI583130, EPI583137, EPI583144, EPI583151, EPI583166, EPI583184, EPI583191, EPI583207, EPI583214, EPI583893, EPI583899, EPI583905, EPI583911, EPI583918, EPI583925, EPI583932, EPI583939, EPI591857, EPI592024, EPI592031, EPI592038, EPI592045, EPI592052, EPI592059, EPI592073, EPI592080, EPI592087, EPI592094, EPI592101, EPI592108, EPI592115, EPI592122, EPI592129, EPI592136, EPI592151, EPI592158, EPI592165, EPI592179, EPI592216, EPI592223, EPI592230, EPI592244, EPI592251, EPI592258, EPI592281, EPI592288, EPI592312, EPI592319, EPI592326, EPI592333, EPI592340, EPI592347, EPI592354, EPI592361, EPI592368, EPI592375, EPI592382, EPI592389, EPI592396, EPI592403, EPI592410, EPI592418, EPI592425, EPI592432, EPI592439, EPI592446, EPI592453, EPI592460, EPI592476, EPI592484, EPI592508, EPI592515, EPI592547, EPI592554, EPI592561, EPI592577, EPI592585, EPI592592, EPI592599, EPI592618, EPI592625, EPI592632, EPI592639, EPI592645, EPI592661, EPI593146, EPI593160, EPI593167, EPI593174, EPI593244, EPI593278, EPI593381, EPI593405, EPI593469, EPI593497, EPI593541, EPI593548, EPI594136, EPI594143, EPI594150, EPI594157, EPI594164, EPI594171, EPI594178, EPI594185, EPI594192, EPI594199, EPI594206, EPI594213, EPI594227, EPI594234, EPI594241, EPI594248, EPI594255, EPI594262, EPI594269, EPI594276, EPI594290, EPI594297, EPI594304, EPI594311, EPI594318, EPI594325, EPI594332, EPI594339, EPI594346, EPI594353, EPI597387, EPI597393, EPI597399, EPI597421, EPI597427, EPI597496, EPI440095, EPI443635, EPI443643, EPI443651, EPI443659, EPI443667, EPI443675, EPI450842, EPI450850, EPI545795, EPI442710, EPI442713, EPI442716, EPI573429, EPI490971, EPI490979, EPI502373, EPI507087, EPI509888, EPI490882, EPI498800, EPI553470, EPI559417, EPI566672, EPI535131, EPI535139, EPI535147, EPI535155, EPI535163, EPI535171, EPI535179, EPI535187, EPI535195, EPI535203, EPI576564, EPI576565, EPI576566, EPI576567, EPI576568, EPI576569, EPI467305, EPI467313, EPI467345, EPI468967, EPI469524, EPI580267, EPI580275, EPI580283, EPI580291, EPI580299, EPI580313, EPI580327, EPI580337, EPI580347, EPI580355, EPI580363, EPI580371, EPI580379, EPI580387, EPI471834, EPI471835, EPI521915, EPI521917, EPI541774, EPI530824, EPI477307, EPI477308, EPI531468, EPI507147, EPI542311, EPI448936, EPI639587, EPI443034, EPI443042, EPI477410, EPI552399, EPI439486, EPI439502, EPI439507, EPI443022, EPI443025, EPI443028, EPI447596, EPI447598, EPI447599, EPI447600, EPI447601, EPI447602, EPI447603, EPI447604, EPI447605, EPI447607, EPI447609, EPI447610, EPI447611, EPI447612, EPI447613, EPI447614, EPI447615, EPI447617, EPI447618, EPI447619, EPI447620, EPI447622, EPI447623, EPI447624, EPI447625, EPI447626, EPI447628, EPI447629, EPI447630, EPI509062, EPI509070, EPI509071, EPI509086, EPI509087, EPI509102, EPI509103, EPI509118, EPI509120, EPI509135, EPI509136, EPI509151, EPI510157, EPI528298, EPI528338, EPI528346, EPI528354, EPI528362, EPI528370, EPI457433, EPI457436, EPI528378, EPI566036, EPI566052, EPI566060, EPI566068, EPI566076, EPI566084, EPI566092, EPI566100, EPI566108, EPI566116, EPI566124, EPI626985, EPI626993, EPI627001, EPI627009, EPI627017, EPI627025, EPI627033, EPI627041, EPI627049, EPI627081, EPI627089, EPI627097, EPI627105, EPI627121, EPI627129, EPI627137, EPI627145, EPI627153, EPI627161, EPI627169, EPI627177, EPI627185, EPI627193, EPI627201, EPI627209, EPI627217, EPI627225, EPI627233, EPI627241, EPI627249, EPI627257, EPI627273, EPI627297, EPI627305, EPI627313, EPI627321, EPI627329, EPI627345, EPI627353, EPI627361, EPI627369, EPI627385, EPI627393, EPI627401, EPI627409, EPI627417, EPI627425, EPI627433, EPI627441, EPI627449, EPI627457, EPI627465, EPI627473, EPI627497, EPI627505, EPI627513, EPI627521, EPI627529, EPI627537, EPI627545, EPI627553, EPI627561, EPI627569, EPI627577, EPI627585, EPI627593, EPI627601, EPI627609, EPI627617, EPI627641, EPI627657, EPI627673, EPI627681, EPI627697, EPI627705, EPI627713, EPI627729, EPI627737, EPI627745, EPI627753, EPI627769, EPI627777, EPI627792, EPI627800, EPI627808, EPI627816, EPI627824, EPI627832, EPI627840, EPI627864, EPI627872, EPI627880, EPI627888, EPI627896, EPI627904, EPI627912, EPI627920, EPI627928, EPI627936, EPI627944, EPI627952, EPI627960, EPI627968, EPI627976, EPI627984, EPI627992, EPI628000, EPI628008, EPI628016, EPI628024, EPI628032, EPI628040, EPI628048, EPI628056, EPI628064, EPI628072, EPI628080, EPI628088, EPI628104, EPI628112, EPI628120, EPI628128, EPI628136, EPI628144, EPI628152, EPI628160, EPI628168, EPI628176, EPI628184, EPI628192, EPI628200, EPI628208, EPI628216, EPI628224, EPI628232, EPI628240, EPI628248, EPI628264, EPI628272, EPI628280, EPI628288, EPI628296, EPI628304, EPI628312, EPI628320, EPI628328, EPI628336, EPI628344, EPI628352, EPI628360, EPI628368, EPI628376, EPI628384, EPI628392, EPI628400, EPI628408, EPI628416, EPI628424, EPI628432, EPI628440, EPI628448, EPI628456, EPI628464, EPI628472, EPI628480, EPI628488, EPI628504, EPI628512, EPI628520, EPI628528, EPI628544, EPI628552, EPI628560, EPI628576, EPI628584, EPI628592, EPI628600, EPI628608, EPI628624, EPI628632, EPI628640, EPI628648, EPI628656, EPI628664, EPI628672, EPI628680, EPI628688, EPI628696, EPI628704, EPI628712, EPI628720, EPI628728, EPI628736, EPI628744, EPI628760, EPI628768, EPI628776, EPI628784, EPI628792, EPI628808, EPI628816, EPI628824, EPI628832, EPI531118, EPI613769, EPI613776, EPI613783, EPI613790, EPI613807, EPI613814, EPI613821, EPI613828, EPI613835, EPI620025, EPI620032, EPI620039, EPI620046, EPI620053, EPI620060, EPI620064, EPI620071, EPI620078, EPI620085, EPI620092, EPI620104, EPI620111, EPI620118, EPI620128, EPI620135, EPI620142, EPI620149, EPI620153, EPI620160, EPI620167, EPI620174, EPI620178, |
