Abstract
The follicular helper T cell (TFH) are established regulators of germinal center (GC) B cells, whether TFH have pathogenic potential independent of B cells is unknown. Based on in vitro TFH cell differentiation, in vivo T cell transfer animal colitis model, and intestinal tissues of inflammatory bowel disease (IBD) patients, TFH and its functions in colitis development were analyzed by FACS, ChIP, ChIP-sequencing, WB, ELISA and PCR. Herein we demonstrate that intestinal tissues of patients and colon tissues obtained from Rag1−/− recipients of naïve CD4+ T cells with colitis, each over-express TFHassociated gene products. Adoptive transfer of naïve Bcl6−/− CD4+ T cells into Rag1−/− recipient mice abrogated development of colitis and limited TFH differentiation in vivo, demonstrating a mechanistic link. In contrast, T cell deficiency of interferon regulatory factor 8 (IRF8) resulted in augmentation of TFH induction in vitro and in vivo. Functional studies showed that adoptive transfer of IRF8 deficient CD4+ T cells into Rag1−/− recipients exacerbated colitis development associated with increased gut TFH-related gene expression, while Irf8−/−/Bcl6−/− CD4+ T cells abrogated colitis, together indicating that IRF8-regulated TFH can directly cause colon inflammation. Molecular analyses revealed that IRF8 suppresses TFH differentiation by inhibiting transcription and transactivation of the TF IRF4, which is also known to be essential for TFH induction. Our documentation showed that IRF8-regulated TFH can function as B-cell-independent, pathogenic, mediators of colitis suggests that targeting TFH could be effective for treatment of IBD.
Keywords: IRF8, TFH, Colitis, IRF4
1. Introduction
T cell help to B cells is a fundamental mechanism for the generation of protective humoral immunity, but over-activation of B cells by T cells can result in excessive humoral immune responses, pathologic inflammation, and autoimmunity1–3. A subset CD4+ T cells, termed follicular helper T cells (TFH), are specialized regulators of T cell help to B cells and are required for induction of germinal center (GC) B cell responses4–5.
TFH stably express C-X-C chemokine receptor 5 (CXCR5), which mediates chemotaxis toward GCs upon ligation by C-X-C chemokine ligand 13 (CXCL13) expressed by follicular dendritic cells (fDCs)6–8. Expression of transcription factor (TF) B-cell lymphoma 6 (Bcl-6), among other TFs (e.g. IRF4, c-MAF, Ascl2), requisitely orchestrates TFH differentiation9–16. TFH also express an elaborate network of cell surface molecules that promote T-B cell collaboration in GCs 7–8, including the co-stimulatory receptor inducible co-stimulatory molecule (ICOS) which engages ICOS ligand (ICOSL) on GC B cells17–18. ICOS expression is essential for the generation and maintenance of TFH cells including production of the cytokine interleukin-21 (IL-21)19–20, which promotes GC B-cell proliferation, class switch recombination (CSR), memory B cell formation and plasma cell differentiation21–22. Antigen-experienced TFH also rapidly up-regulate the expression of CD154 (CD40L)23–24, which ligates the B cell surface receptor CD40 to induce B cell activation, proliferation, somatic hypermutation (SHM) and class switch recombination (CSR). Engagement of GC B cell-expressed programmed cell death 1 ligand 1 (PD-L1) and/or PD-L225–26 to PD-1 on TFH cells negatively regulates the size and function of the induced TFH response. Activated TFH express the highest levels of CD40L, ICOS, PD-1 and IL-21 among T cell subsets27–29.
Through decades of research by many investigators, T helper cells including TH1 and TH2 cells were initially implicated in the pathogenesis of inflammatory bowel diseases (IBD). Following the discovery of TH17 cells, which specifically produce TH17 family cytokines (e.g. IL-17A, IL-17F, and IL-22), investigators reconsidered the TH1/TH2 cytokine balance hypothesis, and posited that TH17 cells are potentially instrumental in IBD pathogenesis. Despite the strong associative evidence between TH17-associated genes/mutations and IBD from murine and human studies, clinical trials targeting IL-17A proved ineffective in Crohn’s disease patients and in fact paradoxically worsened disease in a set of patients. In addition, naïve CD4+ T cells from IL-17 deficient mice induced more severe colitis in recipient mice. Thus, the mechanism through which T helper cells mediate inflammatory bowel diseases remains elusive. Interestingly, TFH signature genes have been expressed in several inflammatory diseases including IBD, suggesting that TFH may contribute to the development of inflammatory diseases.
Multiple transcription factors, including C-Maf, Batf, Irf4, STAT1, STAT3, and Ascl2, are actively involved in the development and function of TFH9–16, but maintenance and full differentiation of TFH critically requires expression of Bcl-69–10. In addition, the TFH differentiation pathway is also opposed by other factors including Blimp-1, Foxo1 and Foxp130–31. However, the molecular mechanism for the regulation of TFH is incompletely understood, especially how TFH are negatively regulated.
IRF8 is a member of the evolutionarily conserved IRF family of transcription factors with diverse and important regulatory roles in the growth, differentiation, and function of innate and adaptive immune cells. IRF8 is expressed by a wide spectrum of immature and mature hematopoietic cells including B cells, dendritic cells (DCs), macrophages, and activated T cells 32–33. It has an N-terminal DNA-binding domain (DBD) and a C-terminal IRF association domain (IAD), the latter of which is responsible for heterodimerization with other transcription factors33–34. IRF8 can function as either a transcriptional repressor or an activator, depending on the specific heterodimeric DNA-binding complexes produced with its varied partners35–41. Previously published work showed that germline Irf8−/− mice develop a chronic myeloid leukemia-like syndrome with impaired TH1 immunity, but how IRF8 controls T cell function was seldom discussed33–35,40. We previously reported that IRF8 negatively regulates TH17 cell differentiation42 raising the possibility that this TF could serve as a negative regulator of other TH subsets including TFH.
Herein, we provide new evidence that TFH can mediate intestinal pathology independent of B cells and show that this pathogenic function is regulated by IRF8 inhibition of IRF4. In addition to providing paradigm shifting mechanistic insight into the functions of TFH our new findings raise the possibility that CD4+ T cell-intrinsic IRF8 expression critically regulates other pathogenic autoimmune responses driven by TFH.
2. Materials and Methods
2.1. Mice
C57BL/6J (B6, stock#000664), Rag1‒/‒ (on B6 background, B6.129S7-Rag1tm1Mom/J, stock#002216), Bcl-6‒/‒ mice (B6.129S(FVB)-Bcl6<tm1.1Dent>/J, stock# 023727) were obtained from the Jackson laboratory. Irf8‒/‒ and Lck-Cre+Irf8fl/fl were maintained in the barrier facility at the Icahn School of Medicine at Mount Sinai. The animal study protocols were approved by the Institutional Animal Care and Use Committees of Icahn School of Medicine at Mount Sinai.
2.2. Human colon tissue
Human colon tissues from Crohn’s disease patients and control patients undergoing resection for cancer screening were obtained from the Mount Sinai Hospital with a protocol approved by the Institutional Review Board of Icahn School of Medicine at Mount Sinai.
2.3. Antibodies
The following antibodies against mouse antigens and conjugated to FITC, PE, PE-Cy5, PerCP-Cy5.5 or APC were purchased from BD Biosciences (CD4 (L3T4), eBioscience: biotin-rat anti-mouse CXCR5 (2G8), plus streptavidin-APC-eFluor 780 or eBioscience: PD-1 (J43), ICOS (7E17G9), Bcl-6 (mG1191E), IL-21 (mhalx21), CD8 (53–6.7), CD3ε (145–2C11), CD62L (MEL-14) and isotype controls.
2.4. Intracellular staining and flow cytometry
Naive CD4+ T cells (CD62L+CD44lo) were prepared by fluorescence-activated cell sorting (FACS) from spleens and lymph nodes of Irf8‒/‒ mice or WT littermates. Anti-mouse CD4 microbeads (L3T4, Miltenyi Biotec) from spleen and lymph nodes of mice using AutoMACS separator (Miltenyi Biotec). The sorted cells were stimulated for 72 hours with plate-bound anti-CD3 (1 μg/ml; 145–2C11; BD Biosciences) and soluble anti-CD28 (1 μg/ml; 37.51; BD Biosciences) with or without IL-21. The cells were then re-stimulated for 5 hours with PMA and ionomycin in the presence of Brefeldin A, cells were fixed with IC Fixation Buffer (BD), incubated with permeabilization buffer, and stained with PE–, APC– or PE–Cy 5.5 anti–mouse antibodies and intracellular cytokines were measured by flow cytometry. Flow cytometry was performed on a FACSCalibur (BD). Cells stimulated under neutral conditions were defined as TH0 cells. Cells were stimulated to differentiate into TFH cells by the supplementation with 10 ng/ml IL-21 (R&D Systems). Flow cytometry was performed on a FACSCalibur or LSR Fortessa analyzer (BD Biosciences).
