Abstract
Breast cancer is one of the most widespread malignancies in females, and the incidence rate has been increasing in recent years in the world. Genetic factors play an important role in the occurrence of breast cancer. Our study aimed to investigate the relationship between single nucleotide polymorphisms (SNPs) of claudin 10 (CLDN10) (rs1325774, rs7333503, rs3751334) and breast cancer and the clinical characteristics of patients.
A total of 104 patients with breast cancer and 118 healthy controls were recruited in our study between 2013 and 2015. The SNPscan system was used for genotyping. Demographic information, health status, anthropometric parameters and clinical data were considered in analysis. Statistical analysis of the data was carried out using the Student t test, the Chi-square test (X2) or Fisher exact test and unconditional logistic regression analysis. The Gene Expression Profiling Interactive Analyses application (GEPIA) was used to analysis the expression of CLDN10 in breast cancer tissues and normal samples.
The polymorphism of rs1325774 was significantly associated with an increased risk of breast cancer (T/G vs T/T: OR = 2.073, 95% CI = 1.095–3.927, P = .025). After adjusting for age, the association remained statically significant (T/G vs T/T: OR = 2.067, 95% CI = 1.070–3.867, P = .026). Furthermore, harbouring G allele in rs1325774 position was significantly associated with increased risk of breast cancer (OR = 1.993, 95% CI = 1.107–3.589, P = .022). However, no significant association among rs7333503, rs3751334, and breast cancer. The expression level of CLDN10 was reduced in breast cancer tissues compared with normal breast tissues according to the analysis of The Cancer Genome Atlas (TCGA) data through GEPIA.
Our results suggest that the polymorphism of rs1325774 associate with increase the breast cancer risk. No significant relationship between rs1325774 polymorphism and clinical as well as pathological characteristics in patients.
Keywords: breast cancer, CLDN10, clinical characteristic, polymorphisms
1. Introduction
Breast cancer is a worldwide health problem that affects a larger number of women. Global Cancer Statistics reported that more than 1.6 million women were diagnosed with breast cancer in 2012 [1]. The incidence and mortality of breast cancer are increasing in China, where it is now the leading cause of cancer death in women younger than 45 years [2]. Although the causes of breast cancer are unclear, some studies have reported the pathogenesis of the disease to be influenced by genetic factors. Known genetic factors include rare variants; moderate to high penetrance of Breast Cancer gene 1(BRCA1), Breast Cancer gene 2 (BRCA2), partner and localizer of BRCA2 (PALB2), ataxia-telangiectasia mutated (ATM), and checkpoint kinase 2 (CHEK2) and more than 90 common polymorphisms with low penetrance [3–10].
The claudin 10 (CLDN10) gene has the cytogenetic location 13q32.1 and encodes a member of the claudin family. Claudins are integral membrane proteins comprising tight junction strands. Tight junction strands serve as physical barriers to prevent solutes and water from passing through epithelial and endothelial cell sheets and play a key role in maintaining cell polarity and signal transductions. Previous studies have shown that the expression level of CLDN10 is associated with thyroid carcinoma, primary hepatocellular carcinoma and lung adenocarcinoma [11–13]. In addition, Na He et al reported a case-control study in China which demonstrated that a CLDN10 polymorphism (rs1325774) is associated with stage I breast cancer risk, although they failed to verify this relationship in the stage II case-control study [14]. The genetic effect of the rs1325774 polymorphism with regards to breast cancer susceptibility is still unclear.
Our study aimed to investigate the association between CLDN10 polymorphisms (rs1325774, rs7333503 and rs3751334) and breast cancer risk in South Chinese women, and to analyse the relationship between its mutation and the clinical and pathological features of patients.
2. Materials and methods
2.1. Study design
The present study design was based on the case–control study and general characteristics of participants as described in a previously published paper. [15]. The case–control study was conducted in the First Affiliated Hospital of Guangxi Medical University from 2013 to 2015. Participants were Chinese women aged 21 to 74 years. Breast cancer was confirmed by a pathologist, based on immunohistochemical staining. To reduce the potential confounding factors and biases, the following exclusion criteria were used:
-
(1)
patients with secondary tumours or other malignant neoplasms and
-
(2)
the presence of severe infectious diseases.
During the same period, healthy individuals without breast cancer who had attended a check-up at the hospital were enrolled as controls and matched with cases. Both breast cancer patients and healthy controls were randomly selected. A total of 104 breast cancer patients and 118 healthy controls were recruited in this study.
Before recruitment, written consent was obtained from each study subject and the study was approved by the Ethical Review Committee of Guangxi Medical University.
2.2. Data collection
Data collection was carried out at the Medical Centre in the First Affiliated Hospital of Guangxi Medical University. Demographic information (age, ethnicity) and health statuses (menarche age and menopause status) were collected through face-to-face interviews. Anthropometric parameters (height and weight) were measured by trained personnel based on standardised protocols. Body mass indices (BMI) were calculated as weight divided by height squared (kg/m2). Clinical data (metastasis distance, molecular classification and pathology type) of patients were obtained from medical records. The intrinsic subtypes of breast cancer included Luminal A subtypes, Luminal B1, Luminal B2, HER-2 overexpressing, and triple-negative. The pathology type was classified into ductal, lobular and other based on the pathological result.
2.3. DNA extraction and genotyping
Genomic DNA was extracted from whole blood samples using a genomic DNA isolation kit (Sangon Biotech, Shanghai, China) according to the manufacturer's instructions. The SNPscan system (Genesky Biotechnologies Inc., Shanghai, China) was used for genotyping. This system has been reported in detail elsewhere (http://biotech.geneskies.com/en/index.php/Index/fuwuer/id/29). Briefly, the technology is a multi-gene mutation screening method, which improves the multiplex ligation-dependent probe amplification technology. Four different fluorescent dyes and lengths of ligations were applied in order to detect multiple SNPs simultaneously in 1 reaction. Quality control assessment was performed during genotyping to ensure accuracy. The samples were blinded to laboratory personnel and blank controls were used for all PCR amplifications. Finally, 10% of samples from the subjects were randomly selected for evaluation of quality of genotyping, which showed 100% concordance. The sequences of specific ligase probes are presented in Table 1.
Table 1.
Primers used for this study.

3. Statistical analysis
All data were recorded as Microsoft Excel files and analysed using Statistical Package for Social Sciences program (SPSS) version 22.0 software (IBM, São Paulo, Brazil). The Hardy–Weinberg equilibrium (HWE; P >.05) was performed using SHEsis (http://analysis.bio-x.cn/myAnalysis.php) [16,17]. Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn) was used to analyse the expression of CLDN10 in breast cancer tissues and normal sample [18]. Briefly, GEPIA is a web-based tool that provides fast and customisable functionalities for research based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data. Differential expression analysis, profiling plotting, correlation analysis, patient survival analysis, similar gene detection and dimensionality reduction analysis can be carried out using GEPIA.
The mean and standard deviation (SD) were used to describe the age variable, and the Student t test was used to analyse the difference in ages between the case and control groups. The association between the CLDN10 polymorphisms (rs1325774, rs7333503, and rs3751334) and breast cancer risk, as well as the patients’ demographic information and health statuses, were analysed using simple unconditional logistic regression and multiple logistic regression analysis. The Chi-square test (X2) or Fisher exact test was applied to calculate the relationship between the rs1325774 polymorphism and tumour characteristics. A P value <.05 was considered statistically significant.
4. Result
4.1. The association between CLDN10 rs1325774, rs7333503 and rs3751334 polymorphisms and the susceptibility to breast cancer
Of the selected study participants, 6 control subjects were removed due to genotyping failure. The genotype frequencies of the controls were in accordance with the Hardy-Weinberg equilibrium. The association between the 3 CLDN10 polymorphisms studied and breast cancer risk is shown in Table 2. A frequency of disease of 24.1% was observed for the rs1325774 T/G genotype, and the distribution of the T/G genotype in patients and healthy controls was 32 and 20 respectively. The G/G genotype was not detected in healthy controls. According to simple unconditional logistic regression, the rs1325774 T/G genotype was significantly associated with an increased risk of breast cancer (T/G vs. T/T: OR = 2.073, 95% CI = 1.095–3.927, P = .025).
Table 2.
Association between CLDN10 polymorphisms (rs1325774, rs7333503, rs3751334) and breast cancer risk.