|
| EPI620185, EPI620192, EPI620199. | ||
| Critical Commercial Assays | ||
| QIAamp Viral RNA Mini Kit | Qiagen | Cat#: 52904 |
| Reverse Transcriptase M-MLV (RNase H-) | TaKaRa | Cat#: 2641B |
| EasyTaq DNA Polymerase | Transgen | Cat#: AP111 |
| Cycle Pure Kit | Omega | Cat#: D6492 |
| BigDye Terminator v3.1 Cycle Sequencing Kit | Thermo-Fisher Scientific | Cat#: 4337457 |
| BigDye Sequencing Clean Up Kit | MCLAB | Cat#: BCB-300 |
| Oligonucleotides | ||
| Reverse transcription primer: 5’-AGCRAAAGCAGG | This study | N/A |
| Amplification Primers | ||
| NA-F-M13: 5’-GTAAAACGACGGCCAGTGRACHCA RGARTCIKMRTG-3’ | Alvarez et al., 2010 | N/A |
| NA-R-M13: 5’-CAGGAAACAGCTATGACCCIIKCCA RTTRTCYCTRCA-3’ | Alvarez et al., 2010 | N/A |
| H7N9-HA-F: 5’-AGCAAAAGCAGGGGATACAAA-3’ | This study | N/A |
| H7N9-HA-R: 5’-AGTAGAAACAAGGGTGTTTTTTYC-3’ | This study | N/A |
| H7N9-N9NA-F: 5’-AAAAGCAGGGTCAAGATGAAT-3’ | This study | N/A |
| H7N9-N9NA-R: 5’-CTTTTTTCTGCGTCTTAGAGGAAG-3’ | This study | N/A |
| H7N2-N2-F: 5’-AGCAAAAGCAGGAGTAAAAATG-3’ | This study | N/A |
| H7N2-N2-R: 5’-TTAGTAGAAACAAGGAGTTTTTTC TAAA-3’ | This study | N/A |
| Other primers: see Table S3 and Table S4 | This study | N/A |
| Software and Algorithms | ||
| Dnastar 6 | DNASTAR, Inc. | http://www.dnastar.com/ |
| Mega 6.0.6 | Mega | https://www.megasoftware.net/mega6/ |
| Animals | ||
| Mice: BALB/c | Vital River Laboratories, Beijing, China | Certificate #: 11400700254408 and 11400700282889 |
| Specific-pathogen-free (SPF) chickens (White Leghorn) | Experimental Animal Division of Harbin Veterinary Research Institute (EAD of HVRI), | Bunch #: 20170317 and 20180224 |
| Harbin, China | ||
| SPF ducks (Shaoxin shelduck) | EAD of HVRI Harbin, China | Bunch #: 20180126 |
| Others | ||
| H5/H7 bivalent inactivated vaccine | Harbin Weike Biotechnology Development Company (HWBDC), Harbin, China | Lot #: 2017002 |
| Embryonated chicken eggs | HWBDC Harbin, China | N/A |
| Chicken red blood cells | HWBDC Harbin, China | N/A |
| H5N1 antigen | HWBDC Harbin, China | Lot #: 2017012 |
| H7N9 antigen | HWBDC Harbin, China | Lot #: 2017001 |
Highlights.
In 2017 across China, H7N9 HPAI viruses evolved into different genotypes.
H7N9 and H7N2 HPAI reassortants are well adapted and lethal in ducks.
An H5/H7 vaccine induced solid protection against H7 viruses in poultry.
