Abstract
Candidatus Neoehrlichia mikurensis (Ca. N. mikurensis; family Anaplasmataceae) is an emerging tick-borne pathogen that causes a systemic inflammatory syndrome with thrombotic complications. We report here the first identification of Ca. N. mikurensis in organ samples from small mammals captured in southwest South Korea. Nested PCR of groEL and 16S rRNA genes was used to confirm the identity of the bacteria present, and successfully amplified fragments were sequenced. All captured animals were identified as striped field mice (Apodemus agrarius), approximately 28.6% (4/14) and 21.4% (3/14) of which were found to be PCR-positive for Ca. N. mikurensis and Anaplasma phagocytophilum, respectively. The detection of Ca. N. mikurensis in these animals represents the first evidence of this pathogen in South Korea. Carriage of this bacterium by rodents highlights the need for more detailed investigation of their role in its transmission to humans.
Introduction
Candidatus Neoehrlichia mikurensis (Ca. N. mikurensis) is an emerging tick-borne pathogen that causes a systemic inflammatory syndrome principally affecting individuals with preexisting hematologic or autoimmune diseases. As it is neither well-known nor well-recognized, Ca. N. mikurensis infection may be misdiagnosed as a recurrence of the underlying disease or an unrelated arteriosclerotic vascular event. This pathogen is transmitted by hard ticks of the genus Ixodes and is closely associated with rodents, in which transplacental transmission occurs [1].
Ca. N. mikurensis was first identified in the late 1990s as a novel α-proteobacterial pathogen (of the family Anaplasmataceae) isolated from I. ricinus in the Netherlands and Italy and a Norway rat (Rattus norvegicus) in China. It was initially termed Ehrlichia-like (or Schotti variant, E. walkeri, Rattus-strain) due to a divergent 16S rRNA gene sequence [2–4], but following further reports of its presence in rats and I. ovatus ticks in Japan and its passaging in laboratory rats, was described as a new species in 2004 [5]. Ca. N. mikurensis has been shown to be a human pathogen, and has been isolated from the blood of febrile patients across Europe and Asia. In most cases, these patients were immunocompromised due to splenectomy or immunosuppressive therapy and exhibited severe symptoms, including thrombotic events, recurrent fever lasting up to 8 months, and even death [6–8].
Several studies have identified Ca. N. mikurensis in questing and host-attached I. ricinus ticks in Europe [4, 9, 10]. However, potential reservoirs and vectors of this bacterium in South Korea have not yet been assessed, despite its documented presence in humans, rodents, and vectors in neighboring countries (China and Japan). Our aim was to evaluate the occurrence of this novel bacterium in the city of Gwangju, South Korea. The current work provides new data concerning the presence of this human pathogen in rodents, and constitutes the first report of Ca. N. mikurensis in South Korea.
Materials and methods
Study site and collection of rodents
Wild rodents were captured using live traps in a sylvatic habitat within an area of farmland in the west of the city of Gwangju, southwest South Korea (34°10′N, 126°55′E), during Autumn 2016 (from October through November). The sampling area consisted of a mixed stand with well-developed leaf litter layers. The live traps were placed along 5 transects (each 45 × 7 cm) spaced 150–200 m apart, depending on location, and were checked the following morning. Any small mammals caught were euthanized by inhalation of 5% isoflurane and organ samples were stored at −20°C until needed in experiments. All of the 14 wild rodents captured were identified as striped field mice (Apodemus agrarius). Twelve were captured in October and two in November. Each mouse was numbered for convenience during experiments and data interpretation.
Ethics statement
This study was been approved by Chosun University Institute of Animal Care and Use Committee (CIACUC2016- S0003). It adheres to Korean Animal Protection Act (2007) Institutional Animal Care and Use (IACUC) committee guidelines and use protocol. The study was carried out on private farmland in the west of the city of Gwangju, southwest South Korea and we obtained informed verbal consent from the owner. The study only involved rodents (wild type mice; Apodemus agrarius) which is not an endangered or protected species in South Korea. Live traps were used to collect the rodents. The sampling area consisted of a mixed stand with well-developed leaf litter layers. The live traps were placed along 5 transects (each 45 × 7 cm) spaced 150–200 m apart, depending on location, and were checked the following morning. The mice were euthanized by inhalant anesthetics, carbon dioxide (CO2). Any small mammals caught were euthanized, and organ samples were stored at −20°C until needed in experiments. All the sampling procedures and experimental manipulations were closely monitored by IACUC committee members.