2.5. T cell transfer model and histology
T cell transfer experiment was performed as previously described42. In brief, purified CD4+CD45RBhi T cells from WT, Irf8‒/‒ and Bcl6‒/‒ mice with or without CD19+B220+ B cells (1×106 cells per mouse in 200 μl sterile PBS) from WT mice were transferred intraperitoneally into Rag1‒/‒ recipients (6×105 cells per mouse in 200 μl sterile PBS). Mice were weighed every week throughout the course of experiments. After 5 weeks, mice were sacrificed, their spleens and mesenteric lymph nodes excised then and analyzed by flow cytometry. Spleens and colon tissues from WT, Irf8‒/‒ or bcl6‒/‒ mice were fixed in 10% neutral buffered formalin and embedded in paraffin or in frozen section. 5μm sections of tissue were stained with fluorochrome-conjugated antibodies. The degree of inflammation in the epithelium, submucosa and muscularis propria of colon tissue was scored separately as described by Totsuka et al43.
2.6. Mice immunization
Mixing anti-mouse CD3 antibody (BD Biosciences, San Jose, CA) 5μg/mouse and anti-mouse CD28 antibody (BD Biosciences, San Jose, CA) 2μg/mouse in 100μL PBS, injection intraperitoneally two times interval three days. NP(40)-OVA/alum was prepared by mixing NP(40)-OVA (Biosearch Technologies, Petaluma, CA) in PBS with alum (Pierce, Rockford, IL) at a 1:1 ratio. NP-OVA/alum immunizations consisted of 100 μg given intradermal injection (i.d) four times interval one week.
2.7. RNA isolation and quantitative real-time RT-PCR
Total RNA was extracted using an RNeasy plus kit (QIAGEN) and cDNA was transcribed using the Superscript II system (Invitrogen) with an oligo-dT primer followed by analysis using iCycler PCR with SYBR Green PCR master Mix (Applied Biosystems) using the primers in Table S1 and S2. Results were normalized based on the expression of ubiquitin.
2.8. Immunoblot
Cells were washed with cold phosphate-buffered saline and lysed for 15 min on ice in 0.5 ml of lysis buffer (50 mM Tris-HCl, pH 8.0, 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol and 1 mM dithiothreitol) containing protease inhibitors. Cell lysates were performed for immunoblotting. Anti-IRF4 and anti-IRF8 (SantaCruz Biotechnology) and anti-β-actin (Sigma) antibodies were used according to the manufactures’ instructions. Secondary antibodies were purchased from SantaCruz Biotechnology.
2.9. Chromatin immunoprecipitation (ChIP)
ChIP was performed using a kit following the manufacturers’ instruction (Upstate Biotechnology). Briefly, cells were fixed by 1% formaldehyde for 10 min at 37 °C. Nuclei were purified and sonicated to obtain DNA fragments. Chromatin fractions were pre-cleared with protein A-conjugated agarose beads followed by immunoprecipitation overnight at 4°C with 3 μg of anti-IRF8 or control antibody. Crosslinking was reversed at 65 °C for 4 h, followed by proteinase K digestion. DNA was purified and subjected to qPCR. The input DNA was diluted 200 times before PCR amplification. The input and immunoprecipitated DNAs were amplified by qPCR using the primers targeting.
2.10. Chromatin immunoprecipitation sequence (ChIP-Seq)
Purified CD4+ T cells from WT mice were stimulated with or without anti-CD3 and anti-CD28 for 48h in the presence of IL-21. The cell pellet was cross-linked and sonicated, the chromatin was ready for MOWChIP-seq. The mixed beads of protein A beads and protein G beads were incubated with IRF8 antibody. After washing, the IRF8-beads were loaded to chamber and incubate with sonicated chromatin samples. After ChIP, the washed immune complexes were collected and resuspended in the reverse crosslinking buffer to incubate. The DNA was extracted and precipitation. Sequencing libraries were prepared by Accel-NGS 2S Plus DNA Library Kit (Swift). The libraries were sequenced on an Illumina HiSeq 4000 with single-end 50-nt reads.
2.11. Statistical analysis
Statistical analysis was performed using Student’s t-test for most of the experiments. P<0.05 was considered statistically significant.
3. Results
3.1. TFH are critically involved in intestinal inflammation
While the primary known function of TFH is to provide helper signals that drive GB B cell differentiation5–8, it is possible that TFH can directly function as pathogenic mediators of autoimmune injury independent of their helper functions. To investigate the possibility that TFH cells contribute to intestinal inflammation, we first examined the expression of TFH signature genes in the intestinal tissues of Crohn’s disease patients. These analyses revealed that TFH-associated gene products including IL-21, CXCR5, ICOS, PD1 and Bcl-6 were significantly up-regulated in intestinal tissues of Crohn’s disease patients compared to normal controls (Fig. S1). Similarly, when we analyzed colon tissue of a murine model in which adoptive transfer of CD4+ T cells together with or without B cells into syngeneic Rag1‒/‒ hosts reproducibly results in colitis42–43 (Fig. S2A and data not shown), we observed significantly higher expression of the same TFH-related gene products (Fig. S2B). Histological staining showed that Bcl-6 was highly expressed in the intestinal tissues with colitis (Fig. S2C) and flow cytometry analysis of spleen cells showed detectable CXCR5+ICOS+PD1+BCL-6+ TFH only in Rag1‒/‒ recipients of WT naïve CD4+ T cells (Fig. S2D).
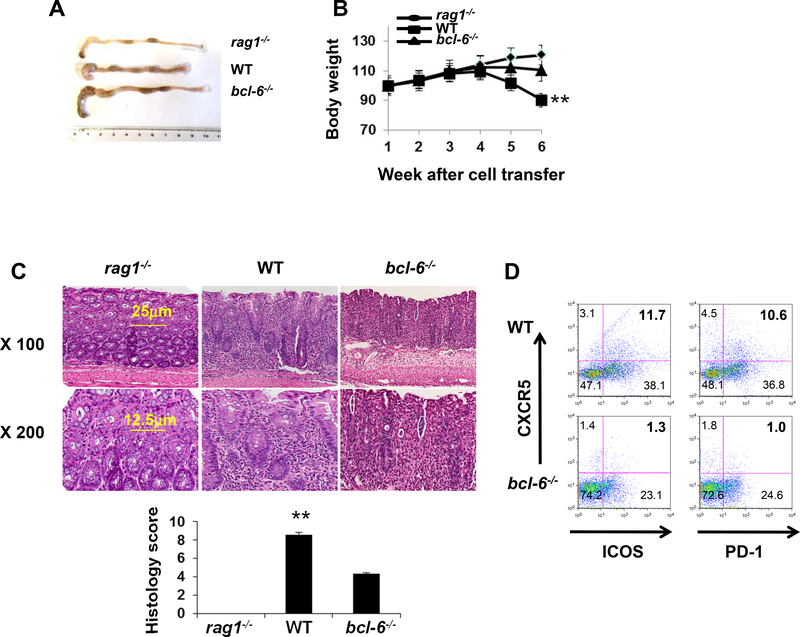
As Bcl-6 is one of several TFs required for TFH differentiation9–10 we directly tested the pathogenicity of TFH by adoptively transferring naïve Bcl-6−/−CD4+ T cells into Rag1−/− recipients (Fig. 1A-B). Distinct from recipients of WT CD4+ T cells, recipients of Bcl-6−/− CD4+ CD45Rbhi cells appeared phenotypically normal and maintained rather than lost weight. Histological analyses of the intestinal tissue 6 weeks post-transfer revealed less inflammatory cell infiltration and lower pathology scores in the Rag1−/− recipients of Bcl-6−/− CD4+ cells (Fig. 1C). When we analyzed T cell phenotypes of mesenteric lymph nodes at 6 weeks we observed significantly fewer TFH in adoptive recipients of Bcl-6−/− CD4+ cells (Fig. 1D). Taken together, the results support the conclusion that that TFH rather than TH1 or TH17 cells (data not shown) are pathogenic mediators of intestinal inflammation in this model.