Acknowledgments
We thank Susan Watson and Bruce Yi Bu for editing the manuscript. This work was supported by the National Key R&D Program of China (2016YFD0500201, 2016YFD0500203, and 2016YFC1202402), the National Natural Science Foundation of China (31521005), the China Agriculture Research System (CARS-41-G12), and the U.S. NIH CEIRS contract HHSN272201400004C. Virus sequence data from this study were deposited in GenBank with the accession numbers MH209256 to MH209575.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DJ, and Brown IH (2009). History of highly pathogenic avian influenza. Rev Sci Tech Oie. 28(1), 19–38. DOI: 10.20506/Rst.28.1.1856. [DOI] [PubMed] [Google Scholar]
- Alvarez AC, Boyd V, Lai R, Pineda S, Bletchly C, Heine HG, Barnard R (2010). Detection of Influenza A Virus Neuraminidase and PB2 Gene Segments by One Step Reverse Transcription Polymerase Chain Reaction In RT-PCR Protocols: Second Edition, Series Title Methods in Molecular Biology, King N, ed. (Springer Science + Business Media, LLC; ), pp. 65–81. [DOI] [PubMed] [Google Scholar]
- Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, et al. (2013). Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 501(7468), 556–559. DOI: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey KA, Bousse TL, Desmet EA, Kim B, and Takimoto T (2010). PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. Journal of virology. 84(9), 4395–4406. DOI: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H (2009). H5N1 avian influenza in China. Science in China. Series C, Life sciences Chinese Academy of Sciences. 52(5), 419–427. DOI: 10.1007/s11427-009-0068-6. [DOI] [PubMed] [Google Scholar]
- Chen H, and Bu Z (2009). Development and application of avian influenza vaccines in China. Current topics in microbiology and immunology. 333, 153–162. DOI: 10.1007/978-3-540-92165-3_7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, et al. (2013). Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 381(9881), 1916–1925. DOI: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Deng G, Song J, Tian G, Suo Y, Jiang Y, Guan Y, Bu Z, Kawaoka Y, and Chen H (2009). Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology. 384(1), 28–32. DOI: 10.1016/j.virol.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Feng X, Wang Z, Shi J, Deng G, Kong H, Tao S, Li C, Liu L, Guan Y, and Chen H (2016). Glycine at Position 622 in PB1 Contributes to the Virulence of H5N1 Avian Influenza Virus in Mice. Journal of virology. 90(4), 1872–1879. DOI: 10.1128/JVI.02387-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, et al. (2004). Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proceedings of the National Academy of Sciences of the United States of America. 101(5), 1356–1361. Published online 2004/01/28 DOI: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, and Kawaoka Y (1999). Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. Journal of virology. 73(4), 3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, McCullers JA, Bethell RC, and Webster RG (1998). Characterization of influenza A/HongKong/156/97 (H5N1) virus in a mouse model and protective effect of zanamivir on H5N1 infection in mice. J Infect Dis. 178(6), 1592–1596. DOI: 10.1086/314515. [DOI] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, and Kawaoka Y (2001). Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 293(5536), 1840–1842. DOI: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Hu J, Hu Z, Song Q, Gu M, Liu X, Wang X, Hu S, Chen C, Liu H, Liu W, et al. (2013). The PA-gene-mediated lethal dissemination and excessive innate immune response contribute to the high virulence of H5N1 avian influenza virus in mice. Journal of virology. 87(5), 2660–2672. DOI: 10.1128/JVI.02891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Kiso M, Nakajima N, Yamayoshi S, Iwatsuki-Horimoto K, Hatta M, Yamada S, Ito M, Sakai-Tagawa Y, et al. (2017). A Highly Pathogenic Avian H7N9 Influenza Virus Isolated from A Human Is Lethal in Some Ferrets Infected via Respiratory Droplets. Cell host & microbe. 22(5), 615–626 e618 DOI: 10.1016/j.chom.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, and Chen H (2008). A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. Journal of virology. 