Abstract
Adult-onset Still's disease (AOSD) is a rare systemic inflammatory disorder in which inflammasome activation plays a pathophysiological role. In view of the inflammatory nature of AOSD, we investigated whether serum amyloid A (SAA) gene polymorphisms affect the susceptibility of patients with AOSD.
Eighty-seven Japanese patients with AOSD and 200 healthy Japanese subjects were recruited in this study. The genotypes of the -13C/T SNP in the 5′-flanking region of the SAA1 gene (rs12218) and two SNPs within exon 3 of SAA1 (2995C/T and 3010C/T polymorphisms) were determined using polymerase chain reaction fragment length polymorphism (PCR-RFLP) assay in all subjects. In AOSD patients, exons 1, 2, 3, and 10 of the MEFV gene were also genotyped by direct sequencing.
The frequency of the SAA1.3 allele was increased in AOSD patients compared with that in healthy subjects (43.1% versus 37.5%), but the difference was not significant. The −13T allele was more frequently observed in AOSD patients than in healthy subjects (50.6% versus 41.0%, P = .0336). AOSD patients with the −13T allele had been treated with immunosuppressants more frequently than those without this allele. MEFV mutations were detected in 49 patients with AOSD (49/87, 57.3%). AOSD patients with MEFV variants frequently exhibit macrophage activation syndrome, but the difference was not significant (34.7% versus 18.4%, P = .081). Also, there was no significant difference in SAA1 -13C/T allele frequency between AOSD patients with and without MEFV mutations.
Our data shows a significant association between T allele of rs12218 and AOSD in Japanese population.
Keywords: adult-onset still's disease, inflammasome, MEFV, polymorphisms, serum amyloid A
1. Introduction
Adult-onset Still's disease (AOSD) is a systemic inflammatory disease characterized by spiking fever, skin rash, and arthritis.[1] Although its pathogenesis is largely unknown,[2] a growing number of studies support the hypothesis that dysregulation of inflammasome activation and the related overproduction of cytokines play a pivotal role in it, similar to other autoinflammatory diseases.[3] Serum amyloid A (SAA) is an acute-phase reactant that is synthesized in the liver by inflammatory cytokines, the level of which can increase 1000-fold during inflammatory conditions.[4] Within the SAA gene cluster, only the SAA1 and SAA2 genes encode acute-phase SAA.[5] The presence of two single-nucleotide polymorphisms (SNPs) within exon 3 of the SAA1 gene, 2995C/T and 3010C/T, define 3 haplotypes that correspond to SAA1.1, SAA1.3, and SAA1.5.[6] Several studies have shown that the development of AA amyloidosis is positively related to the frequency of SAA1.3 alleles in the Japanese population.[7,8] Subsequent investigations indicated that another SNP (rs12218) in the SAA1 gene, at position -13 in the 5′ regulatory region of this gene, is in linkage disequilibrium with the SAA1.3 allele and is closely associated with the occurrence of AA amyloidosis in Japanese and Caucasian rheumatoid arthritis patients.[9,10] Moreover, a functional study suggested that the T variant at this SNP is associated with greater transcriptional activity of the SAA1 gene.[11]
G-protein-coupled formyl peptide receptor 2 (FPR2) is one of the receptors for SAA.[12] In addition to FPR2, several receptors for SAA have been identified.[13] Recent studies have suggested that SAA may play a role in the modulation of inflammation and immunity through these SAA receptors.[14] In this study, we investigated the relationship between SAA1 gene polymorphism and AOSD.