Figure 1. TFH signature is up-regulated in autoimmune and inflammatory diseases.
(A and B) Colon morphology and body weight of Rag1‒/‒ recipient mice after receiving 6×105 purified WT (n = 5) or Bcl-6‒/‒ (n = 5) CD4+ T cells. (C) H&E staining and histology score of colon tissues of Rag1‒/‒ recipient mice after receiving 6×105 purified WT (n = 5) or Bcl-6‒/‒ (n = 5) CD4+ T cells. (D) The percentages of CXCR5+ICOS+ and CXCR5+PD-1+ CD4+ T cells were compared between Rag1‒/‒ recipients of WT CD4+ T cells (n = 5) and those of Bcl-6‒/‒ CD4+ T cells (n = 5).
3.2. IRF8-deficient mice display enhanced TFH cell differentiation
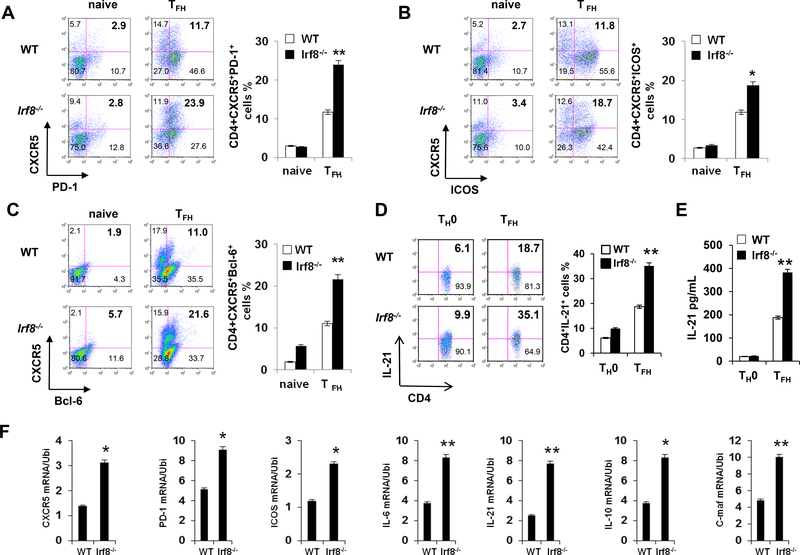
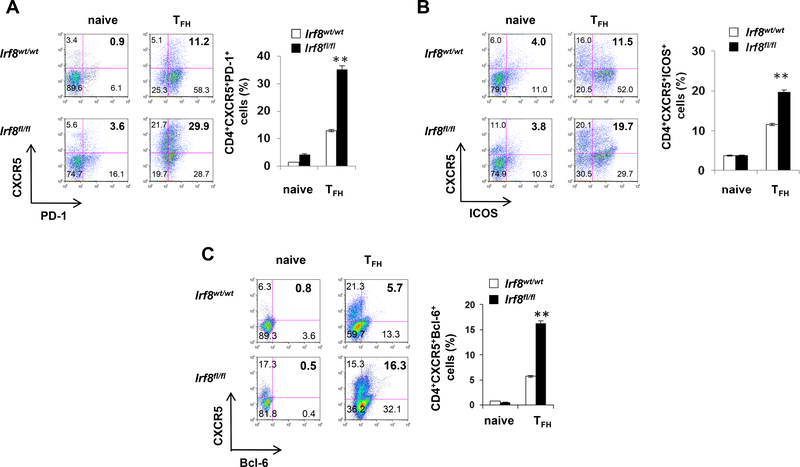
Building upon our previous observation that IRF8 inhibits TH17 differentiation42 we tested the impact of IRF8 on differentiation of naïve CD4+ T cells toward the TFH phenotype using an in vitro culture system (Fig. 2). Three days after stimulating purified naïve Irf8‒/‒ or WT CD4+ T cells with anti-CD3/CD28 under TFH-inducing conditions, we analyzed cytokine, surface marker and TF profiles in the responding T cells. These assays showed higher proportions of CXCR5+PD-1+, CXCR5+ICOS+ and CXCR5+Bcl-6+ cells (Fig. 2A‒C) and more IL-21, within the Irf8‒/‒ CD4+ T cells vs. WT controls (Fig. 2D and 2E). Quantitative PCR assays showed higher Icos, Pdcd1, Cxcr5 and Il21 gene expression in the Irf8‒/‒ CD4+ T cells (Fig. 2F). Analysis of CD4+ T cells obtained from mice with IRF8 deficiency restricted to T cells (Lck-Cre+Irf8fl/fl) confirmed enhanced TFH cell differentiation (Fig. 3A, 3B and 3C). Proliferative responses of WT and IRF8−/− CD4+ T cells cultured under TFH-inducing conditions did not differ (Fig. S3), and IRF8 deficiency did not alter in vitro induction of IFNγ+ TH1, IL-4+ TH2 or Foxp3+ Treg cells (Fig. S4)42, together demonstrating that IRF8 specifically inhibits the TFH differentiation program.
Figure 2. IRF8-deficient CD4+ T cells show enhanced TFH polarization after ex vivo stimulation.
Purified CD4+ T cells from WT or Irf8−/− mice (A, B, and C) were stimulated with or without anti-CD3 and anti-CD28 for 48h in the presence of IL-21. Cell surface expression of PD-1, ICOS, CXCR5 and intracellular expression of Bcl-6 were analyzed by flow cytometry. The percentages of CXCR5+PD1+, CXCR5+ICOS+ and CXCR5+Bcl6+ cells were compared between WT (n = 5) and Irf8−/− (n = 5) CD4+ T cell cultures. (D) Flow cytometry evaluation of IL-21+CD4+ T cells in WT and Irf8‒/‒ CD4+ T cells under TFH condition (n = 5). (E) ELISA of IL-21 levels in culture supernatant of WT and Irf8‒/‒ CD4+ T cells cultured under TFH condition (n = 4). (F) Quantitative real-time RT-PCR analysis of TFH-associated genes in WT and Irf8‒/‒ CD4+ T cells under TFH condition (n = 5). The ubiquitin gene (Ubi) was used as an internal control. Results shown are representative of three independent experiments. Data are given as means ± SEM.
Figure 3. IRF8-deficient in T cells show enhanced TFH polarization after ex vivo stimulation.
Purified CD4+ T cells from Irf8wt/wt or Irf8lck/lck mice (A, B, and C) were stimulated with or without anti-CD3 and anti-CD28 for 48h in the presence of IL-21. Cell surface expression of PD1, ICOS, CXCR5 and intracellular expression of Bcl-6 were analyzed by flow cytometry. The percentages of CXCR5+PD1+, CXCR5+ICOS+ and CXCR5+Bcl6+ cells were compared between Irf8wt/wt (n = 5) and Irf8lck/lck (n = 5) CD4+ T cell cultures. Results shown are representative of three independent experiments. Data are given as means ± SEM.
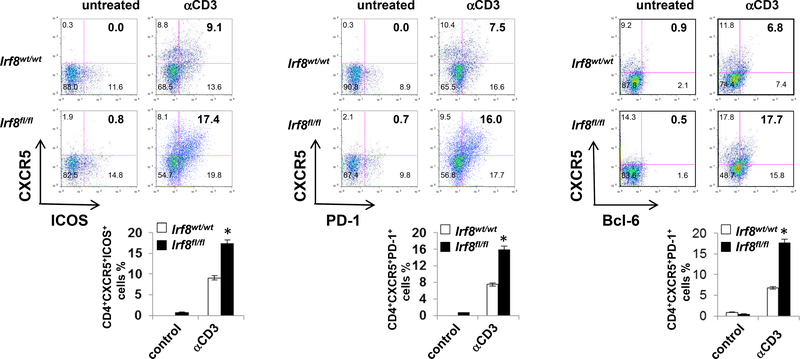
To begin to assess how the above observed effects of T cell IRF8 deficiency apply in vivo, we injected groups of Lck-Cre+Irf8fl/fl and Lck-Cre+Irf8wt/wt mice with anti-CD3 mAb (Fig. 4) and independently we immunized groups of animals with 4-hydroxy-3nitrophenylacetyl-conjugated ovalbumin (NP-OVA, Fig. S5). Under both conditions, flow cytometry analyses revealed ~2-fold higher frequencies of TFH cells in the spleens of treated Lck-Cre+Irf8fl/fl mice (Fig. 4A, and S5). Quantification of TH1, TH2 and Treg did not differ between groups in either set of experiments (Fig. S4), validating that the in vitro findings (Fig. 2) apply in vivo and supporting the concept that IRF8 specifically regulates TFH differentiation42.