82(3), 1146–1154. DOI: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, et al. (2014). Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 Avian Influenza viruses. PLoS pathogens. 10(11), e1004508 DOI: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shi J, Zhong G, Deng G, Tian G, Ge J, Zeng X, Song J, Zhao D, Liu L, et al. (2010). Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. Journal of virology. 84(17), 8389–8397. DOI: 10.1128/JVI.00413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, and Yu K (2005). Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. Journal of virology. 79(18), 12058–12064. DOI: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang Y, Jiao P, Wang A, Zhao F, Tian G, Wang X, Yu K, Bu Z, and Chen H (2006). The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. Journal of virology. 80(22), 11115–11123. DOI: 10.1128/JVI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Zhang Z, He L, Gao Z, Li J, Gu M, Hu J, Wang X, Liu X, and Liu X (2018). Characteristics of the emerging chicken-origin highly pathogenic H7N9 viruses: A new threat to public health and poultry industry. The Journal of infection. 76(2), 217–220. DOI: 10.1016/j.jinf.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Liu L, Zeng X, Chen P, Deng G, Li Y, Shi J, Gu C, Kong H, Suzuki Y, Jiang Y, et al. (2016). Characterization of Clade 7.2 H5 Avian Influenza Viruses That Continue To Circulate in Chickens in China. Journal of virology. 90(21), 9797–9805. DOI: 10.1128/JVI.00855-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, and Katz JM (1999). A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. Journal of virology. 73(7), 5903–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, and Doudna JA (2009). Adaptive strategies of the influenza virus polymerase for replication in humans. Proceedings of the National Academy of Sciences of the United States of America. 106(50), 21312–21316. DOI: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MoA (2017). No. 2541 of the notice of the ministry of agriculture of the People’s Republic of China. The ministry of agriculture; http://www.moa.gov.cn/xzspdtdp/gg/201706/t20170616_25692181.htm. [Google Scholar]
- Mok CKP, Lee HHY, Lestra M, Nicholls JM, Chan MCW, Sia SF, Zhu HC, Poon LLM, Guan Y, and Peiris JSM (2014). Amino Acid Substitutions in Polymerase Basic Protein 2 Gene Contribute to the Pathogenicity of the Novel A/H7N9 Influenza Virus in Mammalian Hosts. Journal of virology. 88(6), 3568–3576. DOI: 10.1128/Jvi.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, and Kawaoka Y (2006). Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis. 12(6), 881–886. Published online 2006/05/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE (2011). Manual of diagnostic tests and vaccines for terrestrial animals. OIE - World Organisation for Animal Health. [Google Scholar]
- OIE (2018). Weekly Disease Information. OIE - World Organisation for Animal Health; http://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/WI/index/newlang/en. [Google Scholar]
- Pantin-Jackwood MJ, Miller PJ, Spackman E, Swayne DE, Susta L, Costa-Hurtado M, and Suarez DL (2014). Role of poultry in the spread of novel H7N9 influenza virus in China. Journal of virology. 88(10), 5381–5390. DOI: 10.1128/JVI.03689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Jia W, Liu D, Li J, Bi Y, Xie S, Li B, Hu T, Du Y, Xing L, et al. (2018). Emergence and Adaptation of a Novel Highly Pathogenic H7N9 Influenza Virus in Birds and Humans from a 2013 Human-Infecting Low-Pathogenic Ancestor. Journal of virology. 92(2), e00921–00917. DOI: 10.1128/JVI.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, et al. (2013). Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 501(7468), 560–563. DOI: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senne DA, Panigrahy B, Kawaoka Y, Pearson JE, Suss J, Lipkind M, Kida H, and Webster RG (1996). Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian diseases. 40(2), 425–437. Published online 1996/04/01. [PubMed] [Google Scholar]