2. Patients and methods
2.1. Design, setting, patients, and measurements
We retrospectively evaluated 87 patients, who were diagnosed as AOSD and be treated at the Rheumatology Units of participating hospital group, between 2010 and 2017. The patients had all been diagnosed with AOSD according to the Yamaguchi criteria,[15] after exclusion of infectious, hematologic, and inflammatory diseases. All patients had undergone laboratory tests, including a complete blood count, biochemistry, urinalysis, erythrocyte sedimentation rate (ESR) and ferritin. The demographic data of the enrolled patients, including clinical manifestations, treatments, prognosis, and complications, were collected using a standardized form. Each patient was assessed for the systemic score proposed by Pouchot et al[26] for AOSD. This score assigns 1 point to each of 12 manifestations: fever, typical rash, pleuritis, pneumonia, pericarditis, hepatomegaly or abnormal liver function tests, splenomegaly, lymphadenopathy, leukocytosis > 15,000/mm3, sore throat, myalgia, and abdominal pain (maximum score: 12 points). This study was approved by the institutional review board of the Sasebo City Hospital institutional review board (No. 2012-A-22) and participating hospitals. Two hundreds of healthy Japanese individuals without pre-existing medical diseases (90 men and 110 women 14 to 64 years, with a mean age of 38.6 ± 13.9 years) from East Japan (n = 86) and West Japan (n = 114) were enrolled in the study after obtaining informed consent.
2.2. Genotyping of SAA1 gene
The SAA1.1, 1.3, and 1.5 alleles, corresponding to the T-C, C-T, and C-C haplotypes of the C2995T (rs1136743) and C3010T (rs1136747) polymorphisms were analyzed by the PCR-RFLP methods.[8] The primers used for the PCR reaction were 5′-GCC AATTACATCGGCCTCAG-3′ (sense) and 5′-TGGCCA AAGAATCTCTGG AT-3′ (antisense). The 518-bp PCR products were digested with restriction enzyme BclI (Promega, San Luis Obispo, CA, USA) and BanI (Promega) and electrophoresed on a agarose gel. The genotypes of the SAA1 -13C/T in the 5′-flanking region of the SAA1 gene, (rs12218) were also determined by the PCR–RFLP method.[8] The primers used for PCR were 5′-ACATCT TGTTCCCTC AGGTTG-3′ (sense) and 5′-GCTGTAGCTGAGCTGCGG-3′ (antisense). The 229-bp PCR products were subjected to the restriction enzyme AciI (BioLabs, Beverly, MA, USA) and electrophoresed on a 12.5% polyacrylamide gel.
2.3. MEFV gene analysis
Peripheral blood samples (collected in vacutainers containing EDTA as anticoagulant) were used for genomic DNA isolation. Genomic DNA was isolated by the Promega Wizard Genomic DNA Purification Kit (Madison, WI, USA). Polymerase chain reaction (PCR) was performed using forward and reverse primers for each exon of the MEFV gene, as described previously.[16] The polymerase chain reaction (PCR) products were purified and directly sequenced using ABI 3100 Genetic Analyzer (Applied Biosystems, Tokyo, Japan). The MEFV genetic analysis was approved by the Ethics Committee of Fukushima Medical University School of Medicine (2016, No. 2920).
2.4. Statistical analysis
Statistical analyses were out using SPSS version 17.0 (SPSS; Chicago, IL, USA). Hardy-Weinberg equilibrium was assessed by the chi-square test. Differences in the distribution of alleles between AOSD patients and control subjects were analyzed using the chi-square test. Differences among AOSD characteristics were analyzed by performing a Mann-Whitney's U test or Chi-squared test using 2 × 2 contingency tables.
3. Results
3.1. Demographic findings of enrolled AOSD patients
A total of 87 Japanese patients (18 men and 69 women) who fulfilled the Yamaguchi criteria for AOSD[15] were recruited into this study. All patients were ethnic Japanese. Routine blood tests including tests of C-reactive protein, serum ferritin, and liver enzymes were performed. The clinical manifestations of the enrolled patients are shown in Table 1. Among the 87 enrolled patients with AOSD, 69 (79.3%) were women and the mean age at diagnosis was 49.3 ± 19.9 years old. Among these 87 patients, 51 patients (58.6%) were treated with immunosppressants (Cyclosporin A 38, Tacrolimus 4, Methotrexate 9).
Table 1.
Demographic findings of enrolled AOSD patients.