Figure 4. TFH-associated signatures were significantly increased in IRF8-deficient T cells in vivo.
Cell surface expression of ICOS, PD-1, CXCR5 and intracellular expression of Bcl-6 in CD4+ T cells of Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice after anti-CD3 administration two times interval three days intraperitoneally, analyzed by flow cytometry. The percentages of CXCR5+ICOS+, CXCR5+PD1+ and CXCR5+Bcl6+ cells in CD4+ T cells of spleen were compared between Lck-Cre+Irf8wt/wt (n = 6) and Lck-Cre+Irf8fl/fl (n = 6) mice. Results shown are representative of three independent experiments. Data are given as means ± SEM.
3.3. IRF8-deficient TFH cells are pathogenic mediators of colitis
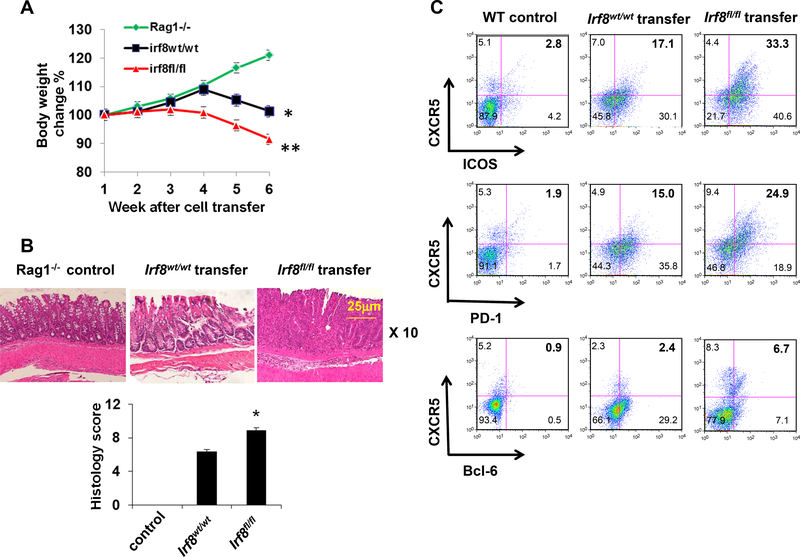
To directly test the hypothesis that IRF8 negatively regulates TFH capable of mediating colonic pathology, we adoptively transferred WT or Irf8‒/‒ CD4+ T cells into groups of Rag1‒/‒ mice and followed the animals for up to 6 weeks, analyzing and comparing clinical disease expression, colon histology and splenic T cell responses between groups. These analyses showed Rag1−/− recipients of Irf8‒/‒ CD4+ T cells exhibited greater and more rapid weight loss with more severe histological changes in the colons (Fig. 5A, 5B) and higher frequencies of splenic TFH (Fig. 5C).
Figure 5. TFH signature is up-regulated in autoimmune and inflammatory diseases.
(A) Body weight of Rag1‒/‒ recipient mice after receiving 6×105 purified CD4+ T cells from LckCre+Irf8wt/wt (n = 5) or Lck-Cre+Irf8fl/fl (n = 5) mice with CD19+B220+ B cells from WT mice. (B) Histological staining of colon tissues and histology score of Rag1‒/‒ recipients of LckCre+Irf8wt/wt CD4+ T cells (n = 5) and those of Lck-Cre+Irf8fl/fl CD4+ T cells (n = 5). (C) The percentages of CXCR5+ICOS+, CXCR5+PD-1+ and CXCR5+Bcl6+ CD4+ T cells were compared between Rag1‒/‒ recipients of Lck-Cre+Irf8wt/wt CD4+ T cells (n = 5) and those of Lck-Cre+Irf8fl/fl CD4+ T cells (n = 5) and WT as control (n=5). Results shown are representative of three independent experiments. Data are given as means ± SD.
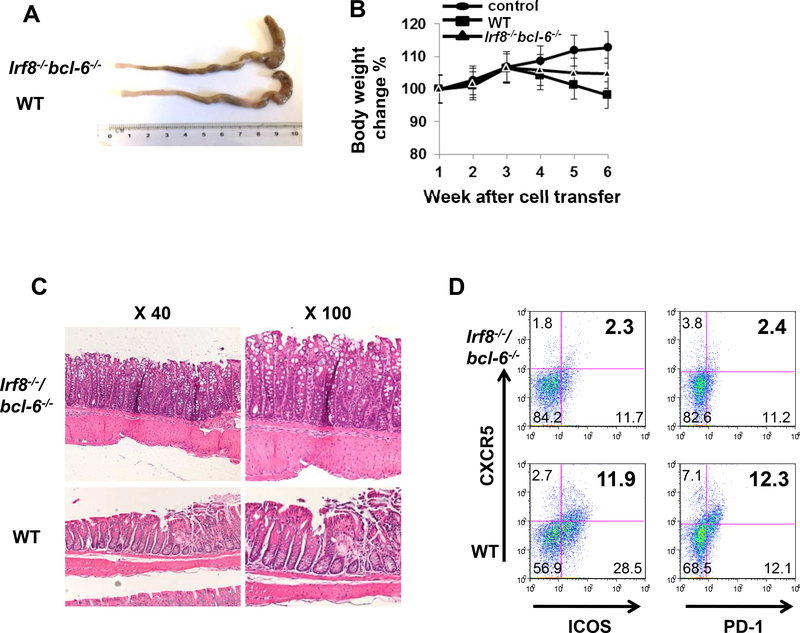
To confirm that absence of T cell IRF8 exacerbates colitis through a TFH–dependent mechanism, we adoptively transferred naïve Irf8−/−Bcl-6−/− CD4+ T cells into Rag1−/− recipients. These experiments showed diminished colitis severity (Fig 6A-C), similar to that observed in the recipients of Bcl-6−/− CD4+ T cells (Fig. 1B). Frequencies of splenic TFH were also reduced in Irf8−/−Bcl-6−/− recipient mice compared to WT controls (Fig. 6D). Taken together, the data support the conclusion that IRF8 suppresses inflammation via the inhibiting differentiation of pathogenic TFH.
Figure 6. TFH signature is up-regulated in autoimmune and inflammatory diseases.
Colon morphology and body weight of Rag1‒/‒ recipient mice after receiving 6×105 purified WT (n = 5) or Irf8‒/‒Bcl6‒/‒ (n = 6) CD4+ T cells with CD19+B220+ B cells from WT mice (A and B). (C) H&E staining of colon tissues of Rag1‒/‒ recipient mice after receiving 6×105 purified WT (n = 5) or Irf8‒/‒Bcl6‒/‒ (n = 6) CD4+ T cells. (D) The percentages of CXCR5+ICOS+ and CXCR5+PD-1+ CD4+ T cells were compared between Rag1‒/‒ recipients of WT CD4+ T cells (n = 5) and those of Irf8‒/‒Bcl6‒/‒ CD4+ T cells (n = 6). Results shown are representative of five mice. Data are given as means ± SD.
