Abstract
Consumption of high fat, high sugar (western) diets is a major contributor to the current high levels of obesity. Here, we used a multidisciplinary approach to gain insight into the molecular mechanisms underlying susceptibility to diet-induced obesity (DIO). Using positron emission tomography (PET), we identified the dorsal striatum as the brain area most altered in DIO-susceptible rats and molecular studies within this region highlighted regulator of G-protein signaling 4 (Rgs4) within laser-capture micro-dissected striatonigral (SN) and striatopallidal (SP) medium spiny neurons (MSNs) as playing a key role. Rgs4 is a GTPase accelerating enzyme implicated in plasticity mechanisms of SP MSNs, which are known to regulate feeding and disturbances of which are associated with obesity. Compared to DIO-resistant rats, DIO-susceptible rats exhibited increased striatal Rgs4 with mRNA expression levels enriched in SP MSNs. siRNA-mediated knockdown of striatal Rgs4 in DIO-susceptible rats decreased food intake to levels comparable to DIO-resistant animals. Finally, we demonstrated that the human Rgs4 gene locus is associated with increased body weight and obesity susceptibility phenotypes, and that overweight humans exhibit increased striatal Rgs4 protein. Our findings highlight a novel role for involvement of Rgs4 in SP MSNs in feeding and DIO-susceptibility.
Introduction
Obesity has reached epidemic proportions1 and yet efficacious treatment options for this disease remain limited. The vast majority of cases are attributed to positive energy balance, which arises from a combination of overeating and lack of physical activity2. Along with such factors, the contribution of underlying behavioral and metabolic disturbances, especially in “susceptible” individuals, likely accentuates obesity risk. Therefore, in addition to policy initiatives3, efforts to decrease obesity prevalence should also be directed at identifying predisposing factors. One approach involves studying populations with known susceptibility or resistance to obesity. Though this type of approach cannot be easily undertaken in humans, it can be used in animal models. Here we examined a well-characterized model of susceptibility to diet-induced obesity (DIO), the Osborne-Mendel (OM) rat, which develops severe obesity and metabolic deficits only after exposure to high-energy diets, and the S5B/Pl (S5B) rat, which is DIO-resistant after similar exposure4–8. In addition to DIO susceptibility, in the absence of high-fat diet exposure, OM and S5B rats exhibit differences in sensory9, 10, anxiety11, hedonic12, arousal13, satiety14, and reward15–17 mechanisms, and abnormalities in such systems are also observed in human obesity18, 19. Furthermore, the OM-S5B model has been used to examine neurobiological mechanisms relevant to obesity susceptibility and related comorbidities, such as depression20.
The unique metabolic, neurobiological, and behavioral sensitivity to DIO in OM and S5B rats renders these strains a relevant laboratory model for investigating neurobiological mechanisms involved in DIO susceptibility and resistance in the absence of obesity manifestation. As such, we characterized behavioral and metabolic profiles of OM and S5B rats and used small animal positron emission tomography (PET) to identify, in an unbiased manner, brain areas where the two strains were characterized by differences in brain metabolic activity. Our efforts revealed that, compared to S5B rats, OM animals exhibited a marked decrease in brain metabolic activity in the dorsal striatum. Transcriptional profiling in this region identified Rgs4, a G-protein signaling regulator, as being upregulated in OM rats and this was paralleled by observations at the level of Rgs4 gene locus-specific histone modifications, striatal cell-specific Rgs4 expression, striatal Rgs4 protein expression, and striatal Rgs4 function. Additionally, striatal Rgs4 knockdown in OM rats decreased food intake to levels comparable to S5B rats. Finally, we extended the relevance of these observations to humans by showing that striatal Rgs4 was associated with increased body weight and obesity susceptibility phenotypes.
Materials and methods
Animals
Male Osborne-Mendel (OM) and S5B/Pl (S5B) rats were bred at Pennington Biomedical Research Center (Baton Rouge, LA). Male Sprague-Dawley (SD) control rats were purchased from Charles River (Wilmington, MA). All rats were individually housed under standard laboratory conditions (22 ± 2 °C, 50 ± 10% relative humidity) with ad libitum access to both normal rat chow (unless otherwise stated) and water and kept in a 12hr/12hr light-dark reverse cycle with the lights off at 0700hr and on at 1900hr. All studies were conducted in agreement with the National Academy of Sciences Guide for the Care and Use of Laboratory Animals and institutional animal care and use committee protocols. For OM and S5B rats fed normal chow or a high fat diet (HF) (Table S1), starting at 2 months of age, food intake (24 hr home cage feeding) was monitored for 8 weeks. Body weight was also assessed. For SD rats, food intake was monitored for 2 weeks. Body weight was assessed at the end of the 2-week period at approximately 4 months of age.
Tissue Harvesting and Preparation
Each animal was deeply anesthetized. The brain was rapidly removed and frozen in an isopentane and dry ice bath and stored in a –80°C freezer and sectioned at 20°C into 14μm thick slices. Slices were mounted onto microscope slides and stored at −80°C. Retroperitoneal and epidydimal fat was dissected from 5-month-old rats, weighed, and the total expressed as percentage of body weight.
Palatable feeding during limited access conditions
We used 5-month-old naïve OM and S5B rats or 4-month-old naïve SD rats. All procedures were conducted 3-5 hours into the dark cycle. Rats were placed in chambers that contained either 5g of Froot Loops® (FL) (3.75 kcal/g; 89.6% carbohydrate (40% sugar), 7.2% fat, and 3.2% protein) or 5g of chow (Lab Diet, St. Louis, MO; laboratory rodent diet 5001: 4.07 kcal/g: 57.9% carbohydrate (5% sugar), 13.5% fat, 28.5% protein) for 20-minute sessions. FL and chow were available on alternate days and amounts consumed were measured for each session.
Intraperitoneal Glucose Tolerance Test (IPGTT)
OM, S5B and SD rats from the above limited access paradigm (1 month later) were fasted overnight and on the next day, at 3-4 hours into their dark cycle were secured in a Decapicone® (Harvard Apparatus, Holliston, MA) and the left lateral tail vein catheterized. Glucose (Sigma-Aldrich, St. Louis, MO), dissolved in sterile water to generate a dose of 2 g/kg, was injected intraperitoneally (IP) and blood glucose (mg/dL) was measured at 0, 30, 60, and 120 minutes post injection using a handheld glucometer (Freestyle Flash, Abbot Diabetes Care, Alameda, CA).
Small animal positron emission tomography (μPET)
OM and S5B rats from the limited access feeding and IPGTT experiments were scanned between 8-9 months of age. Rats were fasted overnight to normalize blood glucose and control for variability in FDG uptake21. Rats were scanned on a μPET R4 tomograph (Concorde CTI Siemens, Knoxville, TN) as previously described22. Awake uptake protocols with [18F]FDG (2-fluoro-2-deoxy-D-glucose) and μPET allow the measurement of brain metabolism and assessment of subtle changes in regional brain-glucose utilization in response to a stimulus or environment. In brief, rats were injected IP with ~1 mCi [18F]FDG 30 minutes prior to image acquisition (uptake period) and then scanned for 20 minutes using a static imaging protocol. This in-vivo IP dosing regimen leads to similar brain glucose uptake as intravenous (IV) administration23 and ex-vivo IP administration of 2-deoxy-D-glucose leads to similar brain glucose uptake at 30 minutes as the original ex-vivo IV technique developed by Sokoloff and colleagues24. All μPET images were processed as previously25 using the Pixel-Wise Modeling Software Suite (PMOD Technologies, Switzerland). Images were analyzed for differences at the voxel-level with statistical parametric mapping (SPM) using methods previously described22.