- Shi J, Deng G, Kong H, Gu C, Ma S, Yin X, Zeng X, Cui P, Chen Y, Yang H, et al. (2017). H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell research. 27, 1409–1421. DOI: 10.1038/cr.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JZ, Deng GH, Liu PH, Zhou JP, Guan LZ, Li WH, Li XY, Guo J, Wang GJ, Fan J, et al. (2013). Isolation and characterization of H7N9 viruses from live poultry markets-Implication of the source of current H7N9 infection in humans. Chinese Sci Bull. 58(16), 1857–1863. DOI: 10.1007/s11434-013-5873-4. [DOI] [Google Scholar]
- Song J, Feng H, Xu J, Zhao D, Shi J, Li Y, Deng G, Jiang Y, Li X, Zhu P, et al. (2011). The PA protein directly contributes to the virulence of H5N1 avian influenza viruses in domestic ducks. Journal of virology. 85(5), 2180–2188. DOI: 10.1128/JVI.01975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, London W, and Murphy BR (1993). A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. Journal of virology. 67(4), 1761–1764. Published online 1993/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne DE (2012). Impact of vaccines and vaccination on global control of avian influenza. Avian diseases. 56(4 Suppl), 818–828. DOI: 10.1637/10183-041012-Review.1. [DOI] [PubMed] [Google Scholar]
- Swayne DE, Pavade G, Hamilton K, Vallat B, and Miyagishima K (2011). Assessment of national strategies for control of high-pathogenicity avian influenza and low-pathogenicity notifiable avian influenza in poultry, with emphasis on vaccines and vaccination. Revue scientifique et technique. 30(3), 839–870. [DOI] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, et al. (2012). A distinct lineage of influenza A virus from bats. Proceedings of the National Academy of Sciences of the United States of America. 109(11), 4269–4274. DOI: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. (2013). New world bats harbor diverse influenza A viruses. PLoS pathogens. 9(10), e1003657 DOI: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot JA, Koch G, de Jong MC, and van Boven M (2005). Quantification of the effect of vaccination on transmission of avian influenza (H7N7) in chickens. Proceedings of the National Academy of Sciences of the United States of America. 102(50), 18141–18146. DOI: 10.1073/pnas.0505098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, et al. (2013). Characterization of H7N9 influenza A viruses isolated from humans. Nature. 501(7468), 551–555. DOI: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO/GIP (2018a). Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2018. World Health Organization; http://www.who.int/influenza/human_animal_interface/2018_2003_2002_tableH2015N2011.pdf?ua=2011. [Google Scholar]
- WHO/GIP (2018b). Monthly Risk Assessment Summary. World Health Organization; http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_02_03_2018.pdf?ua=2011. [Google Scholar]
- Wong SS, and Yuen KY (2006). Avian influenza virus infections in humans. Chest. 129(1), 156–168. DOI: 10.1378/chest.129.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhu W, Li X, Chen M, Wu J, Yu P, Qi S, Huang Y, Shi W, Dong J, et al. (2017). Genesis and Spread of Newly Emerged Highly Pathogenic H7N9 Avian Viruses in Mainland China. Journal of virology. 91(23), e01277–01217. DOI: 10.1128/JVI.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li X, Guo J, Li L, Chang C, Li Y, Bian C, Xu K, Chen H, and Sun B (2014). The PB2 E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. The Journal of general virology. 95(Pt 4), 779–786. DOI: 10.1099/vir.0.061721-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, et al. (2013). H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 341(6144), 410–414. DOI: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- Zhao D, Liang L, Wang S, Nakao T, Li Y, Liu L, Guan Y, Fukuyama S, Bu Z, Kawaoka Y, et al. (2017). Glycosylation of the Hemagglutinin Protein of H5N1 Influenza Virus Increases Its Virulence in Mice by Exacerbating the Host Immune Response. Journal of virology. 91(7), e02215–02216. DOI: 10.1128/JVI.02215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, et al. (2013). Biological features of novel avian influenza A (H7N9) virus. Nature. 499(7459), 500–503. DOI: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Yang H, Chen W, Cao W, Zhong G, Jiao P, Deng G, Yu K, Yang C, Bu Z, et al. (2008). A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. Journal of virology. 82(1), 220–228. DOI: 10.1128/JVI.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.