3.2. SAA1 gene haplotypes in Japanese patients with AOSD
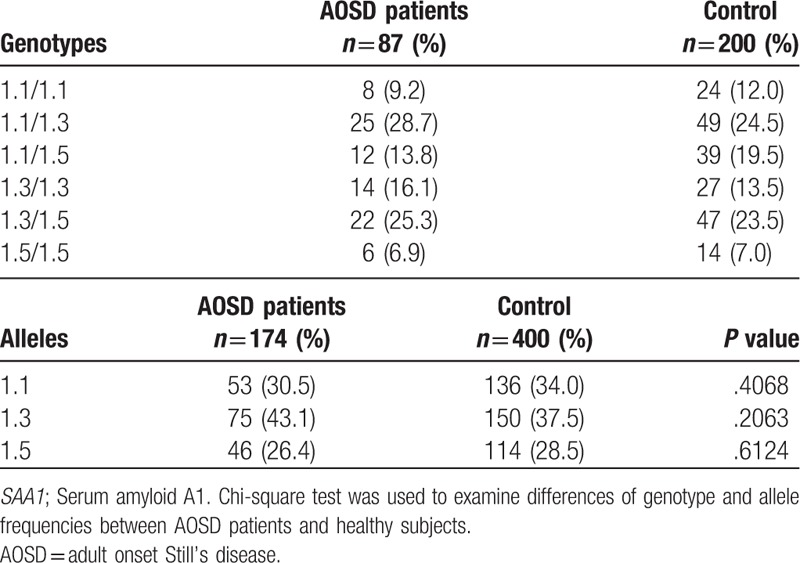
A segment of the SAA1 gene with polymorphic sites was subjected to PCR restriction fragment length polymorphism (PCR-RFLP) analysis. Table 2 shows the frequencies of individuals with various genotypes and alleles at the SAA1 locus in either AOSD patients (n = 87) or healthy Japanese subjects (n = 200). The frequency of the SAA1.3 allele was increased in AOSD patients compared with that in healthy subjects (43.1% versus 37.5%), but the difference was not significant.
Table 2.
SAA1 genotype and allele frequency in the AOSD patients and controls.
3.3. Association of SAA1 −13T allele with AOSD in Japanese
The -13C/T polymorphism, in the 5′-flanking region of the SAA1 gene, which is in linkage disequilibrium with the SAA1.3 allele, has been shown to be associated with susceptibility to AA amyloidosis in Japanese RA patients. We thus analyzed the -13C/T polymorphisms in AOSD patients and healthy Japanese subjects. The observed genotype frequencies did not differ from the expected frequencies of the genotypes under the assumption of Hardy–Weinberg equilibrium (data not shown). Allele frequencies of -13C/T differed between the 2 groups (Table 3), with the −13T allele being significantly more common in AOSD patients than in healthy subjects (50.6% versus 41.0%, P = .0336). This suggests that the −13T allele is associated with susceptibility to AOSD in the Japanese population. To evaluate the potential correlations of -13C/T alleles with the clinical manifestations of AOSD, we stratified the AOSD patients according to the presence of the −13T allele (Table 4). AOSD patients with this allele were found to have been treated more frequently (64.7% versus 36.8%, P = .029) with immunosuppressants (Cyclosporin A 33, Tacrolimus 2, Methotrexate 9), but there was no significant difference in the other clinical manifestations. We also compared the systemic score according to the genotypes of SAA -13C/T among AOSD patients. As demonstrated in Figure 1, we could not find a significant difference in systemic score according to the genotype SAA -13C/T allele in Japanese patients with AOSD.
Table 3.
Genotype and allele frequencies of −13 C/T SAA1 in AOSD patients and the healthy subjects.
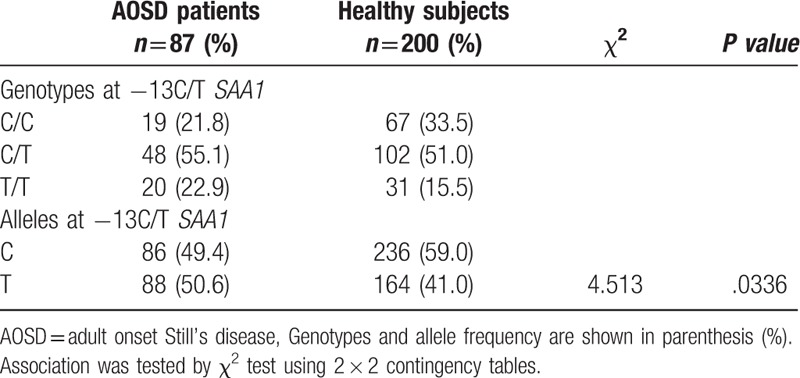
Table 4.