3.4. Mice with T cell-specific IRF8 deficiency exhibit exaggerated B cell differentiation
The data above suggests that IRF8-regulated TFH cells play an important role in T cell-mediated inflammation. To determine whether the enhanced TFH signature functionally affects B cell development, we analyzed mice with germline (Irf8‒/‒) or T cell-specific IRF8 deficiency (Lck-Cre+Irf8fl/fl). These mice had enlarged spleens and lymph nodes (Fig. S6A and B) that harbored increased numbers of CD19+CD138+ plasma cells (Fig. S6C) and germinal center B cells (Fig. S6D). Consistently, Lck-Cre+Irf8fl/fl mice had increased levels of serum IgG and IgM (Fig. S6E). The expansion of plasma cells in secondary lymphoid organs of Irf8‒/‒ mice was not due to infection or autoimmunity, as CD11c+ DCs were not increased in any immune organ or tissue analyzed (Fig. S7A), and there was no autoimmune kidney damage, such as cellular infiltration and glomerular crescents, in Irf8‒/‒ mice (Fig. S7B). There is no significant expansion in population of T cells but as reported data that myeloid cells were obviously expanded (Fig. S7A). To determine whether Irf8‒/‒ CD4+ T cells also induce more B cell proliferation in vivo, we co-transferred WT B cells with either WT or Irf8‒/‒ CD4+ T cells into Rag1−/− mice. Splenomegaly after adoptive transfer was markedly increased in recipients of Irf8−/− CD4+ T cells (Fig. S6F), and elevated percentage of B cells was found in the spleen and lymph nodes (Fig. S6G). These results suggest that IRF8 controls the magnitude of humoral immunity by regulating the differentiation and function of TFH cells. In addition to regulating the function of CD4+ T cells in GC B cell response, IRF8 expression in CD4+ T cells may influence B cell differentiation prior to the GC stage. To test this possibility, we co-transferred C57BL/6 (H-2b) bone marrow cells depleted of T cells with either WT or Irf8‒/‒ C57BL/6 (H-2b) CD4+ T cells into irradiated allogeneic BALB/c (H-2d) recipients. Co-transferred Irf8‒/‒ CD4+ T cells induced more B cell development from H-2b bone marrow cells than co-transferred WT CD4+ T cells (Fig. S8). Therefore, IRF8 in CD4+ T cells also restrains the ability of CD4+ T cells to support B cell development in vivo.
3.5. IRF8 suppresses IRF4 expression in CD4+ T cells
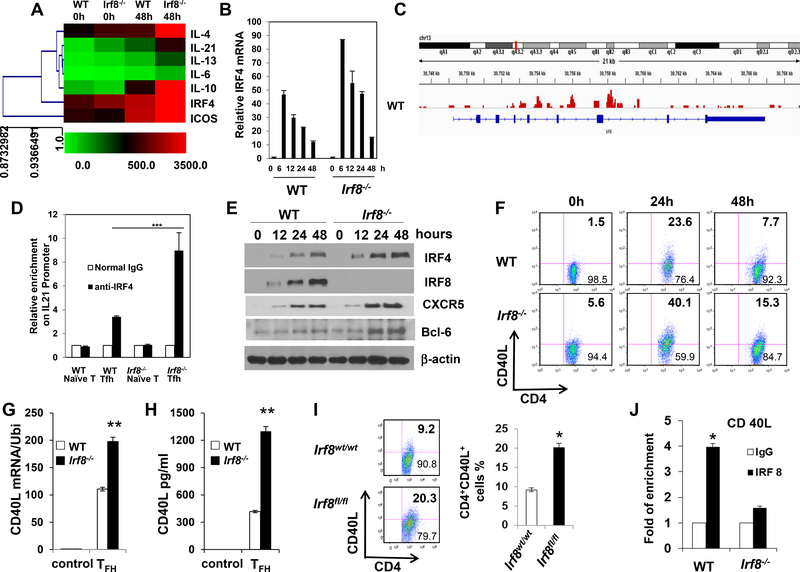
IRF4 is important for the differentiation of various T helper cell subsets, including TFH, TH2, TH9 and TH17 cells11,14,34. IRF4 and IRF8 represent immune-specific members of the interferon regulatory family and co-operate to play major roles in controlling the development and functioning of T cell subsets and other immune cells. To better understand the molecule mechanism of IRF8 in controlling of TFH cell function, we purified CD4+ T cells from WT or Irf8−/− mice and stimulated with or without antiCD3 and anti-CD28 under TFH cells culture condition. The results clearly indicated that Irf4 and Il21 expression levels were significantly increased in Irf8−/− mice compared with WT mice by microarray analysis (Fig. 7A). Furthermore, we evaluated the Irf4 gene by Quantitative real-time-PCR analysis (Fig. 7B) and protein expression levels of CXCR5, Bcl-6, IRF4 and IRF8 by western blotting (Fig. 7E) in WT and Irf8‒/‒ CD4+ T cells under TFH condition for time course. The results showed that both the Irf4 gene and IRF4 protein were significantly up-regulated in Irf8−/− mice compared with WT mice (Fig. 7A, B and E). We performed ChIP-seq assay to understand whether IRF8 binds to the promoter region of IRF4 gene in TFH cell condition. The result showed that IRF8 indeed bound to the promoter region of IRF4 gene (Fig. 7C). Furthermore, the ChIP assays also showed more IRF4 protein bound to the Il21 promoter in CD4+ T cells of Irf8−/− mice under TFH cell culture condition when compared with WT mice (Fig. 7D). The results suggested that IRF8 suppresses TFH differentiation by inhibiting the DNA biding activity of IRF4 to the promoter region of Il21 gene.
Figure 7. Regulation of IRF4 expression by IRF8.
(A) Purified CD4+ T cells from WT or Irf8−/− mice were stimulated with or without anti-CD3 and anti-CD28 for 48h in the presence of IL-21. Microarray analysis of the gene expression by WT and Irf8‒/‒ CD4+ T cells under TFH condition. (B) Quantitative real-time RT-PCR analysis of irf4 gene level in WT and Irf8‒/‒ CD4+ T cells under TFH condition for time course (n = 5). The ubiquitin gene (Ubi) was used as an internal control. (C) IRF8 ChIP-seq signals at the IRF4 gene locus. Blue line denotes the range of the IRF4 gene locus, and the peak patterns in red represent the ChIP-seq signals of IRF8 binding. Signals represent DNA fragments that were captured by the IRF8 antibody through ChIP. (D) Purified CD4+ T cells from WT or Irf8−/− mice were stimulated with anti-CD3 and anti-CD28 for 48h in the presence of IL-21, followed by ChIP assay. Three micrograms of an anti-IRF4 antibody or isotype-matched IgG as control antibody were used in the immunoprecipitation step. PCR was used to quantify the amount of precipitated DNA with primers flanking the irf4-binding site of the IL-21 promoter region. Each bar represents mean ± S.D. from three independent experiments, unpaired Student’s t-test, *P<0.05, versus WT cells. (E) Western blot analysis of IRF4, IRF8, Bcl-6 and CXCR5 protein levels in WT and Irf8‒/‒ CD4+ T cells under TFH condition for time course. The β-actin was used as an internal control. (F) Purified CD4+ T cells from WT or Irf8‒/‒ mice were stimulated with anti-CD3 and anti-CD28 antibody for 48h. CD40L expression was analyzed by flow cytometry gated on CD4+ T cells and used isotype control. (G) Cd40lg RNA level was analyzed by real-time RT-PCR (n = 5). (H) Soluble CD40L protein level in culture media was measured by ELISA. (I) The experiment was repeated by using purified CD4+ T cells of Lck-Cre+Irf8wt/wt (n = 6) or Lck-Cre+Irf8fl/fl (n = 6) mice. (J) ChIP assay of CD4+ T cells from WT and Irf8‒/‒ mice was analyzed for binding of IRF8 to the Cd40lg promoter (n = 4). Results shown are representative of three independent experiments. Data are given as means ± SEM.
We also sought to determine if IRF8 controls the expression of CD40 ligand (CD40L), a molecule crucial to the function of TFH cells. Compared to WT CD4+ T cells, Irf8‒/‒ CD4+ T cells expressed higher protein and mRNA levels of CD40L following ex vivo stimulation in TFH conditions (Fig. 7F, 7G, and 7H). The same results for protein expression were also seen in ex vivo stimulated CD4+ T cells from Lck-Cre+Irf8fl/fl mice (Fig. 7I). Upon TCR stimulation by anti-CD3 Ab in vivo, CD40L expression was higher on CD4+ T cells of Lck-Cre+Irf8fl/fl mice than those of WT control mice in mesenteric lymph nodes (Fig. S9A). Considering that IRF8 can function as a transcriptional repressor, we performed a chromatin immunoprecipitation (ChIP) assay to determine if IRF8 can repress the Cd40lg gene in CD4+ T cells. IRF8 bound to three of four putative IRF8-binding sites in the Cd40lg promoter and one experiment of ChIP data shown as Fig. 7J. Consistently, IRF8 suppressed CD40L expression in CD4+ T cells in a cell-intrinsic fashion, as evidenced by reduced cell surface CD40L levels in CD4+ T cells transduced by an Irf8-encoding retrovirus (Fig. S9B), as compared to marked increases in CD40L staining in cells transfected with empty virus. Taken together, these results suggest that IRF8 represses CD40L expression at both the transcriptional and protein levels in TFH cells.
4. Discussion
Our data newly and uniquely demonstrate a proinflammatory role for TFH as mediators of colitis independent of their ability to provide helper signals to B cells. We show that intestinal tissues of inflammatory bowel disease (IBD) patients and colon tissues obtained from Rag1−/− recipients of naïve CD4+ T cells with colitis, each over-express TFH-associated gene products. Adoptive transfer of naïve Bcl6−/− CD4+ T cells into Rag1−/− recipient mice abrogated development of colitis and limited TFH differentiation in vivo, demonstrating a mechanistic link. In contrast, T cell deficiency of interferon regulatory factor 8 (IRF8) resulted in augmentation of TFH induction in vitro and in vivo. Functional studies showed that adoptive transfer of Irf8−/− CD4+ T cells into Rag1−/− recipients exacerbated colitis development associated with increased gut TFH-related gene expression, while Irf8−/−/Bcl6−/− CD4+ T cells abrogated colitis, together indicating that IRF8-regulated TFH can directly cause colon inflammation. Molecular analyses revealed that IRF8 suppresses TFH differentiation by inhibiting transcription and transactivation of the TF IRF4, which is also known to be essential for TFH induction.
TFH cells are a distinct T helper cell subset shown to coordinate generation of the germinal center (GC) responses by initiating help for antigen specific B cells. Through this B cell help mechanism, TFH have been shown to be pathogenic in a number of autoimmune diseases. Increased frequencies of TFH-like cells in peripheral blood are observed in subsets of patients with Sjogren’s syndrome, juvenile dermatomyositis, and systemic lupus erythematosus44–45, each of which is associated extensive autoantibody production. However, several observations in the literature hint that TFH have other functions beyond providing helper signals for B cell differentiation and antibody switching. TFH are found within injured organs and tissues in subjects with lupus nephritis46, multiple sclerosis47, inflammatory arthritis, type 1 diabetes48, and intestinal tissues of patients with IBD49. Our data using adoptive transfer of Bcl-6 deficient naïve CD4+ T cells into Rag1−/− recipients provide direct evidence in support of this hypothesis linking TFH to intestinal inflammation.
TFH-produced IL-21, which is the key cytokine shown to drive B cell differentiation in GCs19–22. Early studies revealed that IL-21 is markedly overproduced in inflamed guts of patients with IBD compared to non-inflamed controls49. Mice lacking IL-21 are protected against chemically induced colitis; moreover, wild type mice given a neutralizing IL-21R/Fc fusion protein exhibit less experimental colitis as compared to control mice. IL-21 has been shown to activate macrophages, participate in granuloma formation, and inhibit induction of regulatory T cells (Treg) in murine graft vs host diseases. These findings together raise the possibility that TFH/IL-21 have pathogenic potential. In the present study, we demonstrate that IL-21 is highly expressed in intestinal tissues of IBD patients and also in the colon tissues of recipient mice transferred with naïve CD4+ T cells with colitis. Based these published findings and our new data, including the strong association of TFH-related genes to human IBD (Fig S1), we speculate that TFH-produced IL-21 is one pathological mediator of intestinal inflammation.
IRF8 plays critical roles in the differentiation of myeloid cells, B cells, dendritic cells and T cells, and hence the regulation of both innate and adoptive immune responses11,32–42. A recent genome-wide association (GWA) study has strongly implicated a variant near the IRF8 gene in SLE susceptibility in Europeans50. In addition, a different genetic variation in the IRF8 gene has been implicated in multiple sclerosis51. A GWA study identified the IRF8 gene as strongly associated with development of Crohn’s disease52, but provided no mechanistic explanation. Our findings in which we demonstrate that IRF8 deficiency a) favors T cell differentiation toward the TFH lineage and b) exacerbates CD4+ T cell-mediated colitis provide a potential explanation to account for this GWAS association. The observations support the testable hypothesis that IRF8 mutations that diminish production or function of IRF8 lead to augmented TFH differentiation which in turn contributes to the development of IBD.
Published evidence demonstrates that IRF8 cooperates with other TFs including Bcl-6, IRF4, and PU.1 to regulate immune cell function11,33–42. IRF8 and IRF4 are immune-specific members of the IRF TF family and have evolved not only to interact with specific members of the Ets superfamily, e.g. PU.1 and Spi-B, but also with particular members of the AP-1 superfamily, e.g. BATF-containing heterodimers40–41. IRF4 is important for the differentiation of various T helper cell subsets, including TFH, TH2, TH9 and TH17 cells11,14,34. Adding to this literature, in the present study, we provide new insight into mechanistic links among IRF8, IRF4 and IL-21 producing TFH. We demonstrate that IRF8 binds to the promoter region of IRF4 gene and that in the absence of IRF8, T cells express more IRF4. In addition, IRF8 deficiency significantly enhanced the IRF4 binding to the promoter region of IL-21 gene, together supporting the conclusion that IRF8 suppresses TFH differentiation by inhibiting transcription and transactivation of the TF IRF4.
Taken together, our murine and human data has uncovered potentially important pathogenic role for TFH cells in inflammatory diseases, and suggests that the suppressive function of IRF8 in TFH differentiation and the function may be potentially targeted to treat these diseases.
Supplementary Material
TFH signatures were increased in colon tissues of CD patients. Quantitative real-time RT-PCR analysis of the expression of TFH cell signatures in colon tissues of patients with IBD (n = 10) and in healthy subjects (n = 10). Data are given as means ± SEM.
TFH signatures were increased in colon tissues of T cell-mediated mouse model of colitis. CD4+ T cells were purified from WT mice and adoptively transferred to Rag1‒/‒ mice (n = 5 per group). Mice were analyzed 49 days after transfer. Colon morphology and body weight is shown in (A). Quantitative real-time RT-PCR analysis of TFH cell signatures in colon tissues is showed in (B). Histological staining for Bcl-6 in colon tissue was performed as (C). Signatures of TFH cells in mesenteric lymph nodes from the recipient of T cells transfer or control WT mice were analyzed for CD4+CXCR5+PD-1+, CD4+CXCR5+ICOS+ and CD4+CXCR5+Bcl-6+ cells by flow cytometry (D). Results shown are representative of five mice. Data are given as means ± SEM.
T cell proliferation is not different in IRF8-deficiency and WT CD4+ T cells. (A and B) Purified CD4+ T cells from WT or Irf8‒/‒ mice were stimulated under TFH culture condition for 3 days, cells number and proliferation and early or later stage apoptosis were analyzed by flow cytometry. Results shown are representative of three independent experiments.
IRF8-deficient mice show comparable Th1, Th2 and Treg cell subsets with WT mice in vivo. Irf8wt/wt or Irf8lck/lck mice were stimulated with anti-CD3 and antiCD28 antibody for 48h every two days for two times by peritoneal injection. And cells from spleen were analyzed by flow cytometry. The percentages of CD4+IFN-γ+, CD4+IL4+ and CD4+CD25+Foxp3+ cells were compared between Irf8wt/wt (n=5) and Irf8lck/lck (n=5) mice. Results shown are representative of three independent experiments.
TFH-associated signatures were significantly increased in IRF8-deficient T cells in vivo. Flow cytometry analysis of the expression of CXCR5, ICOS and PD-1 in CD4+ T cells of spleen in Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice after intradermal injection of NP-OVA with adjuvant for three times interval one week for evaluation of TFH-polarizing conditions in vivo. Results shown are representative of three independent experiments. Data are given as means ± SEM
IRF8-deficient mice show enhanced B cell differentiation in vivo. (A and C) The size of lymph nodes and spleens of age-matched WT or Irf8‒/‒, Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice (> 3 months old). (B) The percentage of CD19+CD138+ plasma cells in spleen of Lck-Cre+Irf8wt/wt (n = 6), Irf8‒/‒ (n = 6), or Lck-Cre+Irf8fl/fl mice (n = 6), gated on CD19+ B cells and analyzed by flow cytometry. (D) ELISA analysis of serum IgG and IgM levels in Lck-Cre+Irf8wt/wt (n = 6) and Lck-Cre+Irf8fl/fl mice (n = 6). (E and F) Spleen morphology and the percentage of CD19+B220+ in spleen and lymph node of Rag1‒/‒ mice 16 days after adoptive transfer of purified WT CD19+ B cells and either WT or Irf8‒/‒ CD4+ T cells. (G) Splenocytes from WT and Irf8‒/‒ mice were staining for GC B cells and were analyzed by flow cytometry. The percentages of GL-7+Fas+ GC B cells were compared between WT (n=5) and Irf8‒/‒ (n=5) mice. Results shown are representative of three independent experiments. Data are given as means ± SEM.
IRF8-deficient mice do not exhibit autoimmune organ damage. (A) CD11c+CD11b+ dendritic cells in immune organs and tissues were evaluated by flow cytometry. (B) Histology staining of kidney of WT and Irf8‒/‒ mice by Periodic Acid– Schiff (PAS) staining.
Irf8‒/‒ CD4+ T cells have enhanced ability to promote B cell development in vivo. BALB/c mice (H-2d) were irradiated with 9 Gy from a 137Cs source and injected i.v. with 2×106 T cell–depleted BM cells alone (H-2kb+), or combined with 3×105 CD4+ T cells isolated from WT or Irf8‒/‒ mice of the C57BL/6 background (H-2kb+). The H2kb+CD19+ B cells in the indicated chimera mice were analyzed by flow cytometry in spleens 11 and 14 days after donor cell transfer. Results shown are representative of five mice. Data are given as means ± SEM.
CD40 ligand was significantly increased in IRF8 deficient in T cells in vivo. (A) Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice were intraperitoneally injected with anti-CD3 antibody. Cell surface expression of CD40L in CD4+ T cells of mesenteric lymph nodes were analyzed by flow cytometry. The expression levels of CD40L in CD4+ T cells was compared between Lck-Cre+Irf8wt/wt (n = 6) and Lck-Cre+Irf8fl/fl (n = 6) mice. (B) Naive CD4+ T cells from WT mice were infected with retrovirus encoding Irf8 or empty vector and were activated by anti-CD3 antibody for 48h. Surface CD40L expression was analyzed by flow cytometry (n = 5). Results shown are representative of three independent experiments.
our study reveals that TFH cells are critically involved in the development of T cell-mediated inflammatory diseases and defines IRF8 as an important intrinsic suppressor of TFH differentiation, and our studies are really novel and will lead to more exciting investigations concerning the contributions of TFH cells in the development of various T cell-mediated inflammatory diseases in future.
Acknowledgments
We are grateful for Dr. Feng Hong for technical support, Mr. Anthony Bonito and Mr. Zihang Zheng for reading the manuscript. Huabao Xiong was supported by the NIH grant R01AI104688. Peter. S Heeger was supported by R01AI132405 and R01AI071185. This work was supported in part by the Intramural Research Program of NICHD and NIAID.
Footnotes
Competing Interest Statement
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Craft JE Follicular helper T cells in immunity and systemic autoimmunity. Nature Rev Rheumatol. 2012;8:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King C, Tangye SG, and Mackay CR T Follicular Helper (TFH) Cells in Normal and Dysregulated Immune Responses. Annu Rev Immunol. 2008;26:741–66. [DOI] [PubMed] [Google Scholar]
- 3.Tangye SG, Ma CS, Brink R, and Deenick EK The good, the bad and the ugly - TFH cells in human health and disease. Nat Rev Immunol. 2013;13:412–426. [DOI] [PubMed] [Google Scholar]
- 4.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, and Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non Th1/Th2 effector cells that provide help for B cells. J. Immunol 2004;173:68–78. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea JJ and Paul WE Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, and Förster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J. Exp. Med 2001;193:1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, and Sabzghabaei N. Human blood CXCR5+CD4+ T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, and Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. [DOI] [PubMed] [Google Scholar]
- 10.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, and Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang RH, Chen, Peng L, and Xiong HB. Regulation of T helper cell differentiation by interferon regulatory factor family members. Immunol Res. 2012;54:1:169–176. [DOI] [PubMed] [Google Scholar]
- 12.Johnston RJ, Choi YS, Diamond JA, Yang JA, and Crotty S STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012:209:243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, and Crotty S. Bcl6 and maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol 2012;188:3734–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bollig Nadine, Anne Brüstle Kerstin Kellner, Ackermann Waltraud, Abass Elfadil, rtmann Raifer Bärbel Camara, Brendel Cornelia, Giel Gavin, Bothur Evita, agdalena Huber Christoph Paul, Elli Alexandra, Kroczek Richard A., Nuriev Roza, Dong Chen, Jacob Ralf, Mak Tak W. and Lohoff Michael. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci. 2012;109:22:8664–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Xindong, Chen Xin, Zhong Bo, AiboWang XiaohuWang, Chu Fuliang, Nurieva Roza I., Yan Xiaowei, Chen Ping,van der Flier Laurens G., Nakatsukasa Hiroko, Neelapu Sattva S.,Chen Wanjun, Clevers Hans, Tian Qiang, Qi Hai, Wei Lai. and Dong Chen. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:27:513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, and Dobles M. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. [DOI] [PubMed] [Google Scholar]
- 17.Bossaller L, Burger J, Draeger R, Grimbacher B, Knoth R, Plebani A, Durandy A, Baumann U, Schlesier M, and Welcher AA. ICOS deficiency is associated with a severe reduction of CXCR5+CD4+ germinal center Th cells. J. Immunol 2006;177:4927–4932. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, Wu X, Jin W, Chang M, Cheng X, and Sun SC.. Noncanonical NF-κB regulates inducible costimulator (ICOS) ligand expression and T follicular helper cell development. Proc Natl Acad Sci. 2011;108:12827–12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lüthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, and Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. [DOI] [PubMed] [Google Scholar]
- 20.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, Chan TD, Palendira U, Bustamante J, and Boisson-Dupuis S. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207:155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, Johnston J, Madden K, Xu W, and West J. Interleukin-21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 2000:408:57–63. [DOI] [PubMed] [Google Scholar]
- 22.Pène J, Gauchat JF, Lécart S, Drouet E, Guglielmi P, Boulay V, Delwail A, Foster D, Lecron JC, and Yssel H. Cutting edge: IL-21 is a switch factor for the production of IgG1 and IgG3 by human B cells. J. Immunol 2004;172:5154–5157. [DOI] [PubMed] [Google Scholar]
- 23.Grewal IS and Flavell RA CD40 and CD154 in cell-mediated immunity. Annu Rev Immunol. 1998;16:111–135. [DOI] [PubMed] [Google Scholar]
- 24.Armitage RJ, Macduff BM, Spriggs MK, and Fanslow WC Human B cell proliferation and Ig secretion induced by recombinant CD40 ligand are modulated by soluble cytokines. J. Immunol 1993;150:3671–3680. [PubMed] [Google Scholar]
- 25.Butler NS, Moebius J, Pewe LL, Traore B, Doumbo OK, Tygrett LT, Waldschmidt TJ, Crompton PD, and Harty JT. Therapeutic blockade of PD-L1 and LAG-3 rapidly clears established blood-stage Plasmodium infection. Nat Immunol. 2012;13:188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, and Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma CS, Deenick EK, Batten M, and Tangye SG. The origins, function, and regulation of T follicular helper cells. J. Exp. Med 2012;209:7:1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crotty S Follicular helper CD4 T cells (TFH). Annu Rev Immunol. 2011;29:621–63. [DOI] [PubMed] [Google Scholar]
- 29.Cannons JL, Lu KT, and Schwartzberg PL T follicular helper cell diversity and plasticity. Trends Immunol. 2013;34:5:200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, and Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, Katayama CD, and Hedrick SM. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Yan M, Sun J, Jain S, Yoshimi R, Abolfath M, Ozato K, Coleman G Jr, Ng P, and Metcalf D, et al. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. Immunol. 2014;15;193(4):1766–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honda Kenya and Taniguchi Tadatsugu. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. [DOI] [PubMed] [Google Scholar]
- 34.Lohoff Michael and Tak W. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat Rev Immunol. 2005;5:125–135. [DOI] [PubMed] [Google Scholar]