Quantitative PCR (qPCR)
We used 5-month old naïve OM and S5B rats and 4-month old naïve SD rats. Rats were sacrificed and their brains removed and frozen. Brains were sectioned (20 μm thickness) at the striatum level using a cryostat (Microm HM560, Thermo Scientific, Rockford, IL) and a dissecting microscope and sterile scalpel blades were used to dissect out dorsal striatum. Slides were kept on dry ice throughout the tissue isolation procedure. Total RNA was isolated with the RNAGEM Tissue Plus extraction kit (ZyGEM, Hamilton, New Zealand) following this kit’s instructions. After RNA extraction, cDNA was first synthesized from 5 μl RNA using qScript cDNA Supermix reagent (Quanta BioSciences, Gaithersburg, MD) according to the manufacturer’s instructions then diluted 1:2 with PCR-grade water for a final volume of 40 μl. First-strand synthesis was performed in a MyCycler thermal cycler (Bio Rad, Hercules, CA) set to: (i) 25°C for 5 min, (ii) 42°C for 30 min and (iii) 85°C for 5 min, and then kept at 4°C until collected. Synthesized cDNA was stored at −30°C and extracted RNA at −80°C. TaqMan gene expression FAM-labeled assays (Applied Biosystems, Carlsbad, CA) were used to quantify mRNA expression. All assays were performed in triplicate. The ddCt method was used to determine relative mRNA expression. To calculate relative expression, 18S rRNA was simultaneously measured in each well using a VIC-labeled probe (4319413E, Applied Biosystems). If either probes’ Ct value for a given replicate was identified as an outlier, using Grubb’s method, then this animal’s values were removed from the analysis.
Receptor autoradiography
We used 5-month-old naïve OM and S5B rats. Slides with mounted brain sections at the level of dorsal striatum that were stored at −80°C were gradually brought back to room temperature and then exposed to receptor autoradiography procedures using [3H]spiperone, [3H]SCH23390, [3H]MPEP, and [3H]SR141716A as previously described21, 26–28. Radioactivity concentrations were verified via liquid scintillation counting. For [3H]spiperone and [3H]SCH23390 experiments, slides were placed in BAS-TR 2025 (GE Healthcare, Piscataway, NJ) phosphor imaging plates for 7 days along with [14C] standards (GE Healthcare, Piscataway, NJ) (calibrated against 3H-impregnated brain paste standards). Imaging plates were developed using an FLA-7000 phosphorimager (GE Healthcare, Piscataway NJ). Using Multigauge® software (GE Healthcare, Piscataway, NJ), regions of interest (ROIs) were drawn on the dorsolateral (DLST) and dorsomedial striatum (DMST) of each section. Values were averaged and initially expressed as PSL/mm2 and subsequently converted to dpm/mg with the use of [14C] standards. For [3H]MPEP and [3H]SR141716A experiments, slides were imaged using a β-imager (Biospace Lab, France). Scanning and image analysis methods were performed as previously described21.
Western Blotting
We used 5-month old naïve OM and S5B rats. Approximately 50 mg of pulverized dorsal striatum punches per animal were used to generate total protein extracts using 20 ml/g RIPA buffer (Thermo Scientific, Rockford, IL, USA) containing protease (cOmplete Mini, EDTA-free protease inhibitor cocktail, Roche, Basel, Switzerland) and phosphatase inhibitors (Halt phosphatase inhibitor cocktail, Thermo Scientific). The protein was diluted in Laemmli buffer, denatured (for 5 min at 95°C), subjected to electrophoresis, and transferred onto nitrocellulose membranes. The membranes were blocked in Odyssey blocking buffer (LI-COR, Lincoln, NE, USA) and incubated at 4°C overnight with primary antibodies. We used a validated29 rabbit polyclonal antibody to target Rgs4 (ABT17; 1:1000, Millipore, Billerica, MA) and a monoclonal mouse antibody against Gapdh (1:5000, Millipore). Membranes were subsequently incubated with goat anti-rabbit IRDye 800 (LI-COR) and goat anti-mouse IRDye 680 secondary antibodies (LI-COR) at room temperature for 1h. Membranes were imaged on the LI-COR infrared imaging system (LI-COR) and quantified using average integrated density values. Gapdh levels were used to control for total protein content.
Formalin-fixed human postmortem brain tissue sections (putamen) were sourced from the Banner Sun Health Research Institute (Sun City, AZ). Samples were exposed to protein isolation procedures as previously described30 but with minor modifications. Briefly, samples were homogenized in buffer (100 mM Tris-HCl, ph 6.8, 2% w/v SDS, 20% v/v glycerol, 4% β-mercaptoethanol) containing protease (cOmplete Mini, EDTA-free protease inhibitor cocktail, Roche, Basel, Switzerland) and phosphatase inhibitors (Halt phosphatase inhibitor cocktail, Thermo Scientific) followed by heating of samples at 105°C for 20 min and then ice cooling for 5 min. Protein content was quantified using the EZQ Protein quantification kit (Molecular Probes/Invitrogen, Grand Island, NY) and then flash frozen on dry ice and stored at −80°C. For western blotting we used an RGS4 antibody (sc-6204, 1:500, Santa Cruz Biotech, Dallas, TX) previously validated for use in human postmortem brain tissue31 along with procedures as described above (the ABT17 antibody used in rat did not react with human RGS4).
Chromatin immunoprecipitation (ChIP)
We used 5-month-old naïve OM and S5B rats. Fresh tissue was prepared for ChIP as previously described32. Briefly, two bilateral dorsal striatum punches/rat (2 punches per subject) were collected and processed. Chromatin were immunoprecipitated with either anti-H3K9me3 (ab8898) or anti-H3K4me3 (ab8580) antibody (Abcam, Cambridge, MA) then enrichment was measured using quantitative PCR (SYBR Green; Roche, Basel, Switzerland), normalized to each sample’s non-immunoprecipitated (input) fraction. Each reaction was run in triplicate and analyzed using the ddCT method as above. Primer sequences for various Rgs4 loci were as follows: (i) promoter region (forward: CCAGCGAGTCCTTTGCACAT; reverse: ACAGCTTCTTTGCAGAGCAGAA), (ii) 1.5 kb upstream of promoter (forward: TGCCCTAACCACCCACCTT; reverse: ATACCTATTCCCCTCTTTAACATTTGTC).
[35S]GTPγS autoradiography
We used 5-month-old naïve OM and S5B rats and tissue from adult Rgs4 wild-type (WT) and knockout (KO) mice. [35S]GTPγS autoradiography was assessed using previously published protocols33. Briefly, slides with 20 μm-thick sections were thawed at RT and a pap-pen was used to mark the area around tissue sections to contain fluid and to minimize incubation volume. Slides were then placed horizontally and incubated in pre-incubation buffer (50 mM Tris-HC1 (pH 7.4), 1 mM EDTA, 100 mM NaCl, and 5 mM MgC12) for 20 min at 20°C (0.6 ml per slide), after which the buffer was removed by aspiration. Slides were then incubated for 1 h at 20°C in the above buffer with the addition of 2 mM guanosine diphosphate and 1 μM dipropylxanthine (DPCPX), and then removed by aspiration. Finally, slides were incubated in pre-incubation buffer containing also 80-100 pM [35S]GTPγS, 2 mM GDP, 1 mM dithiothreitol, and 1 μM DPCPX in combination with either excess buffer or CCG-63802 (500 nM) for 90 min at 20°C. Nonspecific binding was assessed in parallel in the presence of 10 pM GTPγS. The incubation cocktail was removed by aspiration, and sections were washed twice at 0°C for 5 min each time in washing buffer (50 mM TrisHCl and 5 mM MgCl2, pH 7.4), rinsed in Millipore water for 30 s, air-dried, and apposed to BAS-SR 2040 (GE Healthcare, Piscataway, NJ) phosphorimaging plates for 3 days. [35S]GTPγS binding was assessed as described in the autoradiography section.
siRNA-mediated transcriptional silencing
Adult male SD rats were placed with bilateral steel cannulas directed at dorsal striatum (bregma: AP +1.7 mm, DV: −3.5 mm, ML: ±2.4 mm,) and injected into this region with either Rgs4-targeted (L-087936-02-0005) or non-targeted control (D-001810-10-05) siRNAs (On-targetplus SMARTpool, Dharmacon, GE Life Sciences, Lafayette, CO) and sacrificed 7 days later to verify Rgs4 downregulation. OM and S5B rats received the same siRNA into the dorsal striatum but without the use of cannulas. Delivery of siRNAs was achieved using the in vivo jetSI 10 mM delivery reagent (403-05, Polyplus transfection, Illkrich, France). Two (2 μl each) injections per hemisphere (4 μl total/hemisphere) in a dorsoventral gradient were performed.