DNA extraction from mouse spleen and kidney samples
Spleen and kidney samples (10 mg) from each of the 14 mice were taken from storage at −20°C, homogenized by grinding, and filtered with a sterile nylon cell strainer (70 μm; Falcon, Corning, NY, USA), before being completely lysed by proteinase K treatment and overnight incubation in a water bath. Genomic DNA was then extracted using a QIAamp DNA Blood & Tissue Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions.
PCR amplification
Tissue samples were tested for Ca. N. mikurensis using nested PCR targeting a region of the groEL gene, which encodes a 60-kDa heat shock protein. These results were confirmed by amplification of the 16S rRNA gene. The groEL nested PCR was carried out with the primers GROEL 607F and GROEL 1294R for the initial amplification, and GROEL 667F and GROEL 1121R to generate a final product of 445 bp [11]. The Ca. Neoehrlichia-specific 16S rRNA nested PCR employed the external and internal primer pairs 16S-EC9-F/16S-EC12A-R and 16S-IS58-62f/16S-IS58-594r, respectively, yielding a final product of 488 bp [5]. A separate nested PCR specific to the Anaplasma phagocytophilum 16S rRNA gene was performed using the external primers AE1-F/AE1-R and internal primers AP-F/AP-R to give a final product of 926 bp [12]. Ehrlichia chaffeensis and A. phagocytophilum genomic DNA samples served as positive controls. Both nested PCR rounds were performed in an AB thermal cycler (Applied Biosystems, Foster City, CA, USA) with a 20-μL mixture consisting of 1 μL 10 pmol/μL primers, 10 μL master mix, 2 μL GC enhancer, 4 μL sterile distilled water, and 2 μL genomic DNA (for the first PCR) or 2 μL of the first PCR product (for the second PCR). The primer sequences and annealing temperatures used are shown in the Table 1. Amplicons were separated by electrophoresis on a 1.5% agarose gel and visualized by ethidium bromide staining.
Table 1. Nucleotide sequences of polymerase chain reaction primers and conditions for amplification of Anaplasma and Candidatus Neoehrlichia species genes.
| Species and target genes | PCR primer sequence (5’-3’) | Annealing (°C/min) | PCR product size (bp) |
References |
|---|---|---|---|---|
|
Anaplasma and Ehrlichia spp. groEL (external primer) |
GROEL -607F (5′-GAAGATGCWGTWGGWTGTACKGC-3′) GROEL 1294R (5′-AGMGCTTCWCCTTCWACRTCYTC-3′) |
57 | 688 | 30 |
|
Anaplasma and Ehrlichia spp. groEL (internal primer) |
GROEL- 667F (5′-ATTACTCAGAGTGCTTCTCARTG-3′) GROEL -1121R (5′-TGCATACCRTCAGTYTTTTCAAC-3′) |
57 | 445 | 30 |
|
Ca. Neoehrlichia 16S rRNA (external primer) |
16S-EC9-F (5′-TACCTTGTTACGACTT-3′) 16S-EC12A-R (5′-TGATCCTGGCTCAGAACGAACG-3′) |
41 | 1,462 | 5 |
|
Ca. Neoehrlichia 16S rRNA (internal primer) |
16S-IS58-62f (5′-GGAATAGCTGTTAGAAATGACA-3′) 16S-IS58-594r (5′-CTATCCTCTCTCGATCTCTAGTTT-3′) |
54 | 488 | 5 |
|
A. phagocytophilum 16S rRNA (external primer) |
AE1-F (5′-AAGCTTAACACATGCAAGTCGAA-3′) AE1-R (5′-AGTCACTGA CCCAACCTTAAATG-3′) |
56 | 1,406 | 30 |
|
A. phagocytophilum 16S rRNA (internal primer) |
AP-F (5′-GTCGAACGGATTATTCTTTATAGCTTGC-3′) AP-R (5′-CCCTTCCGTTAAGAAGGATCTAATCTCC-3′) |
56 | 926 | 30 |
Nucleotide sequencing
The groEL and 16S rRNA gene fragments amplified from positive spleen and kidney samples were purified and directly sequenced. The PCR products were visualized by electrophoresis on an ethidium bromide-stained 1.5% agarose gel. A Biosystems Veriti 96-Well Thermal Cycler (Applied Biosystems, Foster City, CA) was used for this experiment. Amplified and purified DNA was prepared for direct sequencing using a QIAquick PCR Purification Kit (Qiagen, Westburg, Netherlands) and was sequenced by dideoxy termination with an automatic sequencer (ABI Prism 3730XL DNA analyzer). Sequence homology analysis was performed by the National Center for Biotechnology Information (National Institutes of Health) BLAST network service. The resulting sequences were used in BLASTN searches of the National Center for Biotechnology Information GenBank database to identify the bacteria present. The nucleotide sequences generated in this study have been deposited in GenBank (Fig 1A and Fig 1B).