Demographic and clinical features of AOSD patients with or without -13 T allele.
Figure 1.
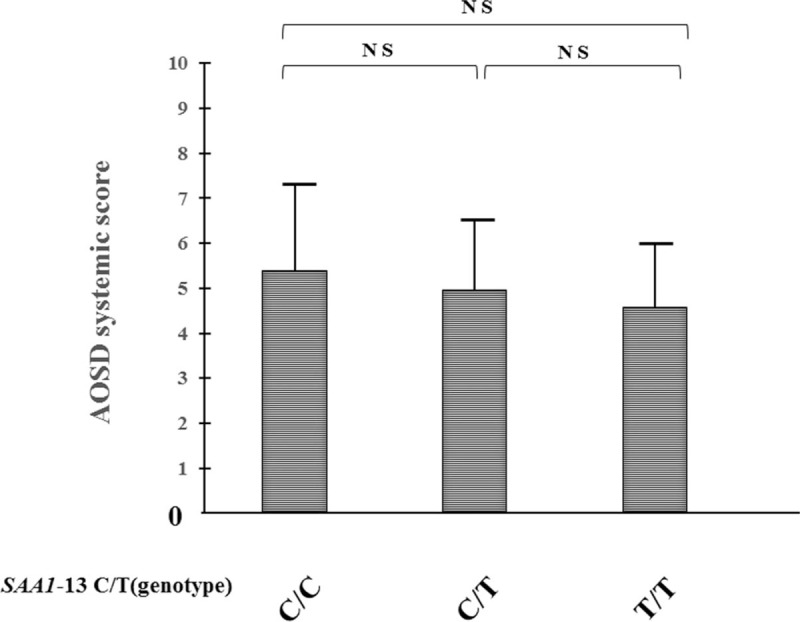
Systemic scores of AOSD patients among the different genotypes of SAA1-13C/T allele. The values of the systemic scores of each AOSD patients stratified by the genotypes of SAA1-13C/T allele are shown. Values are expressed as mean ± standard deviation. NS; not significant. AOSD = adult onset Still's disease.
3.4. MEFV variant analysis
All AOSD patients were successfully genotyped for the MEFV gene. MEFV variants were identified in 46 AOSD patients (57.6%), the distribution of which is shown in Table 5. We stratified the AOSD patients according to the presence or absence of MEFV variants, and then compared the clinical features between these groups (Table 6). Although there was no statistically significant difference, AOSD patients with MEFV variants were more frequently associated with macrophage activation syndrome (34.7% versus 18.4%, P = .092) and the initial doses of prednisolone were higher compared with those without MEFV variants (35.9 ± 12.8 mg/day versus 31.8 ± 11.2 mg/day, P = .081). We also compared the frequencies of -13C/T polymorphisms between the AOSD patients stratified according to the presence of MEFV variants, but no significant differences were found (Table 7).
Table 5.
MEFV genotypes of AOSD patients.
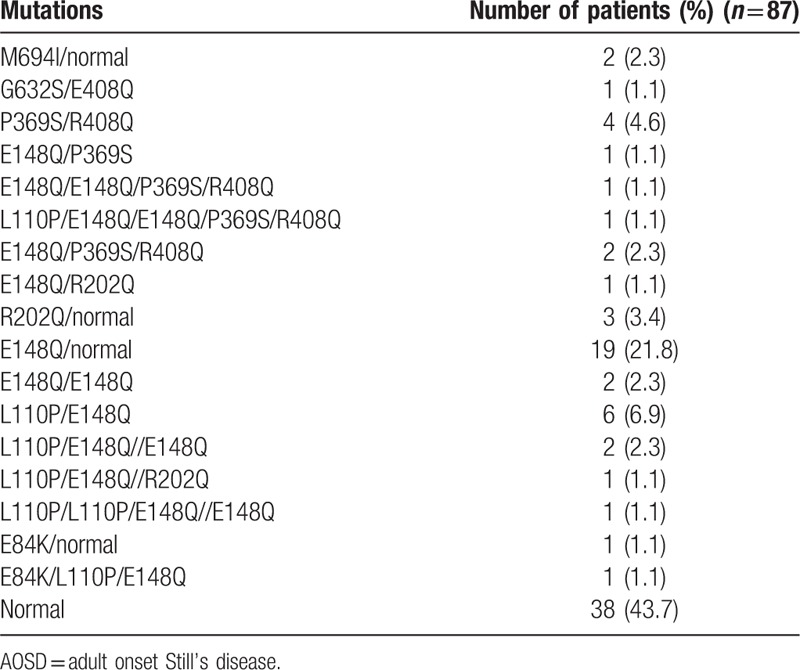
Table 6.
Demographic and clinical features of AOSD patients with or without MEFV variants.
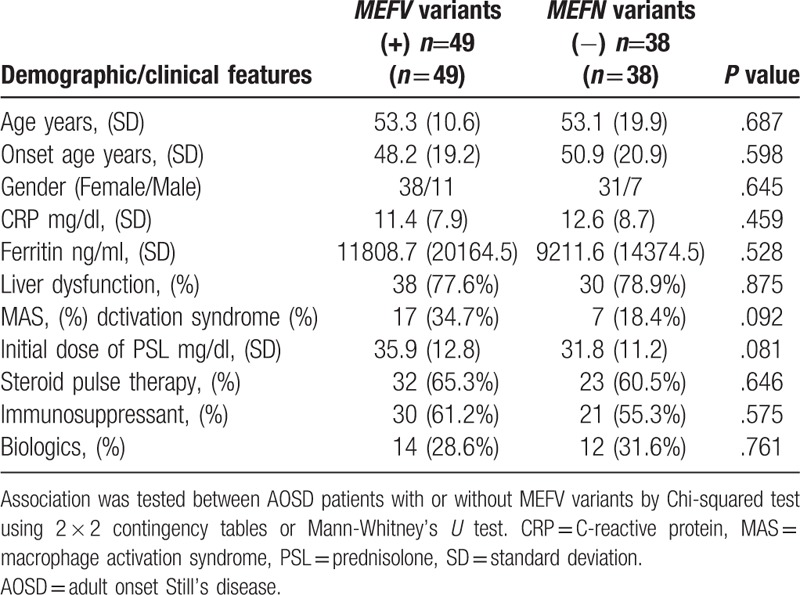
Table 7.
Frequencies at -13C/T alleles in AOSD patients with or without MEFV variants.

4. Discussion
A major acute-phase reactant, SAA, is produced when the immune system senses potential danger signals including trauma, infection, and severe stress.[17] Previous studies accumulated substantial evidence that SAA is involved in inflammatory disorders including autoinflammatory diseases.[18] SAA has proinflammatory activities, including those involving cell migration and phagocytosis.[19] It is also considered to be a danger signal that activates inflammasomes.[20] Specifically, SAA is capable of activating NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome and subsequent IL-1β production,[21] which play important roles in the pathogenesis of AOSD. Therefore, it is possible that SAA is important in the pathogenesis of autoinflammatory diseases including AOSD. Evidence has also been presented that the risk of AOSD is influenced by polymorphisms in proinflammatory genes including cytokines and components of the innate immune system.[22] In this study, we found that polymorphisms of the SAA1 gene were associated with the susceptibility to AOSD in the Japanese population. We demonstrated that the frequency of the T allele at position −13 in the SAA1 gene (rs12218) was significantly increased in AOSD patients compared with that in healthy subjects. Previous studies also demonstrated the strong linkage disequilibrium between the SAA1.3 allele and the SAA1 −13T allele in the Japanese population.[9] Our data also indicate that the allele frequency of SAA1.3 was increased in AOSD patients compared with that in healthy controls, although this did not reach statistical significance.
The overproduction of IL-1β is responsible for a variety of autoinflammatory disorders, including AOSD.[23] Recent studies have shown that SAA activates the NLRP3 inflammasome and subsequent IL-1β production in innate immune cells,[24] suggesting a link between SAA and autoinflammatory diseases. SNPs identified in the promoter region of the SAA1 gene have been shown to be associated with a predisposition to AA amyloidosis.[8,9] The mechanisms by which SAA1 gene polymorphism (-13C/T, rs12218) may be associated with AOSD are unknown. Moriguchi et al demonstrated that a C/T switch at -13 of SAA1 gene (-13C/T) is associated with increased transcription activity of this gene correlating susceptibility to AA amyloidosis in Japanese patients with rheumatoid arthritis. They found 4 major haplotypes of SAA1 promoter based on 3 SNPs at -61, -13, and -2 of the SAA1 promoter region and designed them -14C1 (C-C-G), -13C2 (G-C-G), -13C3 (G-C-A), and -13T (C-T-G). They demonstrated that −13T promoter exhibited greater transcriptional activity compared to other cis-regulating elements promoter haplotypes -13C1, -13C2, and -13C3 in luciferase reporter gene assays under IL-1β or IL-6 stimulation. The serum concentration of A-SAA to C-reactive protein (CRP) were significantly higher in subjects carrying SAA1.5 allele than in those carrying1.3 allele. The data from luciferase reporter assay seem inconsistent with these laboratory data. However, an increased transcriptional ability of A-SAA does not necessarily result in a serum SAA concentrations due to other factors, such as A-SAA stability and degradation may influence the serum concentrations of A-SAA.
It has been reported that SAA1 polymorphisms are associated with the risk of inflammatory disorders.[5] The TT genotype is associated with an increased risk for carotid artery intima-media thickness in obese individuals.[25] Interestingly, AOSD patients with the −13T allele in this study were found to have been treated with immunosuppressants more frequently than those without this allele. These findings suggest that the T allele may confer more proinflammatory phenotypes, including cytokine production, in AOSD patients. In comparison, the CC genotype of vs12218 was found to be more common among patients suffering from coronary artery disease.[26] Because of the presence of multiple receptors, the effects of SAA are pleiotropic and may not be categorized simply as being proinflammatory.
We previously reported that individuals carrying the T allele of -13C/T (rs12218) have increased susceptibility to familial Mediterranean fever (FMF), a hereditary autoinflammatory disease.[27] The significance of the difference in the T-allele frequency compared with that in controls is greater in FMF than in AOSD, a nonhereditary autoimmune disease. We could not provide a plausible explanation for this. We aimed to determine whether any SNPs within the SAA1 gene conferred an increased risk of susceptibility to AOSD. However, collection of AOSD would require collaboration with multiple centers and there is the possibility that the numbers of patients involved would still be small. We assumed that there is a shared genetic basis for AOSD; however, the clinical phenotype of AOSD is heterogeneous.[3] This complexity may have contributed to the less significant association between SAA1 gene polymorphisms and AOSD, compared with that for FMF, a more categorized autoinflammatory disorder.
MEFV gene mutations/polymorphisms have been implicated in more than 1 genetic autoinflammatory disease, FMF.[28] Therefore, we considered that it would be reasonable for the MEFV gene to also be associated with AOSD. The allele frequencies of common polymorphisms in exons 2 and 3 of the MEFV gene in AOSD patients (R408Q, 3.3%; P369S, 3.3%; E148Q, 23.7%) were similar to those of healthy Japanese subjects, as described previously.[29] Therefore, MEFV gene polymorphisms may not be involved in the susceptibility to AOSD. However, these genetic variations may be related to the clinical manifestations of AOSD. Although no statistically significant results were obtained, AOSD patients carrying MEVF variants showed a tendency to be associated with MAS and to be treated with higher doses of steroids than those without these variants. These findings suggest that variation in autoinflammatory genes may modify the disease phenotype of AOSD.
Our study has some limitations, one of which was the relatively small number of studied subjects. This case–control study for SAA1 gene polymorphisms may have been underpowered due to the rarity of patients with AOSD. Another limitation of this study is that the examined subjects were ethnically Japanese; hence, caution should be exercised when extrapolating our results to other ethnic groups. In the present study, we did not examine the differences in circulating SAA levels between each genotype, which was a limitation of our study.
5. Conclusions
In conclusion, we suggest that the SNPs in the promoter region of SAA1 gene (-13C/T) may correlate with AOSD in the Japanese population. Further studies are necessary to determine, which are the predominant SNPs with the susceptibility to AOSD in various ethnic population.
Author contributions
Conceptualization: Atshushi Kawakami.
Data curation: Tomoyuki Asano, Shuzo Sato, Hiroko Kobayashi, Hiroshi Watanabe, Eiji Suzuki, Tadashi Nakamura, Toshimasa Shimizu, Masataka Umeda, and Fumiaki Nonaka.
Formal analysis: Kiyoshi Migita, Tomoyuki Asano, and Tomohiro Koga.
Investigation: Kiyoshi Migita, Hiroshi Furukawa, and Tomohiro Koga.
Methodology: Hiroshi Furukawa and Tomohiro Koga.
Project administration: Kiyoshi Migita, Tomohiro Koga, Yukitaka Ueki, Katsumi Eguchi, and Atshushi Kawakami.
Supervision: Yukitaka Ueki, Katsumi Eguchi, and Atshushi Kawakami.
Validation: Tomohiro Koga.
Writing – original draft: Makiko Yashiro.
Writing – review, and editing: Kiyoshi Migita and Hiroshi Furukawa.
Footnotes
Abbreviations: AOSD = adult onset Still's disease, PCR-RFLP = PCR restriction fragment length polymorphism, SAA = serum amyloid A, SNPs = single-nucleotide polymorphisms.
Ethical approval for this study (No. 21003) was provided by the Ethics Committee of Nagasaki Medical Center and written informed consent was obtained from each individual.
The study was supported by the Practical Research Project for Rare / Intractable Diseases from Japan Agency for Medical Research and Development, AMED.
The authors have no conflicts of interest to disclose.
References
- [1].Castañeda S, Blanco R, González-Gay MA. Adult-onset Still's disease: advances in the treatment. Best Pract Res Clin Rheumatol 2016;30:222–38. [DOI] [PubMed] [Google Scholar]
- [2].Efthimiou P, Georgy S. Pathogenesis and management of adult-onset Still's disease. Semin Arthritis Rheum 2006;36:144–52. [DOI] [PubMed] [Google Scholar]
- [3].Gerfaud-Valentin M, Jamilloux Y, Iwaz J, et al. Adult-onset Still's disease. Autoimmun Rev 2014;13:708–22. [DOI] [PubMed] [Google Scholar]
- [4].Marhaug G, Dowton SB. Serum amyloid A: an acute phase apolipoprotein and precursor of AA amyloid. Baillieres Clin Rheumatol 1994;8:553–73. [DOI] [PubMed] [Google Scholar]
- [5].Sun L, Ye RD. Serum amyloid A1: structure, function and gene polymorphism. Gene 2016;583:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baba S, Masago SA, Takahashi T, et al. A novel allelic variant of serum amyloid A, SAA1 gamma: genomic evidence, evolution, frequency, and implication as a risk factor for reactive systemic AA-amyloidosis. Hum Mol Genet 1995;4:1083–7. [DOI] [PubMed] [Google Scholar]
- [7].Nakamura T, Higashi S, Tomoda K, et al. Significance of SAA1.3 allele genotype in Japanese patients with amyloidosis secondary to rheumatoid arthritis. Rheumatology 2006;45:43–9. [DOI] [PubMed] [Google Scholar]
- [8].Ajiro J, Narita I, Sato F, et al. SAA1 gene polymorphisms and the risk of AA amyloidosis in Japanese patients with rheumatoid arthritis. Mod Rheumatol 2006;16:294–9. [DOI] [PubMed] [Google Scholar]
- [9].Moriguchi M, Terai C, Kaneko H, et al. A novel single-nucleotide polymorphism at the 5′-flanking region of SAA1 associated with risk of type AA amyloidosis secondary to rheumatoid arthritis. Arthritis Rheum 2001;44:1266–72. [DOI] [PubMed] [Google Scholar]
- [10].Yamada T, Okuda Y, Takasugi K, et al. An allele of serum amyloid A1 associated with amyloidosis in both Japanese and Caucasians. Amyloid 2003;10:7–11. [DOI] [PubMed] [Google Scholar]
- [11].Moriguchi M, Kaneko H, Terai C, et al. Relative transcriptional activities of SAA1 promoters polymorphic at position -13(T/C): potential association between increased transcription and amyloidosis. Amyloid 2005;12:26–32. [DOI] [PubMed] [Google Scholar]
- [12].Liang TS, Wang JM, Murphy PM, et al. Serum amyloid A is a chemotactic agonist at FPR2, a low-affinity N-formylpeptide receptor on mouse neutrophils. Biochem Biophys Res Commun 2000;270:331–5. [DOI] [PubMed] [Google Scholar]
- [13].Eklund KK, Niemi K, Kovanen PT. Immune functions of serum amyloid A. Crit Rev Immunol 2012;32:335–48. [DOI] [PubMed] [Google Scholar]
- [14].Ye RD, Sun L. Emerging functions of serum amyloid A in inflammation. J Leukoc Biol 2015;98:923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yamaguchi M, Ohta A, Tsunematsu T, et al. Preliminary criteria for classification of adult Still's disease. J Rheumatol 1992;19:424–30. [PubMed] [Google Scholar]
- [16].Tanaka M, Migita K, Miyashita T, et al. Coexistence of familial Mediterranean fever and Sjögren's syndrome in a Japanese patient. Clin Exp Rheumatol 2007;25:792. [PubMed] [Google Scholar]
- [17].Jensen LE, Whitehead AS. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem J 1998;334:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cunnane G. Amyloid precursors and amyloidosis in inflammatory arthritis. Curr Opin Rheumatol 2001;13:67–73. [DOI] [PubMed] [Google Scholar]
- [19].Gouwy M, De Buck M, Pörtner N, et al. Serum amyloid A chemoattracts immature dendritic cells and indirectly provokes monocyte chemotaxis by induction of cooperating CC and CXC chemokines. Eur J Immunol 2015;45:101–12. [DOI] [PubMed] [Google Scholar]
- [20].Yu N, Liu S, Yi X, et al. Serum amyloid A induces interleukin-1β secretion from keratinocytes via the NACHT, LRR and PYD domains-containing protein 3 inflammasome. Clin Exp Immunol 2015;179:344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Niemi K, Teirilä L, Lappalainen J, et al. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J Immunol 2011;186:6119–28. [DOI] [PubMed] [Google Scholar]
- [22].Chen DY, Chen YM, Chen HH, et al. Functional association of interleukin 18 gene -607 (C/A) promoter polymorphisms with disease course in Chinese patients with adult-onset Still's disease. J Rheumatol 2009;36:2284–9. [DOI] [PubMed] [Google Scholar]
- [23].Efthimiou P, Kontzias A, Ward CM, et al. Adult-onset Still's disease: can recent advances in our understanding of its pathogenesis lead to targeted therapy? Nat Clin Pract Rheumatol 2007;3:328–35. [DOI] [PubMed] [Google Scholar]
- [24].Migita K, Izumi Y, Jiuchi Y, et al. Serum amyloid A induces NLRP-3-mediated IL-1β secretion in neutrophils. PLoS One 2014;9:e96703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xie X, Ma YT, Yang YN, et al. Polymorphisms in the SAA1/2 gene are associated with carotid intima media thickness in healthy Han Chinese subjects: the cardiovascular risk survey. PLoS One 2010;5:e13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang BY, Hang JY, Zhong Y, et al. Association of genetic polymorphisms of SAA1 (rs12218) with myocardial infarction in a Chinese population. Genet Mol Res 2014;13:3693–6. [DOI] [PubMed] [Google Scholar]
- [27].Migita K, Agematsu K, Masumoto J, et al. The contribution of SAA1 polymorphisms to Familial Mediterranean fever susceptibility in the Japanese population. PLoS One 2013;8:e55227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ben-Chetrit E, Peleg H, Aamar S, et al. The spectrum of MEFV clinical presentations--is it familial Mediterranean fever only? Rheumatology (Oxford) 2009;48:1455–9. [DOI] [PubMed] [Google Scholar]
- [29].Migita K, Nakamura T, Maeda Y, et al. MEFV mutations in Japanese rheumatoid arthritis patients. Clin Exp Rheumatol 2008;26:1091–4. [PubMed] [Google Scholar]