Laser capture microdissection
To measure Rgs4 expression in discrete striatal cells, striatonigral (SN) and striatopallidal (SP) pathways were physically separated using a combination of fluorescent retrograde tracers and laser-capture microdissection (LCM). In brief, red Lumafluor retrobeads (0.5 μL per hemisphere) were targeted to the pallidum (AP −0.24 mm, DV +7.45 mm, ML ±3.45 mm, 10°) while green Lumafluor retrobeads (1.0 μL per hemisphere) were targeted to the midbrain (ventra tegmental area/substantia nigra) (AP −5.02 mm, DV +8.50 mm, ML ±1.00 mm, 0°). About 5-7 days after stereotaxic surgery, OM, S5B or SD rats were euthanized with CO2, and brains were immediately flash-frozen in isopentane then stored at −80°C. Prior to LCM, brains were coronally cryosectioned (10 μm thickness), rinsed in a series of ethanol and xylene to respectively dehydrate and clear the tissue. Immediately after processing the cryosections, neurons were identified under fluorescent light and ~750 cells from each pathway were separately collected. RNA was extracted using TRIzol, cDNA was generated from RNA using Quanta Bioscience’s cDNA synthesis system, and then gene expression was measured with predesigned TaqMan assays (as above) and analyzed using the ddCt method.
In-situ hybridization
Slides containing brain tissue at the level of dorsal striatum and stored at −80°C were gradually brought back to room temperature and then fixed with 4% paraformaldehyde, dehydrated in ethanol washes, delipidated in chloroform, rinsed in ethanol, and air-dried. All solutions were made with autoclaved 0.1% diethylpyrocarbonate-treated water and racks and staining dishes were also treated with this solution to inactivate RNases. An antisense riboprobe (complementary to Rgs4 target mRNA) generated by PCR (forward primer: ATGGCCTTCCCTCCTTTG; reverse primer: GGGAGCTCTGGGGACATT) was transcribed by the addition of the SP6 RNA polymerase enzyme and radiolabeled with [35S]uridine 5′-[α-thio] triphosphate (1000–1500 Ci/mmol; Perkin Elmer, Waltham, MA). Brain sections were hybridized for 16 hrs with radiolabeled probes (~4 × 106 cpm/slide) in a humidified chamber at 55–65 °C (depending on the stringency requirements) under appropriate hybridization buffer conditions. The brain sections were carried through post-hybridization washes and air-dried. Control experiments were carried out using sense RNA probes to validate the specificity of the antisense hybridization signal. Slides were apposed to BAS-SR 2040 (GE Healthcare, Piscataway, NJ) phosphor imaging plates for approximately 3 days along with [14C] standards (GE Healthcare, Piscataway, NJ). Imaging plates were developed using an FLA-7000 phosphorimager (GE Healthcare, Piscataway NJ). Using Multigauge® software (GE Healthcare, Piscataway, NJ), regions of interest (ROIs) were drawn on the dorsal striatum of each section. Mean values were initially expressed as PSL/mm2 and subsequently converted to dpm/mg with the use of the aforementioned [14C] standards.
Brain slice electrophysiology
Male 4-5-month-old OM and S5B rats were anesthetized with isoflurane and decapitated. The brain was rapidly removed and placed in an ice-cold modified artificial cerebrospinal fluid consisting of (in mM): NMDG, 93; KCl, 2.5; NaH2PO4, 1.2; NaHCO3, 30; HEPES, 20; Glucose, 25; Ascorbic acid, 5; Sodium pyruvate, 3; MgCl2, 10; CaCl2, 0.5. Coronal hemisections (280 μm) containing the striatum were cut using a vibratome (Leica VT1200S, Buffalo Grove, IL, USA). Slices were incubated in a HEPES-modified aCSF containing (in mM): in mM: NaCl, 109; KCl, 4.5; NaH2PO4, 1.2; NaHCO3, 35; HEPES, 20; Glucose, 11; Ascorbic acid, 0.4; MgCl2, 1; CaCl2, 2.5, maintained at 34-35 °C for ~20 minutes, and then allowed to stabilize at room temperature for at least 30 minutes prior to initiating recordings. During recordings, slices were placed in a small volume recording chamber (RC-26, Warner Instruments), continuously superfused with aCSF containing (in mM): NaCl 126, KCl 3, MgCl2 1.5, CaCl2 2.4, NaH2PO4 1.2, NaHCO3 26, glucose 11, Ascorbic acid, 0.4, picrotoxin 0.05, at a rate of 2 mL/min using a peristaltic pump and maintained at 30-32°C using an in-line solution heater. During LTD recordings, caffeine (50 μM) was also included in the recording aCSF in order to minimize the influence of endogenous adenosine at both pre and postsynaptic receptors. Extracellular recordings were performed using a GeneClamp 500B amplifier (Molecular Devices), and filtered (high pass, 1 Hz; low pass, 10 kHz) using a signal conditioner (Brownless 440, Pacer Scientific, Los Angeles CA). Recording micropipettes were pulled from borosilicate glass (BF150-86-10, Sutter Instruments, Novato CA) and filled with aCSF. Under stereomicroscopic visualization, the pipette was lowered via a micromanipulator into the dorsal striatum, and a constant current stimulus isolation unit (DS-3, Digitimer, LTD) was used to elicit glutamatergic-driven population spikes (PS) via 0.1 ms duration pulses delivered to a twisted bipolar formvar-insulated nichrome stimulating electrode. Signals were acquired directly to PC using WinLTP software (WinLTP Ltd, Bristol UK) and an A/D board (PCI 6251, National Instruments, Austin TX). Input-output curves were obtained in each slice by averaging 3 responses at each stimulus intensity (10-120 μA). For LTD experiments, baseline intensity was set to 30-50% of the maximum signal response, and responses were obtained every 30s. After obtaining ≥10 minutes of stable baseline, 4 high-frequency stimuli (HFS, 100 Hz, 1s duration) were delivered at 10s intervals. Post HFS recordings were obtained for ≥30 minutes to assess LTD.
Online databases
We used the Generic Genome Browser (v. 2.16) (human data assembly GChr37, build NCBI_77.1) and data from the Rat Genome Database website (http://rgd.mcw.edu)34 to explore quantitative trait loci mapping relevant to Rgs4 and obesity-related traits.
Statistics
Sample sizes for experiments (animal and non-animal) were chosen based on either prior experimental work or power calculations from pilot experiments. Rats and mice were assigned randomly to each experimental group. and experiments were not blinded to experimental conditions. Depending on experiment, we used two-sample paired and unpaired t-tests or single factor and multifactor ANOVAs with Holm-Šídák post-hoc tests, taking repeated measures into account if appropriate. All statistical tests were evaluated at the p≤0.05 level (two tailed). All data are expressed as mean ±SEM.
Results
Behavioral and metabolic abnormalities in DIO-susceptible OM rats coincide with decreased metabolic activity in the dorsal striatum
We exposed OM and S5B rats to either normal chow or a high fat diet (Table S1) for 8 weeks to assess differences in DIO between strains. As expected, high fat diet-fed OM rats consumed significantly more calories (Fig. 1a), gained significantly more weight (Fig. 1b), and exhibited significantly greater adiposity (expressed relative to body weight) than normal chow-fed OM rats (Fig. 1c). In contrast, high fat diet-fed S5B rats did not show any significant increase in body weight (Fig. 1b) or adiposity (Fig. 1c) compared to normal chow-fed S5B rats, even though they consumed significantly more calories (Fig. 1a). As above, chow-fed OM rats exhibited significantly greater body weight (Fig. 1d) and food intake (Fig. 1e) than chow-fed S5B rats but the two strains did not differ in adiposity (Fig. 1f). OM rats also had significantly higher glucose intolerance compared to S5B rats (Fig. 1g).
Figure 1. Behavioral and metabolic abnormalities in DIO-susceptible OM rats coincide with decreased metabolic activity in the dorsal striatum.
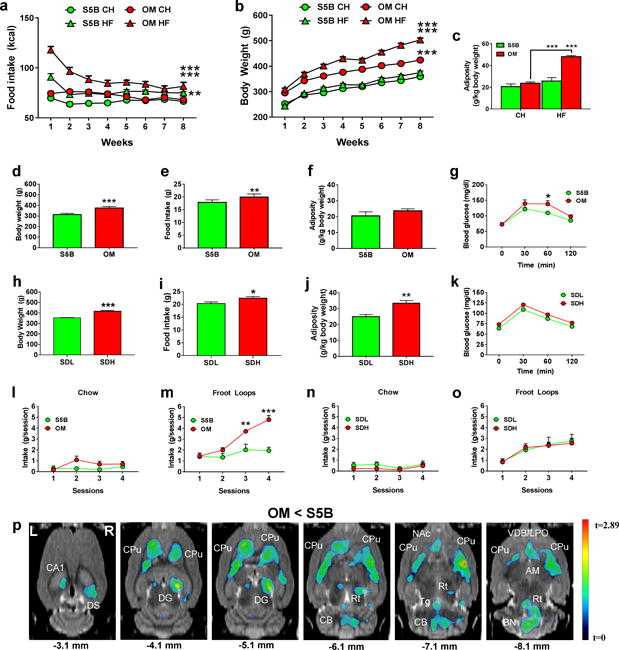
(a) High fat (HF)-fed OM rats (n=8) exhibited significantly increased caloric intake (kcal) relative to both chow (CH)-fed OM rats (n=8) and HF-fed S5B rats (n=8) (two-way repeated measures (RM) ANOVA, interaction effect, F(21, 196)=11.2). (b) HF-fed OM rats (n=10) exhibited significantly increased body weight relative to both CH-fed OM rats (n=10) and HF-fed S5B rats (n=10) (two-way RM ANOVA, interaction effect, F(21, 252)=30.1). (c) HF-fed OM rats (n=4) exhibited significantly increased adiposity compared to CH-fed OM rats (n=4) and to HF-fed S5B rats (n=4) (two-way ANOVA, interaction effect, F(1, 12)=19.04). (d) CH-fed OM rats (n=10) (separate cohort from above) weighed significantly more than S5B (n=10) (separate cohort from above) rats (unpaired t-test; t=10.2, df=18). (e) CH-fed OM (n=8) rats consumed significantly more food than S5B (n=8) rats (unpaired t-test; t=3.59, df=14). (f) CH-fed OM (n=4) and S5B (n=4) rats did not differ in adiposity (n=4) (unpaired t-test). (g) CH-fed OM rats (n=7) exhibited greater glucose intolerance compared to S5B (n=8) rats (two-way RM ANOVA, interaction effect F(3,39)=2.56, p=0.06, 60 min: t=3.1, df=52). (h) SDH rats (n=6) weighed significantly more than SDL rats (n=6) (unpaired t-test, t=6.92, df=10). (i) SDH rats exhibited significantly greater food intake compared to SDL rats (unpaired t-test, t=2.29, df=10). (j) SDH rats exhibited increased adiposity compared to SDL rats (unpaired t-test; t=3.87, df=10). (k) SDH and SDL rats did not differ in glucose tolerance (unpaired t-test). (l) CH-fed OM (n=6) and S5B (n=7) rats did not differ in CH intake during limited access sessions (two-way RM ANOVA). (m) CH-fed OM rats (n=6) exhibited rapid escalation of Froot Loops® (FL) intake and consumed significantly more FL than CH-fed S5B (n=7) rats (two-way RM ANOVA, interaction effect, F(3,33)=9.48, p<0.001, session 3: t=3.49, df=44, session 4: t=5.81, df=44). (n, o) SDL and SDH rats did not differ in CH or FL intake (two-way RM ANOVA). (p) Statistical parametric map from FDG uptake images overlaid onto MRI showing that CH-fed OM rats (n=5) exhibited significantly lower FDG uptake than CH-fed S5B rats (n=5) (unpaired t-test, t=2.71; df=8; p≤0.05, uncorrected) in several regions (hippocampus, CA1, dorsal subiculum, DS, dentate gyrus, DG, cerebellum, CB, tegmental nuclei, Tg, reticular nuclei, Rt, amygdala, AM, brainstem nuclei, BN, nucleus accumbens, NAc, vertical diagonal band, lateral preoptic, VDB/LPO) with the most notable effect in dorsal striatum (caudate putamen, CPu); L=left, R=right.*p≤0.05, **p≤0.01, ***p≤0.001
We next compared normal chow-fed, age-matched SD rats (control) that were stratified into low (SDL) and high body weight (SDH) groups to control for weight differences between the S5B and OM rats. Comparisons between SDL and SDH rats were similar with respect to body weight (Fig. 1h) and food intake (Fig. 1i). In contrast to S5B and OM rats, however, SDL and SDH rats differed in adiposity (Fig. 1j) and did not differ in glucose intolerance (Fig. 1k). Food intake was further examined by measuring intake of a high-energy, sucrose-rich “snack food” (Froot Loops®; FL) or chow during limited access conditions. OM rats consumed significantly more chow (Fig. 1l) and FL than S5B rats (Fig. 1m). In contrast, SDL and SDH rats did not differ in chow or FL intake (Figs. 1n, o).
We performed in vivo whole-brain metabolic mapping using small animal PET (μPET) and [18F]fluoro-2-deoxy-D-glucose (FDG)35, which allowed us to profile, in an unbiased, non-invasive manner, brain-wide differences in metabolic activity patterns between OM and S5B rats during freely-moving conditions. Compared to S5B rats, OM rats exhibited significantly lower FDG uptake in several brain areas including striatum (caudate putamen, CPu), hippocampus/dentate gyrus (CA1/DG), cerebellum (CB) and reticular (Rt) and tegmental (Tg) nuclei (Fig. 1p). Notably, the most widespread and pronounced decrease in FDG uptake was observed in the dorsal striatum bilaterally (Fig. 1p).
Striatal Rgs4 regulates feeding and is associated with DIO-susceptibility
Given the significantly lower FDG uptake in dorsal striatum of OM rats, and the proposed involvement of this region in food craving, ingestive behavior and obesity, we profiled striatal gene expression in OM and S5B rats for transcripts known to influence striatal signaling, plasticity and reward/aversion. Compared to S5B rats, OM rats exhibited significantly greater mRNA expression of Rgs4, Drd1a, Drd2l, Grm5, and Cnr1 (Fig. 2a).
Figure 2. Striatal Rgs4 regulates feeding and is associated with DIO-susceptibility.
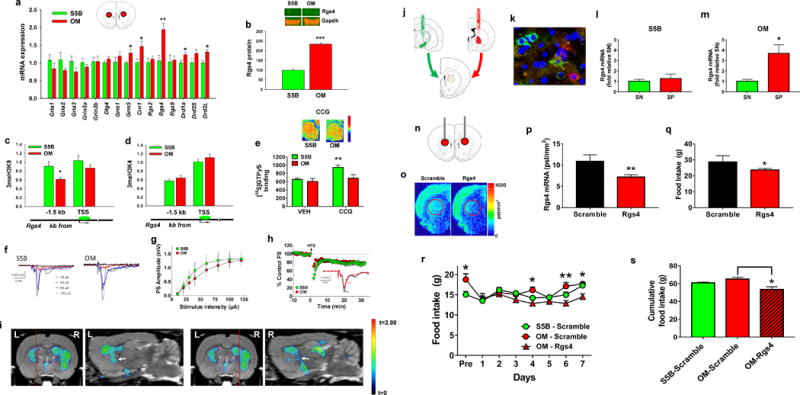
(a) Transcriptional profiling in dorsal striatum demonstrated that OM rats (n=7) exhibited significantly greater mRNA expression (unpaired t-tests) of Rgs4 (t=3.22; df=12), Drd1a (t=2.32; df=12), Drd2L (t=2.46; df=12), Grm5 (t=2.81; df=12), and Cnr1 (t=2.48; df=12) than S5B rats (n=7). (b) OM rats (n=8) had significantly greater Rgs4 protein in dorsal striatum compared to S5B rats (n=8) (unpaired t-test, t=7.74; df=14) (representative blots shown). (c) Compared to S5B (n=8), OM rats (n=8) exhibited significantly lower enrichment of the repressive 3meH3K9 histone mark (unpaired t-test, t=2.28; df=14) but not (d) the permissive 3meH3K4 at a highly conserved genomic region (~1.5 kb upstream of transcriptional start site (TSS)) of Rgs4. (e) OM (n=6) and S5B (n=6) rats did not differ in basal [35S]GTPγS binding but the Rgs4 inhibitor CCG-63802 (0.5 μM) led to a significant increase in [35S]GTPγS binding in dorsal striatum in S5B rats (n=6) (paired t-test, t=6.29; df=5) and to a smaller, marginally significant increase in [35S]GTPγS binding in OM rats (n=6) (paired t-test, t=2.38; df=5; p=0.06). (f, g) Representative extracellular recordings from the dorsal striatum of S5B and OM rats. The glutamatergic-driven population spike (PS) increased with increasing stimulation intensity. Summary input-output (I/O) curves from all slices revealed no significant group differences between S5B (n = 12 slices, 3 rats) and OM rats (n = 12 slices, 3 rats; two-way repeated measures (RM) ANOVA, F(1,22) = 0.87, p = 0.36). (h) Summary time course of long-term depression (LTD) elicited by high frequency stimulation (HFS; 100 Hz, 1s duration trains delivered 4 times at 10 s intervals). Inset shows representative traces from a control (S5B) brain slice, prior to (pre) and 30 minutes following HFS (post). No significant differences in LTD were observed between S5B and OM rats (two-way RM ANOVA, F(1,22) = 0.29, p = 0.59). (i) Coronal and sagittal planes of SPM images shown in Figure 1p indicating that the lower FDG uptake observed in dorsal striatum of OM rats extends to the globus pallidus (white arrow). Red line denotes location of sagittal plane. L=left, R=right. (j) Strategy for dissecting striatonigral (SN) (green) and striatopallidal (SP) (red) projection pathways using retrograding latex beads. (k) Representative microscopy image (20×) of latex bead accumulation in SN and SP MSNs. (l, m) Rgs4 mRNA is selectively enriched in SP compared to SN MSNs (unpaired t-test, t=3.15; df=4) of OM (n=3) but not S5B (n=3) rats. (n) Targeted cannula placement for siRNA delivery to dorsal striatum. (o) Representative in situ hybridization autoradiograms from rats injected with scrambled (n=4) or Rgs4-targeted siRNA (n=4). (p) siRNA-mediated inhibition of striatal Rgs4 in SD rats significantly decreased striatal Rgs4 mRNA (unpaired t-test, t=4.55; df=6) and (q) 24-hr food intake (unpaired t-test, t=2.51; df=6). (r) OM and S5B rats differed in 24-hr food intake (unpaired t-test, Pre; t=3.01; df=40) but siRNA-mediated Rgs4 inhibition decreased food intake (two-way RM ANOVA, interaction effect, F(12,48)=1.45, p=0.17; Session 4 (t=2.60; df=56), Session 6 (t=3.38; df=56), and Session 7 (t=2.52; df=56)) in OM rats (n=4) but not in scramble controls (OM; n=4; S5B; n=3). (s) Cumulative food intake from days 4-7 from adjacent figure (one way ANOVA, F(2, 8)=6.94; p=0.018). *p≤0.05, **p≤0.01, ***p≤0.001
Next, we examined whether the above differentially-expressed transcripts were also altered at the protein/receptor level. In vitro autoradiography with [3H]spiperone, [3H]SCH23390, [3H]MPEP, and [3H]SR141716A was used as previously described21, 26–28 to respectively assess striatal dopamine D2 receptor (D2R), dopamine D1 receptor (D1R), metabotropic glutamate receptor 5 (mGluR5), and cannabinoid receptor 1 (CB1R) levels in dorsal striatum. No significant differences between the two strains were observed for any of the receptors (Fig. S1). In contrast, Rgs4 protein levels in dorsal striatum, determined via western blotting using an anti-Rgs4 antibody for use in rats29, 36, was significantly greater in OM relative to S5B rats (Fig. 2b). There were no differences in dorsal striatal Rgs4 levels between SDH and SDL rats (Fig. S2).
Increased Rgs4 in OM rats was also observed at the epigenetic level using chromatin immunoprecipitation (ChIP)32, which assessed the enrichment of repressive (3meH3K9) and permissive (3meH3K4) histone modifications at the Rgs4 gene locus. Compared to S5B rats, OM rats exhibited significantly lower enrichment of the repressive 3meH3K9 modification at a highly conserved region of the Rgs4 gene (Fig. 2c). In contrast, no significant differences in 3meH3K4 enrichment were observed (Fig. 2d), overall suggesting that repressive epigenetic regulatory mechanisms are associated with the differential regulation of the Rgs4 gene between the two strains.
Rgs4 is a GTPase accelerating enzyme that negatively modulates G-protein activity37. To probe functional differences in Rgs4-dependent G-protein activity in striatum between OM and S5B rats, we used [35S]GTPγS autoradiography and the selective Rgs4 inhibitor CCG-6380238. Since Rgs4 acts to terminate G-protein activity, we expected that Rgs4 inhibition via CCG-63802 would increase striatal [35S]GTPγS binding. First, we validated this assay at measuring Rgs4-specific G-protein activity using brain tissue from Rgs4 knockout (KO) and wild-type (WT) mice. As expected, tissue sections containing dorsal striatum from WT mice exposed to CCG-63802 (0.5 μM; IC50=1.4 μM38) exhibited significantly greater (~69%) striatal [35S]GTPγS binding than vehicle-exposed sections, whereas CCG-63802 did not increase [35S]GTPγS binding (~2.9%) in Rgs4 KO mice (Fig. S3), indicating that CCG-63802 effects on [35S]GTPγS are mediated via Rgs4. As expected, CCG-63802 (0.5 μM) led to a significant increase (~45%) in dorsal striatum [35S]GTPγS binding in S5B rats (Fig. 2e) whereas it led to a much smaller, marginally significant increase in [35S]GTPγS binding in OM rats (~13%) (Fig. 2e). OM rats have significantly higher Rgs4 in dorsal striatum and exhibited significantly lower CCG-63802-stimulated [35S]GTPγS binding in this region compared to S5B rats suggesting that increased striatal Rgs4 mRNA and protein coincide with increased Rgs4 function in OM rats relative to S5B rats, though it is noteworthy to point out that effects of Rgs4 on [35S]GTPγS binding can be complex as previously Rgs4 was shown to increase [35S]GTPγS binding in cells (but not in tissue)39.