Fig 1.
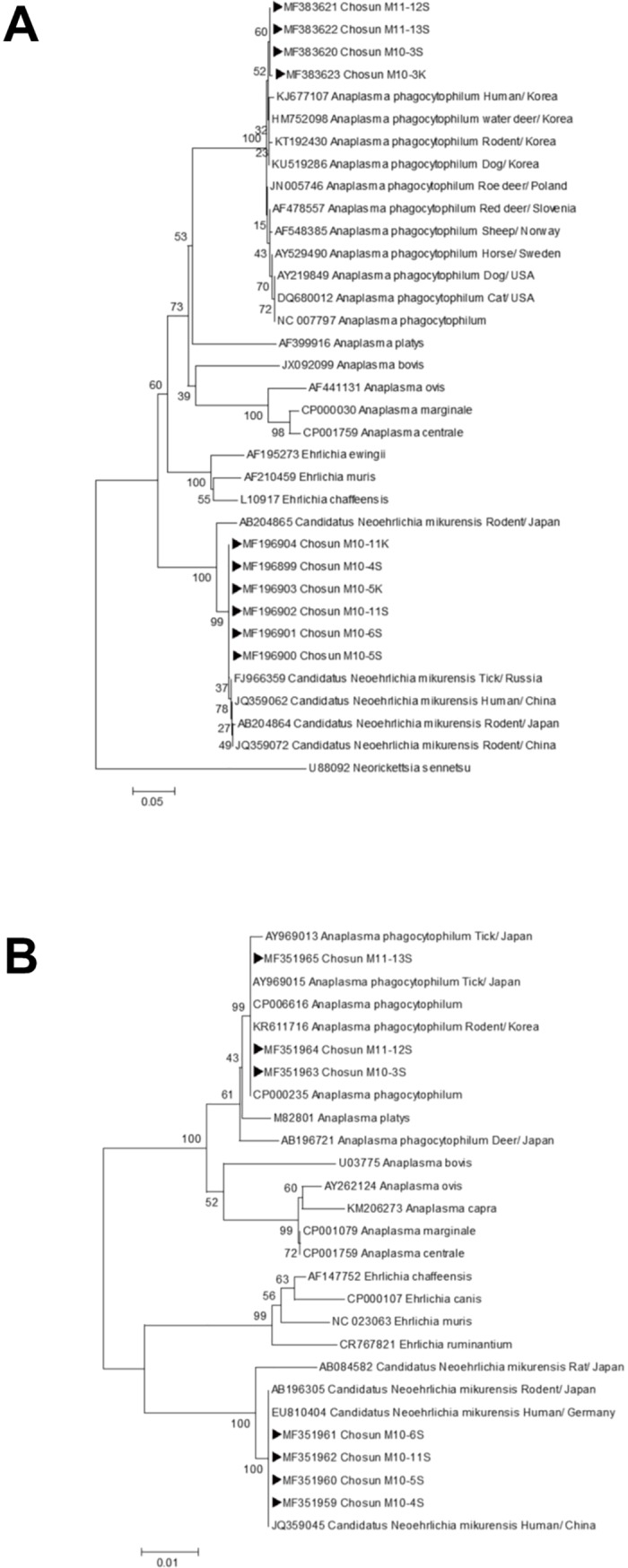
Neighbor-joining trees based on groEL (A, 445 bp) and 16S rRNA (B, 463 bp) gene sequences from GenBank and rodents having tested PCR-positive for Candidatus Neoehrlichia mikurensis and Anaplasma phagocytophilum in this study. The trees were generated in Molecular Evolutionary Genetics Analysis version 4.0 using the maximum composite likelihood method and 1,000 bootstrap replicates. Numbers next to nodes indicate the percentages of replicate trees in which the corresponding clade was recovered. Scale bars indicate 0.05 (A) and 0.01 (B) base substitutions per site. GenBank accession numbers and sources of Ca. N. mikurensis and A. phagocytophilum sequences are shown.