- 35.Wang HS, Lee CH, Qi CF, Tailor P, Feng J, Abbasi S, Atsumi T, and Morse HC 3rd. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:10:4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaoka A, Tamura T, and Taniguchi T. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer sci. 2008;99:3:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida Y, Yoshimi R, Yoshii H, Kim D, Dey A, Xiong H, Munasinghe J, Yazawa I, O’Donovan MJ, Maximova OA, et al. The Transcription Factor IRF8 Activates Integrin-Mediated TGF-β Signaling and Promotes Neuroinflammation. Immunity. 2014;40:20:187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng JX, Wang HS, Shin DM, Masiuk M, Qi CF, and Morse HC 3rd. IFN Regulatory Factor 8 Restricts the Size of the Marginal Zone and Follicular B cell Pools. J Immunol. 2011;186:1458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, and Bacon CM. IRF8 Mutations and Human Dendritic-Cell Immunodeficiency. N Engl J Med. 2011;10:365:2:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, Horvath E, and Eklund EA. PU.1, interferon regulatory factor (IRF) 2, and the interferon consensus sequence-binding protein (ICSBP/IRF8) cooperate to activate NF1 transcription in differentiating myeloid cells. J Biol Chem. 2007;282(9):6629–43. [DOI] [PubMed] [Google Scholar]
- 41.Glasmacher Elke, Agrawal Smita, Chang Abraham B., Murphy Theresa L., Zeng Wenwen, Bryan Vander Lugt Aly A. Khan, Ciofani Maria, Spooner Chauncey J., Rutz Sascha, Hackney Jason, Nurieva Roza, Escalante Carlos R., Ouyang Wenjun, Littman Dan R., Murphy Kenneth M., and Singh Harinder. A Genomic Regulatory Element That Directs Assembly and Function of Immune-Specific AP-1– IRF Complexes. Science. 2012;338:16:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouyang X, Zhang R, Yang J, Li Q, Qin L, Zhu C, Liu J, Ning H, Shin MS, Gupta M, Qi CF, He JC, Lira SA, Morse HC 3rd, Ozato K, Mayer L, and Xiong H. Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat Commun. 2011;2:314. doi: 10.1038/ncomms1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harboura Stacey N., Maynarda Craig L., Zindla Carlene L., Schoebb Trenton R., and Weavera Casey T. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci. 2015;112:22:7061–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson N Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62:234–244. [DOI] [PubMed] [Google Scholar]
- 45.Choi JY, Ho JH, Pasoto SG, Bunin V, Kim ST, Carrasco S, Borba EF, Gonçalves CR, Costa PR, Kallas EG, Bonfa E, and Craft J. Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity. Arthritis Rheumatol. 2015;67:4:988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liarski VM, Kaverina N, Chang A, Brandt D, Yanez D, Talasnik L, Carlesso G, Herbst R, Utset TO, Labno C, Peng Y, Jiang Y, Giger ML, and Clark MR. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med. 2014;2:6(230):230–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt N Role of T Follicular Helper cells in Multiple Sclerosis. J Nat Sci. 2015;1:7:139–148. [PMC free article] [PubMed] [Google Scholar]
- 48.Kenefeck R, Wang CJ, Kapadi T, Wardzinski L, Attridge K, Clough LE, Heuts F, Kogimtzis A, Patel S, and Rosenthal M, Follicular helper T cell signature in type 1 diabetes. J clinic Inves. 2015;125:1:292–303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarra M, Monteleone I, Stolfi C, Fantini MC, Sileri P, Sica G, Tersigni R, Macdonald TT, Pallone F and Monteleone G. Interferon-gamma-expressing cells are a major source of interleukin-21 in inflammatory bowel diseases. Inflamm Bowel Dis. 2010;16:8:1332–1339. [DOI] [PubMed] [Google Scholar]
- 50.Gateva V A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009; 41:1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Jager PL Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2011; 11:4:776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elding H, Lau W, Swallow D M. and Maniatis N. Dissecting the genetics of complex inheritance: linkage disequilibrium mapping provides insight into Crohn disease. Am J Hum Genet. 2011; 89:798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TFH signatures were increased in colon tissues of CD patients. Quantitative real-time RT-PCR analysis of the expression of TFH cell signatures in colon tissues of patients with IBD (n = 10) and in healthy subjects (n = 10). Data are given as means ± SEM.
TFH signatures were increased in colon tissues of T cell-mediated mouse model of colitis. CD4+ T cells were purified from WT mice and adoptively transferred to Rag1‒/‒ mice (n = 5 per group). Mice were analyzed 49 days after transfer. Colon morphology and body weight is shown in (A). Quantitative real-time RT-PCR analysis of TFH cell signatures in colon tissues is showed in (B). Histological staining for Bcl-6 in colon tissue was performed as (C). Signatures of TFH cells in mesenteric lymph nodes from the recipient of T cells transfer or control WT mice were analyzed for CD4+CXCR5+PD-1+, CD4+CXCR5+ICOS+ and CD4+CXCR5+Bcl-6+ cells by flow cytometry (D). Results shown are representative of five mice. Data are given as means ± SEM.
T cell proliferation is not different in IRF8-deficiency and WT CD4+ T cells. (A and B) Purified CD4+ T cells from WT or Irf8‒/‒ mice were stimulated under TFH culture condition for 3 days, cells number and proliferation and early or later stage apoptosis were analyzed by flow cytometry. Results shown are representative of three independent experiments.
IRF8-deficient mice show comparable Th1, Th2 and Treg cell subsets with WT mice in vivo. Irf8wt/wt or Irf8lck/lck mice were stimulated with anti-CD3 and antiCD28 antibody for 48h every two days for two times by peritoneal injection. And cells from spleen were analyzed by flow cytometry. The percentages of CD4+IFN-γ+, CD4+IL4+ and CD4+CD25+Foxp3+ cells were compared between Irf8wt/wt (n=5) and Irf8lck/lck (n=5) mice. Results shown are representative of three independent experiments.
TFH-associated signatures were significantly increased in IRF8-deficient T cells in vivo. Flow cytometry analysis of the expression of CXCR5, ICOS and PD-1 in CD4+ T cells of spleen in Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice after intradermal injection of NP-OVA with adjuvant for three times interval one week for evaluation of TFH-polarizing conditions in vivo. Results shown are representative of three independent experiments. Data are given as means ± SEM
IRF8-deficient mice show enhanced B cell differentiation in vivo. (A and C) The size of lymph nodes and spleens of age-matched WT or Irf8‒/‒, Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice (> 3 months old). (B) The percentage of CD19+CD138+ plasma cells in spleen of Lck-Cre+Irf8wt/wt (n = 6), Irf8‒/‒ (n = 6), or Lck-Cre+Irf8fl/fl mice (n = 6), gated on CD19+ B cells and analyzed by flow cytometry. (D) ELISA analysis of serum IgG and IgM levels in Lck-Cre+Irf8wt/wt (n = 6) and Lck-Cre+Irf8fl/fl mice (n = 6). (E and F) Spleen morphology and the percentage of CD19+B220+ in spleen and lymph node of Rag1‒/‒ mice 16 days after adoptive transfer of purified WT CD19+ B cells and either WT or Irf8‒/‒ CD4+ T cells. (G) Splenocytes from WT and Irf8‒/‒ mice were staining for GC B cells and were analyzed by flow cytometry. The percentages of GL-7+Fas+ GC B cells were compared between WT (n=5) and Irf8‒/‒ (n=5) mice. Results shown are representative of three independent experiments. Data are given as means ± SEM.
IRF8-deficient mice do not exhibit autoimmune organ damage. (A) CD11c+CD11b+ dendritic cells in immune organs and tissues were evaluated by flow cytometry. (B) Histology staining of kidney of WT and Irf8‒/‒ mice by Periodic Acid– Schiff (PAS) staining.
Irf8‒/‒ CD4+ T cells have enhanced ability to promote B cell development in vivo. BALB/c mice (H-2d) were irradiated with 9 Gy from a 137Cs source and injected i.v. with 2×106 T cell–depleted BM cells alone (H-2kb+), or combined with 3×105 CD4+ T cells isolated from WT or Irf8‒/‒ mice of the C57BL/6 background (H-2kb+). The H2kb+CD19+ B cells in the indicated chimera mice were analyzed by flow cytometry in spleens 11 and 14 days after donor cell transfer. Results shown are representative of five mice. Data are given as means ± SEM.
CD40 ligand was significantly increased in IRF8 deficient in T cells in vivo. (A) Lck-Cre+Irf8wt/wt or Lck-Cre+Irf8fl/fl mice were intraperitoneally injected with anti-CD3 antibody. Cell surface expression of CD40L in CD4+ T cells of mesenteric lymph nodes were analyzed by flow cytometry. The expression levels of CD40L in CD4+ T cells was compared between Lck-Cre+Irf8wt/wt (n = 6) and Lck-Cre+Irf8fl/fl (n = 6) mice. (B) Naive CD4+ T cells from WT mice were infected with retrovirus encoding Irf8 or empty vector and were activated by anti-CD3 antibody for 48h. Surface CD40L expression was analyzed by flow cytometry (n = 5). Results shown are representative of three independent experiments.