Rgs4 is necessary for striatal plasticity, and specifically dopamine-mediated regulation of long-term depression (LTD), a marker of presynaptic glutamatergic function, in dorsal striatum40. Given the differences in striatal Rgs4 and FDG uptake in OM rats and S5B rats, we also examined whether the two strains differed in measures of glutamate function in dorsal striatum, as assessed using extracellular recordings of glutamatergic-driven population spikes in OM and S5B brain slices. We found that OM and S5B rats showed no significant differences in baseline glutamatergic transmission, demonstrated using input-output curves of stimulation intensity vs. response (Figs. 2f, g). In addition, LTD elicited by high-frequency stimulation did not significantly differ between groups (Fig. 2h) indicating that general presynaptic glutamate function in the striatum is identical between the two strains.
The lack of presynaptic glutamate functional differences between the two strains indicated that presynaptic glutamate input into dorsal striatum could not account for the significantly lower FDG uptake in OM rats. Instead, it suggested that lower striatal FDG uptake in OM rats may be driven by a postsynaptic mechanism. The vast majority (~95% in rodents) of the dorsal striatum consists of two cell types, striatonigral (SN) and striatopallidal (SP) medium spiny neurons (MSNs), with each differing in their neurochemical makeup, anatomical connectivity (dorsal SN MSNs mainly project to substantia nigra whereas dorsal SP MSNs mainly project to the globus pallidus) and their synergistic and complementary effects on behavior41. The FDG-PET metabolic mapping results indicated that the striatal deficits we had identified in OM rats extended into the globus pallidus region bilaterally, mapping the known anatomical projection pattern of SP MSNs (Fig. 2i). Interestingly, SP MSNs in particular have been implicated in both human and animal obesity42, and Rgs4 has been shown to modulate striatal plasticity mechanisms in SP MSNs40. Nevertheless, the MSN expression profile of Rgs4 has not been previously described. To address this paucity, we first injected latex retrobeads (Lumafluor) into the globus pallidus (red fluorescence) or the substantia nigra (green fluorescence) of SD rats, to respectively label, capture, and transcriptionally profile dorsal striatal SP and SN MSNs (Figs. 2j, k). We found that Rgs4 mRNA (and Drd2 mRNA which is enriched in SP MSNs41 serving as positive control) was selectively enriched in SP compared to SN MSNs (Fig. S4). The same approach was then implemented in OM and S5B rats. Like SD rats, Rgs4 was enriched in SP MSNs of OM rats (Fig. 2l). In contrast, Rgs4 was not enriched in SP MSNs of S5B rats (Fig. 2m).
To examine a mechanistic role for striatal Rgs4 in regulating food intake we used an siRNA approach to inhibit striatal Rgs4 mRNA. We first validated the approach in SD rats by injecting siRNA bilaterally into the dorsal striatum (Fig. 2n). Compared to the scrambled control, the Rgs4-targeting siRNA led to a 40% decrease in Rgs4 mRNA in SD rats as assessed using in situ hybridization32 (Figs. 2o, p). As expected, siRNA-mediated inhibition of Rgs4 in SD rats led to a significant reduction in food intake (Fig. 2q). siRNA-mediated Rgs4 inhibition was then performed in OM rats injected bilaterally into the dorsal striatum with the siRNA constructs (Rgs4-targeting or scrambled). S5B rats were injected with the scrambled siRNA only. As expected, OM rats consumed significantly more food than S5B rats prior to siRNA injection (Fig. 2r) and Rgs4 inhibition significantly decreased their food intake (Figs. 2r, s).
Rgs4 is associated with increased body weight and obesity susceptibility traits in humans
SP MSNs have been implicated in human obesity43, but a role for Rgs4 has not been described. To extend the relevance of our findings to humans, we first explored whether the RGS4 gene was associated with features of obesity in humans using publicly-available databases. We found that the RGS4 genomic locus coincided with several quantitative trait loci (QTL) significantly associated with obesity-related phenotypes such as body weight and adiposity (Fig. 3a). These findings raised the possibility that overweight humans, like the overweight DIO-susceptible OM rats, also exhibited elevated striatal RGS4. To test this hypothesis, we examined RGS4 protein levels in human postmortem striatal tissue from normal, otherwise healthy subjects who according to their body mass index (BMI) were classified as either normal weight (BMI: 18.5-24.9) or overweight (non-obese) (BMI: 25- 29.9) (Fig. 3b & Table S2). The anti-Rgs4 antibody used for our rat experiments did not react with human RGS4 and thus we used an antibody previously reported for use with human postmortem brain tissue31. Our results revealed two distinct bands (at ~25 kDa and ~35 kDa) corresponding to two human RGS4 isoforms previously reported31. As hypothesized from our rat experiments, we found that overweight subjects had significantly greater RGS4 protein levels (~35 kDa band) compared to normal weight controls (Fig. 3c). No difference between the two groups however was detected in the smaller (~25 kDa) band.
Figure 3. Striatal Rgs4 is associated with increased body weight in humans.
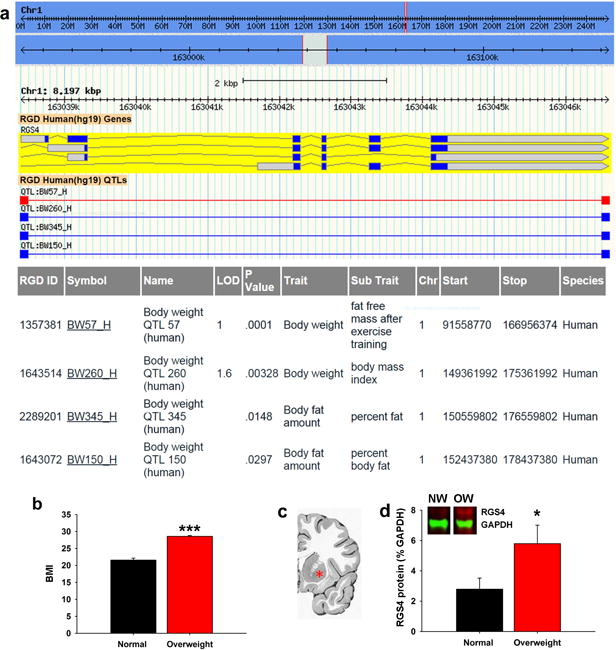
(a) Quantitative trait locus (QTL) data showing that human RGS4 is associated with obesity-related traits including body weight and adiposity. (b) Body mass index (BMI) used to stratify normal weight (NW; n=8) and overweight (OW; n=6) human subjects (unpaired t-test, t=10.81, df=12). (c) Coronal human brain section showing representative region of dorsal striatum sampled for downstream analysis (red asterisk, putamen). (d) Overweight human subjects (n=6) exhibit significantly greater striatal Rgs4 (~35 kDa) protein levels than normal weight controls (n=8) (unpaired t-test; t=2.23; df=12). *p≤0.05, ***p≤0.001
Discussion
The striatum regulates compulsive-like food seeking behavior42 and is implicated in human and animal obesity44. Within this brain region, disturbances in SP MSNs in particular are believed to be critically involved in obesity43. Here we report that striatal Rgs4, an intracellular G protein signaling regulator, mediates feeding in rats. In addition, we report this gene’s association with body weight and DIO susceptibility in rats and humans, providing novel translational evidence that striatal Rgs4 regulates feeding and obesity-related traits.