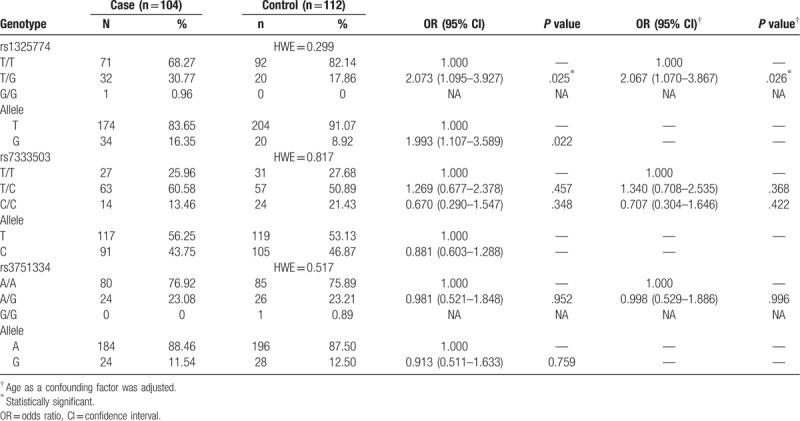
After adjusting for age, associations between the T/G genotype of rs1325774 and breast cancer remained statically significant (OR = 2.067, 95% CI = 1.070–3.867, P = .026). Furthermore, the G allele of rs1325774 was significantly associated with an increased risk of breast cancer (OR = 1.993, 95% CI = 1.107–3.589, P = .022).
In this study, no significant association between rs7333503 or rs3751334 and breast cancer was observed.
4.2. Lack of association between rs1325774 polymorphism of CLDN10 and clinical characteristics in breast cancer patients
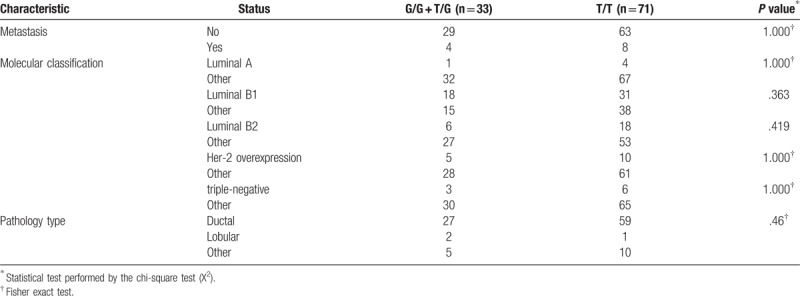
To understand the association between the rs1325774 polymorphism of CLDN10 and clinical risk factors, we analysed the clinical data including demographic information, health status and clinical data. No parameters were significantly associated with the polymorphism and subset analysis did not indicate any significant result (Tables 3 and 4). Therefore, no association was seen between the rs1325774 polymorphism of CLDN10 and clinical characteristics of breast cancer.
Table 3.
Association analysis between CLDN10 rs1325774 and subgroups of 104 patients with breast cancer.
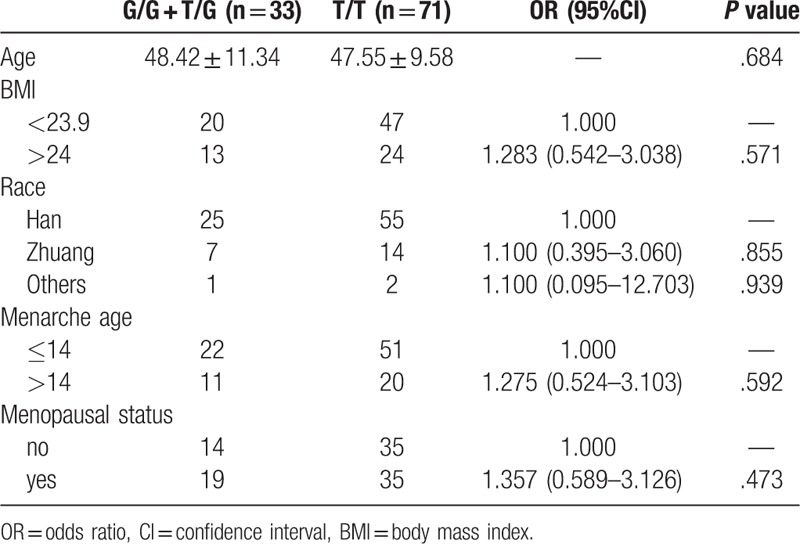
Table 4.
The relationship between rs1325774 polymorphism and tumor characteristic.
4.3. Expression of CLDN10 is reduced in breast cancer tissues
Analysis of matched normal tissue from TCGA database using GEPIA indicated that the expression level of CLDN10 in breast cancer tissues (n = 1085) was reduced compared with normal tissues (n = 112) (Fig. 1A). The expression body map analysed by normalising to transcripts per million (TPM) revealed the median expression of CLDN10 to be reduced in breast cancer tissues compared with normal samples, with numerical values of 0.21 and 0.88 respectively (Fig. 1B).
Figure 1.
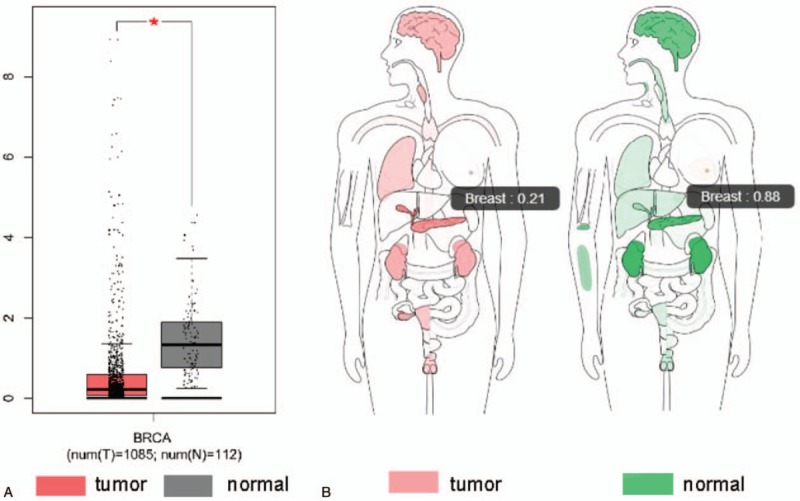
The median expression level of CLDN10 gene in normal and breast cancer tissues by GEPIA. The GEPIA indicated that the expression level of CLDN10 in breast cancer tissues (n = 1085) was reduced compared with normal tissues (n = 112) based on TCGA data (Figure 1A). The median expression of CLDN10 to be reduced in breast cancer tissues compared with normal samples based on the expression body map analysed through normalising to TPM, with numerical values of 0.21 and 0.88 respectively (Figure 1B). GEPIA = Gene Expression Profiling Interaction Analysis, TPM = transcripts per million.
5. Discussion
Breast cancer is one of the most widespread malignancies in females [1,19], and genetic effects can have important influences on the occurrence of the disease. In this study, we aimed to evaluate the relationship between 3 SNPs of CNDL10 and breast cancer, as well as relationship of the SNPs to the clinical characteristics of the patients. Our study involved 104 breast cancer patients and 112 healthy controls and found that the rs1325774 polymorphism was significantly associated with an increased risk of breast cancer. No significant association was observed between the rs7333503 or rs3751334 polymorphisms and breast cancer. Expression of CLDN10 was reduced in breast cancer tissues compared with normal breast tissues according to the analysis of matched normal tissue from TCGA using GEPIA [18].
There are some limitations in our study: First, our study has selection bias because our subjects recruited from the same hospital. Second, with the limited sample size and statistical power (<80%), the P value was only considered significant at the .05 level, which may affect the results of stratified analysis. Further studies are needed to confirm the influence of the rs1325774 polymorphism on breast cancer based on larger populations. Third, our study did not elucidate the mechanism of pathology. Further functional studies are required to reveal the molecular mechanism of rs1325774-promotion of breast cancer.
Previous studies have reported that the level of CLDN10 expression is associated with hepatocellular carcinoma, thyroid carcinoma and lung adenocarcinoma [11–13]. Human CLDN10 expression is associated with cancer progression—when CLDN10 is overexpressed in patients with lung adenocarcinoma, a good prognosis for overall survival is reported [13]. However, it is not clear whether SNPs located in different regions of CLDN10 are associated with the risk and clinical characteristics of breast cancer.
Na He et al previously reported that rs1325774, located in the microRNA binding site in the seed region of the 3’-UTR of CLDN10, is significantly associated with an increased risk of breast cancer in a large case-control study which included genome-wide microRNA binding site SNPs in stage I [14]. The results of the present study are consistent with those of Na He et al.
The study of SNPs at miRNA target sites has attracted attention. It has been reported that genetic changes in the 3’-UTR of transcribed genes can alter the affinity between miRNA and mRNA. Therefore, these single nucleotide mutations can change the levels of protein expression through manipulation of the interactions between miRNA and their target sequences [20–23]. It can be hypothesised that miRNA-related SNPs may be a factor in the occurrence of breast cancer.
Huiyan Sun et al reported that miR-486 functions as a novel tumour suppressor miRNA in hepatocellular carcinoma (HCC) by targeting CLDN10. By regulating the expression of this gene, miR-486 inhibits proliferation and migration of hepatoma cells and functions. It has been reported that CLDN10 dominantly contributes to the suppressive effect of miR-486 on the epithelial-mesenchymal transition (EMT) of hepatocellular carcinoma cell [11]. Currently, 8 miRNAs that can target CLDN10 are detailed in miRTarBase. In regards to the specific function of CLDN10 in breast cancer development and progression, we hypothesise that rs1325774 affects the binding affinity of miRNAs and thus affects the occurrence of breast cancer by regulating the expression of CLDN10.
In conclusion, our results demonstrate the relationship between the rs1325774 polymorphism of CLDN10 and the risk of breast cancer. This finding is consistent with the large case-control study of Han Chinese women with stage I breast cancer. Together, the 2 studies suggest that rs1325774 may be a biomarker for breast cancer predisposition. Further functional studies are needed to identify the exact role of rs1325774 in the occurrence of breast cancer.
Author contributions
Conceptualization: Jinling Liao.
Data curation: zengnan mo, Jinling Liao, Jie Li.
Formal analysis: Hong Cheng, Yang Chen.
Investigation: Jinling Liao, Jie Li, Yang Chen.
Methodology: zengnan mo, Jie Li, Hong Cheng, Yang Chen.
Project administration: zengnan mo.
Writing – original draft: Jinling Liao.
Footnotes
Abbreviations: CLDN10 = claudin 10, GEPIA = Gene Expression Profiling Interactive Analyses application, PALB2 = partner and localizer of BRCA2, SNPs = single nucleotide polymorphisms, TCGA = The Cancer Genome Atlas.
JL and JL contributed equally to this work.
The authors have no conflicts of interest to disclose.
Funding: This study was funded by the Natural Science Foundation of China (grant number: 81370857), the Natural Science Foundation of China (grant number: 81770759).
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [3].Rudolph A, Chang-Claude J, Schmidt MK. Gene-environment interaction and risk of breast cancer. Br J Cancer 2016;114:125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lalloo F, Evans DG. Familial breast cancer. Clin Genet 2012;82:105–14. [DOI] [PubMed] [Google Scholar]
- [5].Tabatabaeian H, Hojati Z. Assessment of HER-2 gene overexpression in Isfahan province breast cancer patients using Real Time RT-PCR and immunohistochemistry. Gene 2013;531:39–43. [DOI] [PubMed] [Google Scholar]
- [6].Sadeghi S, Hojati Z, Tabatabaeian H. Cooverexpression of EpCAM and c-myc genes in malignant breast tumours. J Genet 2017;96:109–18. [DOI] [PubMed] [Google Scholar]
- [7].Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007;447:1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A 2008;105:4340–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 2007;39:865–9. [DOI] [PubMed] [Google Scholar]
- [10].Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 2003;72:1117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sun H, Cui C, Xiao F, et al. miR-486 regulates metastasis and chemosensitivity in hepatocellular carcinoma by targeting CLDN10 and CITRON. Hepatol Res 2015;45:1312–22. [DOI] [PubMed] [Google Scholar]
- [12].Barros-Filho MC, Marchi FA, Pinto CA, et al. High diagnostic accuracy based on CLDN10, HMGA2, and LAMB3 transcripts in papillary thyroid carcinoma. J Clin Endocrinol Metab 2015;100:E890–9. [DOI] [PubMed] [Google Scholar]
- [13].Barros-Filho MC, Marchi FA, Pinto CA, et al. Expression of CLDN1 and CLDN10 in lung adenocarcinoma in situ and invasive lepidic predominant adenocarcinoma. J Cardiothorac Surg 2013;8:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].He N, Zheng H, Li P, et al. miR-485-5p binding site SNP rs8752 in HPGD gene is associated with breast cancer risk. PLoS One 2014;9:e102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liao J, Chen Y, Zhu J, et al. Polymorphisms in the TOX3/LOC643714 and risk of breast cancer in south China. Int J Biol Markers 2018;1724600818755633. [DOI] [PubMed] [Google Scholar]
- [16].Shi YY, He L. SHEsis a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97–8. [DOI] [PubMed] [Google Scholar]
- [17].Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res 2009;19:519–23. [DOI] [PubMed] [Google Scholar]
- [18].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- [20].Chen K, Song F, Calin GA, et al. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis 2008;29:1306–11. [DOI] [PubMed] [Google Scholar]
- [21].Landi D, Gemignani F, Barale R, et al. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol 2008;27:35–43. [DOI] [PubMed] [Google Scholar]
- [22].Turashvili G, Lightbody ED, Tyryshkin K, SenGupta SK, Elliott BE, Madarnas Y, et al. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. FASEB J 2018:fj201800120R. [DOI] [PubMed] [Google Scholar]
- [23].Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res 2010;70:2789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]