Phylogenetic analysis
GroEL and 16S rRNA gene sequences were obtained from GenBank, aligned with ClustalX, and analyzed using Molecular Evolutionary Genetics Analysis version 6.0. Phylogenetic trees were constructed with the neighbor-joining method, and the percentage of replicate trees in which nodes were recovered under the bootstrap test (1,000 replicates) was calculated.
Results
Ca. N. mikurensis was detected in one or more organs of 4 of the 14 mice captured. Three of the 4 Ca. N. mikurensis -positive animals were caught in October, and 1 was caught in November. The groEL nested PCR revealed 7 spleen and 3 kidney samples to be positive for Anaplasma/Ehrlichia. Sequencing identified A. phagocytophilum in 3 of these 7 spleen samples (Chosun M10-3S, M11-12S, and M11-13S) and Ca. N. mikurensis in the other four (Chosun M10-4S, M10-5S, M10-6S, and M10-11S).
A. phagocytophilum and Ca. Neoehrlichia 16S rRNA genes were detected by species-specific nested PCR. Four spleen samples were PCR-positive for Ca. Neoehrlichia (Chosun M10-4S, M10-5S, M10-6S, and M10-11S), and 3 for A. phagocytophilum (Chosun M10-3S, M10-12S, M11-13S). Sequencing of the PCR products confirmed the presence of Ca. N. mikurensis and A. phagocytophilum, respectively.
Phylogenetic trees were inferred from comparisons of groEL (445 bp) and 16S rRNA (463 bp) gene sequences (Fig 1A and Fig 1B). The Ca. N. mikurensis (488 bp) and A. phagocytophilum (926 bp) 16S rRNA gene sequences were trimmed to 463 bp to create a single tree. The Ca. N. mikurensis sequences derived from the spleen and kidney samples in the present study were phylogenetically close to Ca. N. mikurensis sequences previously isolated from rodents in China, Japan, and Russia, being grouped in the same clade. Similarly, in the phylogenies generated, the A. phagocytophilum sequences obtained here neighbored those of A. phagocytophilum previously detected in rodents from Korea.
The trees generated with groEL and 16S rRNA gene sequences had similar topologies and both indicated a close relationship between the bacteria examined in this investigation and other organisms identified as Ca. N. mikurensis. As in previous studies, Ca. N. mikurensis was phylogenetically distinct from other genera in the family Anaplasmataceae and formed a well-supported sister clade to the genus Ehrlichia. All of the currently available sequences from this group have been categorized together under the single candidate species Ca. N. mikurensis. Comparisons between the groEL and 16S rRNA gene sequences generated and those of specific genospecies related to Anaplasmataceae pathogens strongly support the Ca. N. mikurensis identification made here.
Discussion
In this study, we tested small mammals to establish the occurrence of Ca. N. mikurensis in southwestern South Korea. As rodents have been found to harbor Ca. N. mikurensis, it has been suggested that they act as a reservoir of this bacterium [5, 6, 13–16]. The high infection rate observed among striped field mice in the present study corroborates this hypothesis. In a previous investigation carried out in Germany, 48 of the 91 (52.7%) small mammals tested were found to harbor Ca. N. mikurensis in one or more of their organs or body fluids [17]. In contrast, only 68 (8.8%) of the 771 rodents examined in a prior study based in Sweden were infected with this bacterium [15]. Similarly, a Chinese survey of 211 rodents of various species captured with snap traps revealed the rate of Ca. N. mikurensis carriage to be just 3.8% [18]. Nevertheless, such findings indicate that rodents play a role in the natural life cycle of Ca. N. mikurensis and are likely to be competent reservoir hosts of this bacterium. Ca. N. mikurensis has been detected in 6 rodent species in Europe (A. agrarius, Apodemus flavicollis, Apodemus sylvaticus, Myodes glareolus, Microtus agrestis, and Microtus arvalis) and 10 species in Asia, although infection rates vary considerably (8.3%–52.7%) between species. The bank vole M. glareolus has been identified as the most frequently infected species, with 9.1% of individuals testing positive on average. However, estimates of Ca. N. mikurensis prevalence in this species vary from 1.8% (in France) to 52.7% (in Germany) [1].
Ca. N. mikurensis was named in 2004, after its discovery in ticks and rodents on the Japanese island of Mikura-jima by using PCR targeting conserved bacterial genes, including 16S rRNA and groEL [5]. Transmission electron microscopy of infected rat tissue revealed small cocci in the cytoplasm of endothelial cells. Phylogenetic analyses showed this emerging zoonotic intracellular tick-borne pathogen to be a new species of the family Anaplasmataceae, in which it forms a distinct cluster together with the North American bacterium Ca. N. lotoris, which has been detected in raccoons [6, 19, 20]. Moreover, a more recent study comparing 16S rRNA and groEL gene sequences confirmed that organisms initially identified as “Ehrlichia-like” may in fact be members of the novel species Ca. N. mikurensis, or at least very close relations. Related species include E. ruminantium, E. chaffeensis, and A. phagocytophilum [5, 6], all of which are strict intracellular pathogens that can only be cultured in live cells. N. mikurensis retains the status “Candidatus” because its culture in vitro has not yet been reported.
Ca. N. mikurensis was first described as a human pathogen in 2010, and a total of 15 human cases of Ca. N. mikurensis associated disease have been reported to date in Europe and Asia, with just over half concerning apparently healthy individuals [7] and the remainder immunocompromised patients [7, 8, 21]. In a study of human Ca. N. mikurensis infections in China, all 7 of the patients examined exhibited relatively mild symptoms consisting of fever, headache, and malaise, and none had a history of immunocompromising illness [22]. The cells infected by Neoehrlichia bacteria in humans remain to be identified, although polymorphonuclear granulocytes and endothelial cells [6] may be involved. At present, the only diagnostic options comprise pan-bacterial PCR (targeting the 16S rRNA and groEL genes) followed by sequence analysis [8], and specific real-time PCR performed on whole blood, plasma, or bone marrow. Interestingly, Ca. N. mikurensis infection in humans appears to be associated with a high rate of vascular and thromboembolic events. Grankvist et al. reported that more than half of the affected patients (6/11) in their study developed upper- or lower-limb deep vein thrombosis [23].
The first South Korean case of human A. phagocytophilum infection occurred in 2013 [24]. Striped field mice, the dominant rodent species in South Korea and an agricultural pest, can be latently infected with various Ehrlichia and Anaplasma spp. [25, 26], and A. phagocytophilum has been identified in Haemaphysalis longicornis, I. nipponensis, and I. persulcatus ticks in this country [27, 28]. Moreover, previous molecular epidemiologic studies in South Korea have shown this bacterium to be present in 2.6% (5/196) of striped field mice [28, 29] and 63.6% (42/66) of Korean water deer [29]. Notably, the seroprevalence of A. phagocytophilum based on immunofluorescence tests has been estimated to be 1.8% among Korean patients with acute fever [30].
Anaplasma spp. and Ca. N. mikurensis are closely related organisms, and given the high prevalence of the latter among the wild mice examined in the present work, human infection with this pathogen in this region seems likely. Clinicians in South Korea should test for Ca. N. mikurensis in patients with recent tick bites seeking treatment for vascular and thromboembolic events, with a view to establishing its prevalence.
Conclusion
In this study, 28.57% and 21.4% of the mice tested were positive for Ca. N. mikurensis and A. phagocytophilum, respectively. We conclude that Ca. N. mikurensis is widespread in the city of Gwangju, South Korea, and its relatively high prevalence in a common rodent species implies a substantial risk of infection for humans and domestic animals. Our work represents the first identification of this organism in this country, and indicates the need for more specific investigation into its importance as a human pathogen.
Data Availability
Data are from the study "Assessment of Tick-borne bacterial zoonotic diseases in Gwangju City", and all relevant data are within the paper.
Funding Statement
This research was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI16C2118). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Silaghi C, Beck R, Oteo JA, Pfeffer M, Sprong H. Neoehrlichiosis: an emerging tick-borne zoonosis caused by Candidatus Neoehrlichia mikurensis. Exp Appl Acarol. 2016; 68: 279–297. 10.1007/s10493-015-9935-y [DOI] [PubMed] [Google Scholar]
- 2.Schouls LM, Van De Pol I, Rijpkema SG, Schot CS. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J Clin Microbiol. 1999; 37: 2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan H, Liu S, Ma Y, Tong S, Sun Y. Ehrlichia-like organism gene found in small mammals in the Suburban district of Guangzhou of China. Ann N Y Acad Sci. 2003; 990: 107–111. [DOI] [PubMed] [Google Scholar]
- 4.Sanogo YO, Parola P, Shpynov S, Camicas JL, Brouqui P, Caruso G, et al. Genetic diversity of bacterial agents detected in ticks removed from asymptomatic patients in northeastern Italy. Ann N Y Acad Sci. 2003; 990:182–190. [DOI] [PubMed] [Google Scholar]
- 5.Kawahara M, Rikihisa Y, Isogai E, Takahashi M, Misumi H, Suto C, et al. Ultrastructure and phylogenetic analysis of ‘Candidatus Neoehrlichia mikurensis' in the family Anaplasmataceae, isolated from wild rats and found in Ixodes ovatus ticks. Int J Syst Evol Microbiol. 2003; 54: 1837–1843. [DOI] [PubMed] [Google Scholar]
- 6.Rar V, Golovljová I. Anaplasma, Ehrlichia, and "Candidatus Neoehrlichia" bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011; 11:1842–1861. 10.1016/j.meegid.2011.09.019 [DOI] [PubMed] [Google Scholar]
- 7.von Loewenich FD, Geissdörfer W, Disqué C, Matten J, Schett G, Sakka SG,et al. Detection of "Candidatus Neoehrlichia mikurensis" in two patients with severe febrile illnesses: Evidence for a European sequence variant. J Clin Microbiol. 2010; 48:2630–2635. 10.1128/JCM.00588-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welinder-Olsson C, Kjellin E, Vaht K, Jacobsson S, Wenneras C. First case of human "Candidatus Neoehrlichia mikurensis" infection in a febrile patient with chronic lymphocytic leukemia. J Clin Microbiol. 2010; 48: 1956–1959. 10.1128/JCM.02423-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fertner ME, Mølbak L, Boye Pihl TP, Fomsgaard A, Bødker R. First detection of tick-borne "Candidatus Neoehrlichia mikurensis" in Denmark 2011. Euro Surveill. 2012; 17 (8). pii: 20096 [PubMed] [Google Scholar]
- 10.Richter D, Matuschka FR. "Candidatus Neoehrlichia mikurensis," Anaplasma phagocytophilum, and Lyme disease spirochetes in questing European vector ticks and in feeding ticks removed from people. J Clin Microbiol. 2012; 50: 943–947. 10.1128/JCM.05802-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takano A, Ando S, Kishimoto T, Fujita H, Kadosaka T, Nitta Y, et al. Presence of a novel Ehrlichia sp. in Ixodes granulatus found in Okinawa, Japan. Microbiol Immunol. 2009; 53:101–106. 10.1111/j.1348-0421.2008.00093.x [DOI] [PubMed] [Google Scholar]
- 12.Oh JY, Moon BC, Bae BK, Shin EH, Ko YH, Kim YJ, et al. Genetic identification and phylogenetic analysis of Anaplasma and Ehrlichia species in Haemaphysalis longicornis collected from Jeju Island, Korea. J Bacteriol Virol. 2009; 39:257–267. [Google Scholar]
- 13.Beninati T, Piccolo G, Rizzoli A, Genchi C, Bandi C. Anaplasmataceae in wild rodents and roe deer from Trento Province (northern Italy). Eur J Clin Microbiol Infect Dis. 2006; 25:677–678. 10.1007/s10096-006-0196-x [DOI] [PubMed] [Google Scholar]
- 14.Andersson M, Råberg L. Wild rodents and novel human pathogen Candidatus Neoehrlichia mikurensis, Southern Sweden. Emerg Infect Dis. 2011; 17:1716–1718. 10.3201/eid1709.101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rar VA, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, et al. Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 2010; 1:57–65. 10.1016/j.ttbdis.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 16.Pan L, Zhang L, Wang G, Liu Q, Yu Y, Wang S, et al. Rapid, simple, and sensitive detection of Anaplasma phagocytophilum by loop-mediated isothermal amplification of the msp2 gene. J Clin Microbiol. 2011; 49:4117–4120. 10.1128/JCM.01085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silaghi C, Woll D, Mahling M, Pfister K, Pfeffer M. Candidatus Neoehrlichia mikurensis in rodents in an area with sympatric existence of the hard ticks Ixodes ricinus and Dermacentor reticulatus, Germany. Parasites & Vectors. 2012; 5:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson M, Råberg L. Wild Rodents and Novel Human Pathogen Candidatus Neoehrlichia mikurensis, Southern Sweden. Emerg Infect Dis. 2011; 17(9):1716–1718. 10.3201/eid1709.101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yabsley MJ, Murphy SM, Luttrell MP, Wilcox BR, Ruckdeschel C. Raccoons (Procyon lotor), but not rodents, are natural and experimental hosts for an ehrlichial organism related to “Candidatus Neoehrlichia mikurensis”. Vet Microbiol. 2008; 131:301–308. 10.1016/j.vetmic.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 20.Munderloh UG, Yabsley MJ, Murphy SM, Luttrell MP, Howerth EW. Isolation and establishment of the raccoon Ehrlichia-like agent in tick cell culture. Vector Borne Zoonotic Dis. 2007; 7:418–425. 10.1089/vbz.2007.0640 [DOI] [PubMed] [Google Scholar]
- 21.Maurer FP, Keller PM, Beuret C, Joha C, Achermann Y, Gubler J, et al. Close geographic association of human neoehrlichiosis and tick populations carrying “Candidatus Neoehrlichia mikurensis” in eastern Switzerland. J Clin Microbiol. 2013; 51:169–176. 10.1128/JCM.01955-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Jiang JF, Liu W, Zheng YC, Huo QB, Tang K, et al. Human infection with Candidatus Neoehrlichia mikurensis, China. Emerg Infect Dis. 2012; 18:1636–1639. 10.3201/eid1810.120594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grankvist A, Andersson PO, Mattsson M, Sender M, Vaht K, Höper L, et al. Infections with the tick-borne bacterium “Candidatus Neoehrlichia mikurensis” mimic noninfectious conditions in patients with B cell malignancies or autoimmune diseases. Clin Infect Dis. 2014; 58:1716–1722. 10.1093/cid/ciu189 [DOI] [PubMed] [Google Scholar]
- 24.Kim KH, Yi J, Oh WS, Kim NH, Choi SJ, Choe PG, et al. Human granulocytic anaplasmosis, South Korea 2013. Emerg Infect Dis. 2014; 20:1708–1711. 10.3201/eid2010.131680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chae JS, Kim CM, Kim EH, Hur EJ, Klein TA, Kang TK, et al. Molecular epidemiological study for tick-borne disease (Ehrlichia and Anaplasma spp.) surveillance at selected U.S. military training sites/installations in Korea. Ann N Y Acad Sci. 2003; 990: 118–125. [DOI] [PubMed] [Google Scholar]
- 26.Palmer SR, Soulsby L, Torgerson PR, Brown DWG, editors. Oxford textbook of zoonoses: biology, clinical practice, and public health control 2nd ed. New York: Oxford University Press; Part 2, Bacterial, chlamydial, and rickettsial zoonoses; 2011; p. 45–271. [Google Scholar]
- 27.Kim CM, Kim MS, Park MS, Park JH, Chae JS. Identification of Ehrlichia chaffeensis, Anaplasma phagocytophilum, and A. bovis in Haemaphysalis longicornis and Ixodes persulcatus ticks from Korea. Vector Borne Zoonotic Dis. 2003; 3:17–26. 10.1089/153036603765627424 [DOI] [PubMed] [Google Scholar]
- 28.Chae JS, Yu do H, Shringi S, Klein TA, Kim HC, Chong ST, et al. Microbial pathogens in ticks, rodents and a shrew in northern Gyeonggi-do near the DMZ, Korea. J Vet Sci. 2008; 9:285–293. 10.4142/jvs.2008.9.3.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heo EJ, Park JH, Koo JR, Park MS, Park MY, Dumler JS, et al. Serologic and molecular detection of Ehrlichia chaffeensis and Anaplasma phagocytophila (human granulocytic ehrlichiosis agent) in Korean patients. J Clin Microbiol. 2002; 40: 3082–3085. 10.1128/JCM.40.8.3082-3085.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang JG, Ko S, Kim YJ, Yang HJ, Lee H, Shin NS, et al. New genetic variants of Anaplasma phagocytophilum and Anaplasma bovis from Korean water deer (Hydropotes inermis argyropus). Vector Borne Zoonotic Dis. 2011; 11: 929–938. 10.1089/vbz.2010.0214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are from the study "Assessment of Tick-borne bacterial zoonotic diseases in Gwangju City", and all relevant data are within the paper.