We show that DIO-susceptible (but not obese) OM rats exhibit normal adiposity, have enhanced predisposition to overconsumption of a palatable high-energy snack food, and show lower brain metabolic activity in various brain regions but most prominently in the dorsal striatum. The lower striatal brain activity in OM rats did not coincide with a decrease in presynaptic glutamatergic signaling, indicating that the striatal activity deficit in OM rats may be due to decreased postsynaptic metabolic activity in striatal MSNs, as opposed to activity at corticostriatal terminals. Indeed, focusing on this region, we discovered that OM rats exhibited significantly greater striatal Rgs4 levels and striatal Rgs4 function compared to DIO-resistant S5B rats. Notably, striatal Rgs4 expression was predominantly enriched in SP MSNs compared to SN MSNs in OM rats, however, this was not the case in S5B rats. Since normal SD rats, which like OM rats can develop obesity in response to high fat diets45, also exhibited enrichment of Rgs4 in SP MSNs, this finding indicates that discrete MSN Rgs4 enrichment patterns are associated with DIO susceptibility and resistance. Interestingly, we observed increased striatal Rgs4 levels in overweight human subjects and additionally, found evidence of genetic association of Rgs4 with obesity-related human traits. While Rgs4 interacts with several different receptor systems expressed in the striatum, including dopamine receptors46, 47,48–51, mGluR552, 53, adenosine 2a receptors40, opioid receptors54, 55, and muscarinic 4 receptors56, 57 and therefore it is possible that some of our observations may be associated with striatal Rgs4 action at these sites, our collective findings demonstrate the novel and critical role of striatal Rgs4 in feeding and in DIO susceptibility and resistance.
Indeed, the vast majority of human obesity cases arise from DIO, which is characterized by symptoms such as compulsive overeating, impaired inhibitory control and negative emotionality (e.g. anxiety, stress), features which are also prominent in drug addiction, and as such, the concept of food addiction as an etiological factor contributing to DIO has recently gained traction among scientists58. Although not traditionally associated with overeating or obesity, emerging evidence suggests that brain regions such as the striatum, and specifically, imbalances between SP and SN MSNs, which are heavily involved in substance abuse/addiction59, are also implicated in overeating/obesity.43 Thus these circuits may represent a promising therapeutic target for hyperphagia and obesity-related disorders.
Supplementary Material
Acknowledgments
This work was supported by the NIAAA (AA11034, AA07574, AA07611, Y1AA3309), the NIDA (DA006278, DA015446, DA023214, DA030359, ZIA000069) and the NINDS (NS086444, NS093537). MM was supported by DA007135. MLM was supported by GM007280 and DA038954. MM is a cofounder and owns stock in Metis Laboratories.
Footnotes
All authors declare no conflicts of interest.
References
- 1.Hedley AA, Ogden CL, Johnson CL, Carroll MD, Curtin LR, Flegal KM. Prevalence of overweight and obesity among US children, adolescents, and adults, 1999-2002. JAMA. 2004;291(23):2847–2850. doi: 10.1001/jama.291.23.2847. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Baker JL, Hill JO, Dietz WH. Controversies regarding reported trends: has the obesity epidemic leveled off in the United States? Adv Nutr. 2012;3(5):751–752. doi: 10.3945/an.112.002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietz WH. The response of the US Centers for Disease Control and Prevention to the obesity epidemic. Annu Rev Public Health. 2015;36:575–596. doi: 10.1146/annurev-publhealth-031914-122415. [DOI] [PubMed] [Google Scholar]
- 4.Schemmel R, Mickelsen O, Tolgay Z. Dietary obesity in rats: influence of diet, weight, age, and sex on body composition. The American journal of physiology. 1969;216(2):373–379. doi: 10.1152/ajplegacy.1969.216.2.373. [DOI] [PubMed] [Google Scholar]
- 5.Fisler JS, Shimizu H, Bray GA. Brain 3-hydroxybutyrate, glutamate, and GABA in a rat model of dietary obesity. Physiol Behav. 1989;45(3):571–577. doi: 10.1016/0031-9384(89)90075-9. [DOI] [PubMed] [Google Scholar]
- 6.Obst BE, Schemmel RA, Czajka-Narins D, Merkel R. Adipocyte size and number in dietary obesity resistant and susceptible rats. The American journal of physiology. 1981;240(1):E47–53. doi: 10.1152/ajpendo.1981.240.1.E47. [DOI] [PubMed] [Google Scholar]
- 7.Primeaux SD, Barnes MJ, Braymer HD, Bray GA. Sensitivity to the satiating effects of exendin 4 is decreased in obesity-prone Osborne-Mendel rats compared to obesity-resistant S5B/Pl rats. Int J Obes (Lond) 2010;34(9):1427–1433. doi: 10.1038/ijo.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allerton TD, Primeaux SD. High-fat diet differentially regulates metabolic parameters in obesity-resistant S5B/Pl rats and obesity-prone Osborne-Mendel rats. Canadian journal of physiology and pharmacology. 2015:1–10. doi: 10.1139/cjpp-2015-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pittman DW, Smith KR, Crawley ME, Corbin CH, Hansen DR, Watson KJ, et al. Orosensory detection of fatty acids by obesity-prone and obesity-resistant rats: strain and sex differences. Chemical senses. 2008;33(5):449–460. doi: 10.1093/chemse/bjn012. [DOI] [PubMed] [Google Scholar]
- 10.Gilbertson TA, Liu L, Kim I, Burks CA, Hansen DR. Fatty acid responses in taste cells from obesity-prone and -resistant rats. Physiol Behav. 2005;86(5):681–690. doi: 10.1016/j.physbeh.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 11.Schaffhauser AO, Madiehe AM, Braymer HD, Bray GA, York DA. Effects of a high-fat diet and strain on hypothalamic gene expression in rats. Obesity research. 2002;10(11):1188–1196. doi: 10.1038/oby.2002.161. [DOI] [PubMed] [Google Scholar]
- 12.Barnes MJ, Holmes G, Primeaux SD, York DA, Bray GA. Increased expression of mu opioid receptors in animals susceptible to diet-induced obesity. Peptides. 2006;27(12):3292–3298. doi: 10.1016/j.peptides.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 13.White CL, Ishii Y, Mendoza T, Upton N, Stasi LP, Bray GA, et al. Effect of a selective OX1R antagonist on food intake and body weight in two strains of rats that differ in susceptibility to dietary-induced obesity. Peptides. 2005;26(11):2331–2338. doi: 10.1016/j.peptides.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, York DA, Bray GA. Regulation of ghrelin gene expression in stomach and feeding response to a ghrelin analogue in two strains of rats. Peptides. 2004;25(12):2171–2177. doi: 10.1016/j.peptides.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Thanos PK, Kim R, Cho J, Michaelides M, Anderson BJ, Primeaux SD, et al. Obesity-resistant S5B rats showed greater cocaine conditioned place preference than the obesity-prone OM rats. Physiol Behav. 2010;101(5):713–718. doi: 10.1016/j.physbeh.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thanos PK, Cho J, Kim R, Michaelides M, Primeaux S, Bray G, et al. Bromocriptine increased operant responding for high fat food but decreased chow intake in both obesity-prone and resistant rats. Behavioural brain research. 2010;217(1):165–170. doi: 10.1016/j.bbr.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White CL, Ishihara Y, York DA, Bray GA. Effect of meta-chlorophenylpiperazine and cholecystokinin on food intake of Osborne-Mendel and S5B/P1 rats. Obesity (Silver Spring) 2007;15(3):624–631. doi: 10.1038/oby.2007.579. [DOI] [PubMed] [Google Scholar]
- 18.Kenny PJ. Reward mechanisms in obesity: new insights and future directions. Neuron. 2011;69(4):664–679. doi: 10.1016/j.neuron.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tulloch AJ, Murray S, Vaicekonyte R, Avena NM. Neural responses to macronutrients: hedonic and homeostatic mechanisms. Gastroenterology. 2015;148(6):1205–1218. doi: 10.1053/j.gastro.2014.12.058. [DOI] [PubMed] [Google Scholar]
- 20.Primeaux SD, Barnes MJ, Bray GA. Olfactory bulbectomy increases food intake and hypothalamic neuropeptide Y in obesity-prone but not obesity-resistant rats. Behavioural brain research. 2007;180(2):190–196. doi: 10.1016/j.bbr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thanos PK, Michaelides M, Piyis YK, Wang GJ, Volkow ND. Food restriction markedly increases dopamine D2 receptor (D2R) in a rat model of obesity as assessed with in-vivo muPET imaging ([11C] raclopride) and in-vitro ([3H] spiperone) autoradiography. Synapse. 2008;62(1):50–61. doi: 10.1002/syn.20468. [DOI] [PubMed] [Google Scholar]
- 22.Thanos PK, Michaelides M, Gispert JD, Pascau J, Soto-Montenegro ML, Desco M, et al. Differences in response to food stimuli in a rat model of obesity: in-vivo assessment of brain glucose metabolism. Int J Obes (Lond) 2008;32(7):1171–1179. doi: 10.1038/ijo.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiffer WK, Mirrione MM, Dewey SL. Optimizing experimental protocols for quantitative behavioral imaging with 18F-FDG in rodents. J Nucl Med. 2007;48(2):277–287. [PubMed] [Google Scholar]
- 24.Meibach RC, Glick SD, Ross DA, Cox RD, Maayani S. Intraperitoneal administration and other modifications of the 2-deoxy-D-glucose technique. Brain Res. 1980;195(1):167–176. doi: 10.1016/0006-8993(80)90874-4. [DOI] [PubMed] [Google Scholar]
- 25.Schweinhardt P, Fransson P, Olson L, Spenger C, Andersson JL. A template for spatial normalisation of MR images of the rat brain. J Neurosci Methods. 2003;129(2):105–113. doi: 10.1016/s0165-0270(03)00192-4. [DOI] [PubMed] [Google Scholar]
- 26.Nader MA, Daunais JB, Moore T, Nader SH, Moore RJ, Smith HR, et al. Effects of cocaine self-administration on striatal dopamine systems in rhesus monkeys: initial and chronic exposure. Neuropsychopharmacology. 2002;27(1):35–46. doi: 10.1016/S0893-133X(01)00427-4. [DOI] [PubMed] [Google Scholar]
- 27.Samadi P, Gregoire L, Morissette M, Calon F, Hadj Tahar A, Dridi M, et al. mGluR5 metabotropic glutamate receptors and dyskinesias in MPTP monkeys. Neurobiol Aging. 2008;29(7):1040–1051. doi: 10.1016/j.neurobiolaging.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Thanos PK, Ramalhete RC, Michaelides M, Piyis YK, Wang GJ, Volkow ND. Leptin receptor deficiency is associated with upregulation of cannabinoid 1 receptors in limbic brain regions. Synapse. 2008;62(9):637–642. doi: 10.1002/syn.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krumins AM, Barker SA, Huang C, Sunahara RK, Yu K, Wilkie TM, et al. Differentially regulated expression of endogenous RGS4 and RGS7. The Journal of biological chemistry. 2004;279(4):2593–2599. doi: 10.1074/jbc.M311600200. [DOI] [PubMed] [Google Scholar]
- 30.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Development and validation of a novel protein extraction methodology for quantitation of protein expression in formalin-fixed paraffin-embedded tissues using western blotting. J Pathol. 2009;217(4):497–506. doi: 10.1002/path.2504. [DOI] [PubMed] [Google Scholar]
- 31.Rivero G, Gabilondo AM, Garcia-Sevilla JA, La Harpe R, Morentin B, Javier Meana J. Characterization of regulators of G-protein signaling RGS4 and RGS10 proteins in the postmortem human brain. Neurochem Int. 2010;57(7):722–729. doi: 10.1016/j.neuint.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Tomasiewicz HC, Jacobs MM, Wilkinson MB, Wilson SP, Nestler EJ, Hurd YL. Proenkephalin mediates the enduring effects of adolescent cannabis exposure associated with adult opiate vulnerability. Biol Psychiatry. 2012;72(10):803–810. doi: 10.1016/j.biopsych.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laitinen JT, Jokinen M. Guanosine 5′-(gamma-[35S]thio)triphosphate autoradiography allows selective detection of histamine H3 receptor-dependent G protein activation in rat brain tissue sections. Journal of neurochemistry. 1998;71(2):808–816. doi: 10.1046/j.1471-4159.1998.71020808.x. [DOI] [PubMed] [Google Scholar]
- 34.Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R, et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–750. doi: 10.1093/nar/gku1026. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aarons AR, Talan A, Schiffer WK. Experimental protocols for behavioral imaging: seeing animal models of drug abuse in a new light. Curr Top Behav Neurosci. 2012;11:93–115. doi: 10.1007/7854_2012_206. [DOI] [PubMed] [Google Scholar]
- 36.Stratinaki M, Varidaki A, Mitsi V, Ghose S, Magida J, Dias C, et al. Regulator of G protein signaling is a crucial modulator of antidepressant drug action in depression and neuropathic pain models. Proc Natl Acad Sci U S A. 2013;110(20):8254–8259. doi: 10.1073/pnas.1214696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, et al. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24(6):765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 38.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, et al. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol. 2010;78(3):524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong H, Wade SM, Woolf PJ, Linderman JJ, Traynor JR, Neubig RR. A spatial focusing model for G protein signals. Regulator of G protein signaling (RGS) protien-mediated kinetic scaffolding. The Journal of biological chemistry. 2002;278(9):7278–7284. doi: 10.1074/jbc.M208819200. [DOI] [PubMed] [Google Scholar]
- 40.Lerner TN, Kreitzer AC. RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron. 2012;73(2):347–359. doi: 10.1016/j.neuron.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobo MK. Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. Int Rev Neurobiol. 2009;89:1–35. doi: 10.1016/S0074-7742(09)89001-6. [DOI] [PubMed] [Google Scholar]
- 42.Johnson PM, Kenny PJ. Dopamine D2 receptors in addiction-like reward dysfunction and compulsive eating in obese rats. Nat Neurosci. 2010;13(5):635–641. doi: 10.1038/nn.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kenny PJ, Voren G, Johnson PM. Dopamine D2 receptors and striatopallidal transmission in addiction and obesity. Curr Opin Neurobiol. 2013 doi: 10.1016/j.conb.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Val-Laillet D, Aarts E, Weber B, Ferrari M, Quaresima V, Stoeckel LE, et al. Neuroimaging and neuromodulation approaches to study eating behavior and prevent and treat eating disorders and obesity. Neuroimage Clin. 2015;8:1–31. doi: 10.1016/j.nicl.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin BE, Dunn-Meynell AA, Balkan B, Keesey RE. Selective breeding for diet-induced obesity and resistance in Sprague-Dawley rats. The American journal of physiology. 1997;273(2 Pt 2):R725–730. doi: 10.1152/ajpregu.1997.273.2.R725. [DOI] [PubMed] [Google Scholar]
- 46.Burchett SA, Bannon MJ, Granneman JG. RGS mRNA expression in rat striatum: modulation by dopamine receptors and effects of repeated amphetamine administration. