Abstract
DNA-protein cross-links (DPCs) are super-bulky DNA adducts induced by common chemotherapeutic agents, reactive oxygen species, and aldehydes, and also formed endogenously as part of epigenetic regulation. Despite their presence in most cells and tissues, the biological effects of DPCs are poorly understood due to the difficulty of constructing site-specific DNA-protein conjugates. In the present work, a new approach of conjugating proteins to DNA using oxime ligation was used to generate model DPCs structurally analogous to lesions formed in cells. In our approach, proteins and peptides containing an unnatural oxy-Lys amino acid were cross-linked to DNA strands functionalized with 5-formyl-dC or 7-(2-oxoethyl)-7-deaza-dG residues using oxime ligation. The conjugation reaction was site-specific with respect to both protein and DNA, provided excellent reaction yields, and formed stable DPCs amenable to biological evaluation.
Graphical Abstract
Model site-specific DNA-protein cross-link formation by bioorthogonal oxime ligation
Exposure to exogenous and endogenous bis-electrophiles, α,ϐ- unsaturated carbonyls, and free radicals results in irreversible entrapment of cellular proteins on genomic DNA to give DNA-protein cross-links (DPCs).1 DPCs accumulate in the heart and brain tissues and are hypothesized to contribute to aging, cancer, and neurological diseases.2, 3 Interestingly, DPCs involving histone proteins were recently shown to reversibly form in healthy cells as part of epigenetic regulation.4, 5 Owing to their unusually bulky size, DPC lesions are expected to block the majority of DNA transactions including replication, transcription, and repair, leading to cell death and genomic instability.6, 7 Our understanding of the structural consequences and biological outcomes of DPC formation is incomplete due to the paucity of synthetic methodologies to generate site-specific model DNA-protein cross-links for biological studies.
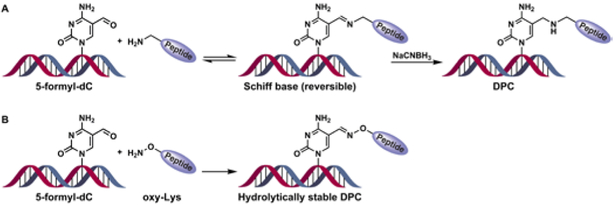
Our previous studies have employed a reductive amination strategy to attach proteins and peptides to DNA containing 5-formyl-dC and 7-(2-oxoethyl)-7-deaza-dG.s In that approach, Schiff base conjugates between basic side chains of proteins and DNA are reduced with NaCNBH3 to produce a stable amine-linked conjugate (Scheme 1A).8 A major limitation of the reductive amination methodology is that a mixture of products is often formed due to the participation of multiple Lys or Arg protein side chains in the cross-linking reaction.8 Furthermore, the use of strong reducing agents may lead to protein denaturation, precluding structural studies of the resulting DNA-protein conjugates.
Scheme 1.
DPC formation at the C5-position of dC by reductive amination (A) and oxime ligation (B).
In the present study, we developed a new methodology for site specific conjugation of proteins to DNA by taking advantage of the ability of substituted alkoxyamines to form stable oxime conjugates with aldehydes under mild conditions (Scheme 1B).9, 10 A structural analogue of lysine, where the ε-CH2 group was replaced with oxygen (oxy-Lys), was site-specifically incorporated into peptides using solid phase synthesis. Oxy-Lys containing peptides were ligated with proteins using sortase A mediated ligation. DNA strands incorporating site-specific 5-formyl-dC (5fC) were prepared by phosphoramidite chemistry and conjugated to oxy-Lys-containing peptides and proteins to obtain structurally defined DNA-peptide and DNA-protein cross-links (Scheme 1B), which were characterized by gel electrophoresis and mass spectrometry.
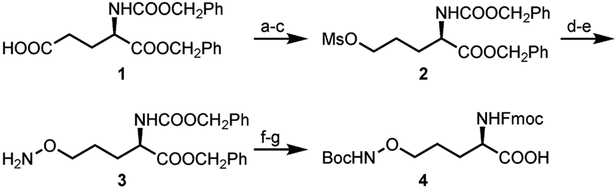
To synthesize the oxy-Lys building block for polypeptide assembly, glutamic acid derivative 1 was chain-extended and reduced with NaBH4 to give the corresponding alcohol, which was protected with mesyl chloride to give 2 (Scheme 2). Treatment of 2 with N-hydroxyphthalimide in presence of DBU and subsequent hydrazinolysis gave oxy-amine 3 in a good yield (40% over two steps).11 The free amine of 3 was further protected with Boc anhydride and Fmoc-succinate to yield compound 4, a monomer suitable for peptide synthesis. Detailed experimental procedures, reaction yields, and structural characterization details are given in Scheme S1 and Supplementary Methods (SI, S2).
Scheme 2.
Synthesis of protected oxy-lysine. a) ethylchloroformate, N-methylmorpholine, THF, −10°C; b) NaBH4, MeOH, 0°C; c) methanesulfonylchloride, Et3N, DCM, 0°C; d) N-hydroxyphthalimide, DBU, DMF, 0°C-rt; e) methylhydrazine, DCM, 0°C; f) Boc2O, Et3N, THF, rt; g) H2/Pd/C, rt, FmocOsu, NaHCO3, dioxane, H2O, rt.
A panel of oxy-Lys containing peptides 4-11 amino acids in length was prepared by standard solid phase peptide synthesis and structurally characterized by HPLC-ESI-MS (Pep 1-4 in Table 1). A DNA 12-mer containing 5-formyl-dC (DNA-1 in Table 1) was prepared by solid phase synthesis using commercially available 5-formyl-deoxycytidine phosphoramidite.
Table 1.
Synthetic ODNs and polypeptides prepared for the conjugation studies. G* = DHP-dG, C* = 5-formyl-dC, K* = oxy-Lys.a
| Sequences | m/z Calc. |
m/z Obs. | |
|---|---|---|---|
| Pep-1 | NH2-K*GG A | 333.3 | 334.2 |
| Pep-2 | NH2-GGK GLG K*GG A | 802.9 | 803.6 |
| Pep-3 | NH2-GGG KGL GKG GA | 857.9 | 858.4 |
| Pep-4 | NH2-GGG KGL GK*G GA | 859.7 | 860.4 |
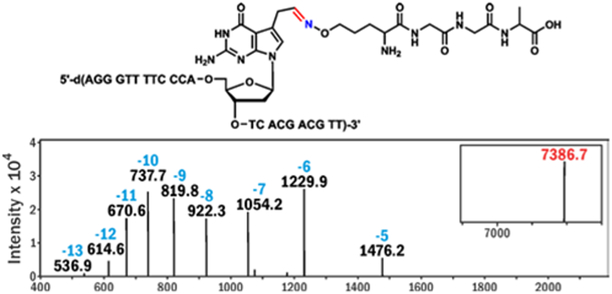
| DNA-1 | 5'-d(ATG GCG TGC* TAT)-3’ | 3704.4 | 3704.0 |
| DNA-2 | 5' -d(AG3T4C3AG*TCACGACGTT)-3' | 7102.6 | 7103.4 |
| DNA-3 | 5'-d(ATG GCG TGC TAT)-3' | 3676.4 | 3675.6 |
| DNA-4 | 5'-d(AG3T4C3AGTCACGACG TT)-3' | 7030.6 | 7029.1 |
| DPC-1 | 4019.5 | 4019.1 | |
| DPC-2 | 4490.5 | 4489.0 | |
| DPC-3 | 4547.3 | 4546.2 | |
| DPC-4 | 7387.9 | 7386.7 | |
| DPC-5 | 7856.9 | 7853.2 | |
| DPC-6 | 7914.5 | 7913.0 |
Peptides 1-4 were measured in +ve and DNA and DPCs are in −ve mode.
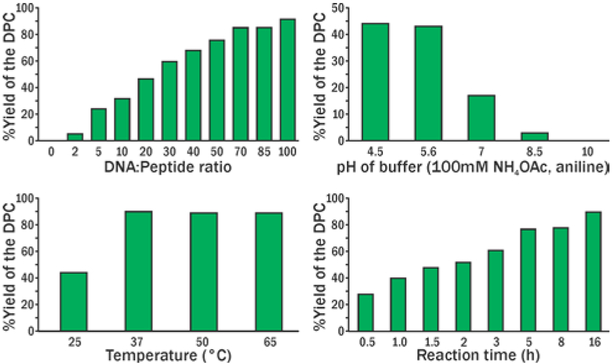
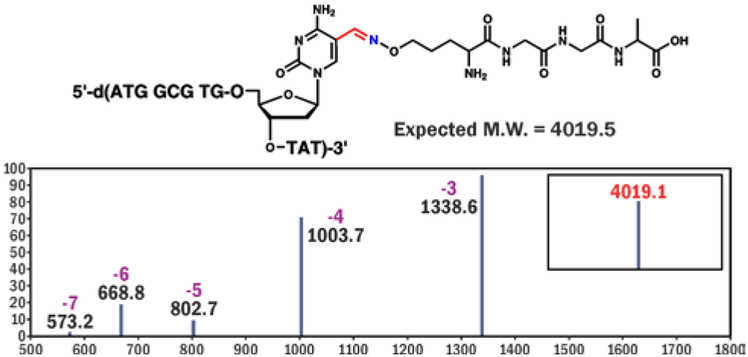
The conjugation reactions between 5-formyl-dC-containing DNA and oxy-Lys-containing peptides (Scheme 1B) were monitored by denaturing gel electrophoresis (SI, Figures S1-S3). Aniline (100 mM) was used to catalyze the oxime bond formation.12 A range of reaction conditions with varying temperature, reaction time, pH, and molar concentrations of the reactants were tested (Figure 1, S1-S3). The best reaction yields (up to 95%) were achieved for reactions performed in NH4OAc buffer pH 4.5 containing 100 mM aniline, at 37°C for 24 h. The structure of the conjugate was confirmed by HPLC-ESI-MS/MS (Figure 2, Table 1). Conjugation reaction works efficiently on ds-DNA as well (SI, Figure S4).
Figure 1.
Optimization of reaction conditions for DPC formation between 5-formyl-dC containing DNA-1 and oxy-Lys containing Pep-1 (Table 1).
Figure 2.
ESI+ mass spectrum and deconvoluted MS (insert) of DPC between 5-formyl-dC containing DNA-1 and oxy-Lys containing Pep-1.
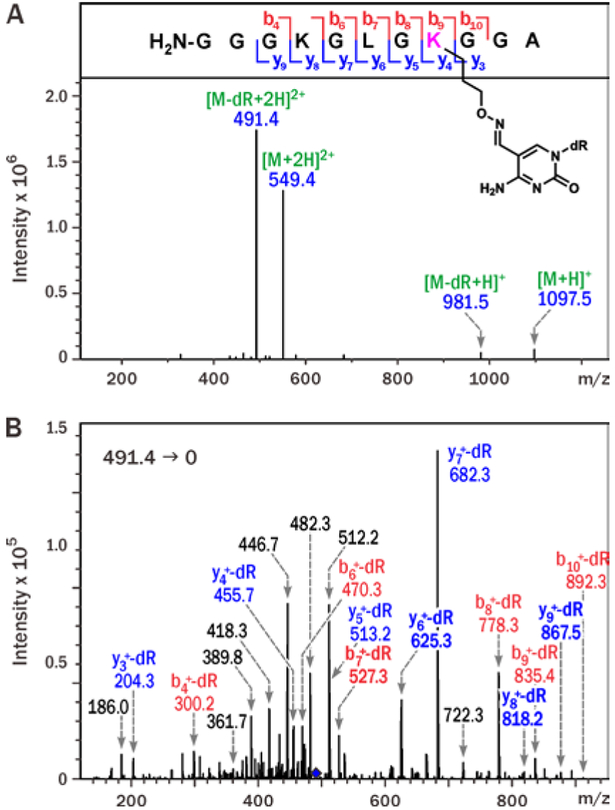
To confirm the exact chemical structure of the 5fC-mediated DNA-peptide conjugates, Pep-4 (Table 1) was incubated with excess of 5-formyl-dC nucleoside in the presence of 100 mM aniline (SI, S5). The resulting peptide-nucleoside conjugates were sequenced by tandem mass spectrometry (Figure 3). ESI MS signal at m/z 1097.5 matched the molecular weight of the expected conjugate between the 11-mer peptide and 5-formyl-dC (Figure 3A). MS/MS fragmentation of the doubly charged peptide–nucleoside conjugate [M + 2H]2+ (m/z 549.4) in an ion trap mass spectrometer was dominated by product ions at m/z 491.4, corresponding to the loss of deoxyribose from dC, while the b and y ion series were consistent with modification of the K3 lysine residue (Figure 3B).
Figure 3.
ESI+ MS (A) and MS2 spectra of 11-mer Pep-4 conjugated to 5-formyl-dC nucleoside via oxy-Lys (B).
These results are in agreement with the cross-linking mechanism13 depicted in (Scheme 1, in which DPC formation between 5fC and oxy-Lys involves oxime formation at the modified Lys.
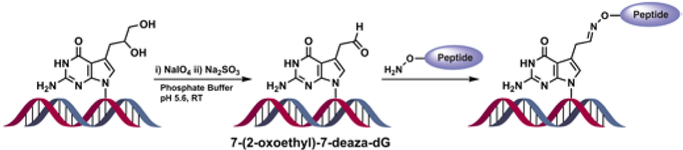
A similar methodology was employed to introduce DPC lesions at the C7 position of 7-deaza-dG residues of DNA (Scheme 3).7, 14 The latter conjugates serve as hydrolytically stable mimics of N7-dG lesions formed in cells treated with bis-electrophiles such as nitrogen mustards, platinum compounds, and 1,2,3,4-diepoxybutane.15-18 In this case, the reactive aldehyde was masked as a vicinal diol (Scheme 3) by introducing 7-deaza-7-(2,3-dihydroprop-1-yl)-dG into DNA. The N7 nitrogen of guanine was replaced with carbon in order to prevent spontaneous depurination.19 Following DNA synthesis by solid phase methodologies and cleavage from the solid support, the 7-deaza-7-(2,3-dihydroprop-1-yl)-dG containing DNA-2 was oxidized with NaIO4 to give the corresponding 7-deaza-7-(2-oxoethyl)-dG - containing DNA and characterized by ESI-MS (SI S4, Scheme S3, Figure S7). The conjugation reactions between DNA 23-mer containing site-specific 7-deaza-7-(2-oxoethyl)-dG and aminoxy-Lys containing peptides (Scheme 3) were conducted in phosphate buffer pH=5.6 at room temperature, resulting in reaction yields of 60-80% (SI S4). The reaction conditions were modified because methods employed for conjugation at 5fdC did not yield acceptable DPC yields at 7-deaza-dG. The reaction products were isolated by denaturing PAGE and characterized by HPLC-ESI-MS (Table 1, (Figures 4, S5; for results of conjugation reactions on ds-DNA see SI, Figure S6).
Scheme 3.
DPC generation at the C7 position of 7-deaza-dG via oxime ligation.
Figure 4.
ESI+ mass spectrum and deconvoluted spectrum (insert) of DPC formed between 7-deaza-7-(2-oxoethyl)-dG containing DNA-2 and oxy-Lys containing Pep-1.
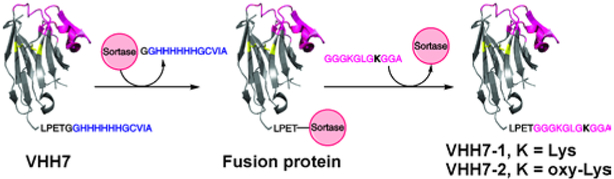
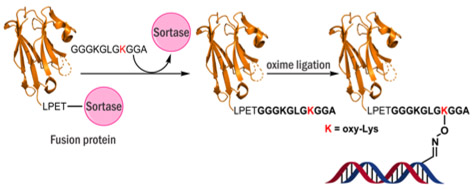
In order to use the novel oxime ligation chemistry for the creation of DNA-protein conjugates, the unnatural oxy-Lys residue was incorporated into VHH7 protein, a single domain antibody, using sortase A mediated ligation (Scheme 4).20 In this protein, the C-terminus-engineered with the sortase A recognition sequence (LPETG) is distant from the antigen-binding site and hence incorporation of oxy-Lys containing peptide should not compromise its antigen-binding capacity.21 We chose VHH7 since this is a small antigen binding protein and is easily obtainable via bacterial expression.22
Scheme 4.
Sortase A mediated ligation of of Lys and oxy-Lys containing peptides 3 and 4 (Table 1) with VHH7 protein.
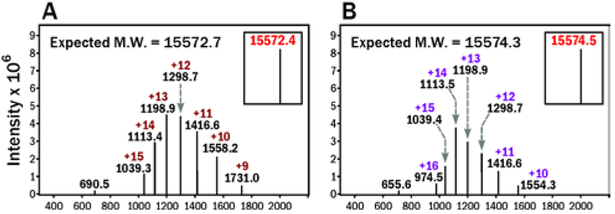
To create a suitable substrate for sortase A mediated ligation, solid phase synthesis was used to generate peptides with N-terminal triglycine segment and containing either native Lys or oxy-Lys as part of their amino acid sequence (Peptides 3 and 4 in Table 1). Peptides 3 and 4 were used as substrates for sortase A mediated ligation (Scheme 4, SI S6, S7). Two protein species were prepared using this approach: VHH7-1 containing native Lys and VHH7-2 containing oxy-Lys at the same position and are characterized by ESI-MS (Figure 5).
Figure 5.
ESI mass spectra of sortase mediated ligation products VHH7-1 (A) and VHH7-2 (B).
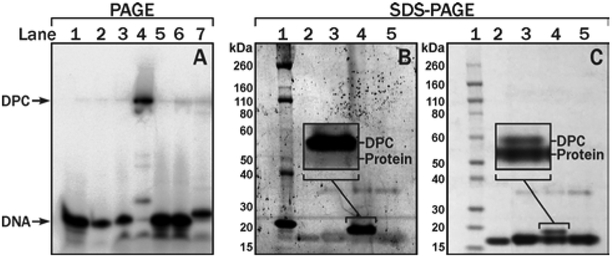
The resulting fusion proteins (VHH7-1 and VHH7-2) were subjected to oxime ligation with 5-formyl-dC containing DNA (DNA-1 in Table 1), and the reaction products were resolved by denaturing PAGE (SI S8). DNA-protein cross-links were observed as a slowly migrating band (Lane 4 in (Figure 6A), which reverted back to a species similar to the starting DNA upon treatment with proteinase K (Lane 7, (Figure 6). The reaction of VHH7-2 with 5-formyl-dC DNA was performed in NH4OAc buffer at pH 4.5 in the presence of aniline, leading to the formation of DPC conjugates (Lane 4 in (Figure 6A). In contrast, no reaction was observed for the corresponding reaction with the VHH7-1 protein lacking the oxy-Lys residue (Lane 5 in (Figure 6A), confirming that the reaction is specific for the oxy-Lys site. The reaction yields increased as the molar equivalents of the protein were increased (SI Figure S8).
Figure 6.
Denaturing PAGE (A) and SDS-PAGE (B,C) analyses of 5-formyl-dC-mediated DNA-VHH7 protein cross-links (A) Lane 1, DNA-3. 2. DNA-1. 3. VHH7-1 + DNA-1. 4. VHH7-2 + DNA-1. 5. VHH7-1 + DNA-3. 6. VHH7-2+DNA-3. 7. VHH7-2 + DNA-1 after Proteinase K treatment (B) SYBR green staining of DNA; (C) Simply blue staining of proteins. Lane 1. Protein ladder. 2. VHH7-1 protein. 3. VHH7-2. 4. VHH7-2 + DNA-1. 5. VHH7-2 + DNA-3.
DPC formation upon reaction of DNA-1 with VHH7-2 was further confirmed by SDS-PAGE analysis (Figure 6B and C). Protein bands with reduced mobility (M ~ 19 kDa) were identified as DPC because they could be visualised by both DNA (Figure 6B) and protein stains (Figure 6C). Similarly, VHH7 proteins were conjugated to 7-deaza-7-(2-oxoethyl)-dG containing DNA (DNA-2) resulting in covalent DPCs in ~60% yield for VHH7-2 (SI, Lane 4 in Figure S9).
To identify amino acid residue(s) of VHH7 participating in cross-linking, DNA portion of gel purified DPC was digested to nucleosides, and the resulting protein-nucleoside conjugate was then subjected to tryptic digestion (SI S9). Tryptic peptides were detected by nanoLC-ESI+-MS/MS analysis using an Orbitrap Elite mass spectrometer. Only the oxy-Lys residue was found to be cross-linked to 5-formyl-dC (SI Figure S10), while four other Lys and 10 Arg residues were intact (Table S1). Therefore, oxime ligation was site-specific for oxy-Lys within VHH7-2 and 5-formyl-dC in DNA. In summary, we have developed a new methodology to create covalent DNA-protein conjugates using a bioorthogonal reaction between aldehyde groups present in DNA and oxy-Lys residues within proteins and peptides. The reaction proceeds spontaneously in good yield and does not require a reducing agent to generate a stable conjugate. The resulting DPCs are structurally analogous to DPCs endogenously generated in chromatin at 5-formyl-dC (Scheme 1 and Figure S10)4, 5 and those created in the genome by the action of common chemotherapeutic agents such as nitrogen mustards and cisplatin (Scheme 2) 1, 17, 18, 23 They differ only by the substitution of an oxygen atom in place of the ε-CH2 group normally found in a natural lysine residue. We anticipate that this new methodology will find broad utility in the areas of chemical carcinogenesis, DNA repair, and epigenetic regulation.
We thank Dr. Guru S. Madugundu and Yingchun Zhao for their help with MS analysis of DPCs, Dr. M. Rashidian and H. Ploegh for providing recombinant Sortase A and pHEN6, and Robert Carlson (UMN Masonic Cancer Center) for help with editing and the graphics for the manuscript. This research was funded by NIH grants ES-023350 and GM-084152.
Supplementary Material
Footnotes
Conflicts of interest
There are no conflicts of interest to declare
Notes and references
- 1.Tretyakova NY, Groehler A and Ji S, Acc. Chem. Res, 2015, 48, 1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izzotti A, Cartiglia C, Taningher M, De Flora S and Balansky R, Mutat. Res, 1999, 446, 215–223. [DOI] [PubMed] [Google Scholar]
- 3.Wu F-Y, Lee Y-J, Chen D-R and Kuo H-W, Mutat. Res, 2002, 501, 69–78. [DOI] [PubMed] [Google Scholar]
- 4.Ji S, Shao H, Han Q, Seiler CL and Tretyakova NY, Angew. Chem. Int. Ed, 2017, 56, 14130–14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, Zhang Y, Bai J, Greenberg MM, Xi Z and Zhou C, J. Am. Chem. Soc, 2017, 139, 10617–10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M and Salem AM, Mutat. Res, 2011, 711,113–122. [DOI] [PubMed] [Google Scholar]
- 7.Wickramaratne S, Ji S, Mukherjee S, Su Y, Pence MG, Lior-Hoffmann L, Fu I, Broyde S, Guengerich FP, Distefano M, Schärer OD, Sham YY and Tretyakova N, Biol. Chem, 2016, 291, 23589–23603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickramaratne S, Mukherjee S, Villalta PW, Scharer OD and Tretyakova NY, Bioconjug. Chem, 2013, 24,1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kölmel DK and Kool ET, Chem. Rev, 2017, 117,10358–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich S, Boturyn D, Marra A, Renaudet O and Dumy P, Chemistry - A European Journal, 2014, 20, 34–41. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Thomas J and Burke TR Jr., Synthesis (Stuttg), 2008, 15, 2432–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirksen A, Hackeng TM and Dawson PE, Angew. Chem. Int. Ed, 2006, 45, 7581–7584. [DOI] [PubMed] [Google Scholar]
- 13.Guo P, Yan S, Hu J, Xing X, Wang C, Xu X, Qiu X, Ma W, Lu C, Weng X and Zhou X, Org. Lett, 2013, 15, 3266–3269. [DOI] [PubMed] [Google Scholar]
- 14.Roy U, Mukherjee S, Sharma A, Frank EG Schärer OD, Nucleic Acids Res, 2016, 44, 7281–7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE and Tretyakova N, Chem. Res. Toxicol, 2008, 21, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeber R, Rajesh M, Fang Q, Pegg AE and Tretyakova N, Chem. Res. Toxicol, 2006, 19, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groehler A, Villalta PW, Campbell C and Tretyakova N, Chem. Res. Toxicol, 2016, 29,190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ming X, 2011. MS Thesis, University of Minnesota. [Google Scholar]
- 19.Angelov T, Guainazzi and A Schärer OD, Org. Lett, 2009, 11, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorr BM, Ham HO, An C, Chaikof EL and Liu DR, Proc. Natl. Acad. Sci. USA, 2014, 111, 13343–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S and Wyns L, Proc. Natl. Acad. Sci. USA, 2006, 103, 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashidian M, Keliher EJ, Bilate AM, Duarte JN, Wojtkiewicz GR, Jacobsen JT, Cragnolini J, Swee LK, Victora GD, Weissleder R and Ploegh HL, Proc. Natl. Acad. Sci. USA, 2015, 112, 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelson-Richie ED, Ming X, Codreanu SG, Loeber RL, Liebler DC, Campbell C and Tretyakova NY, J. Proteome. Res, 2011, 10, 2785–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.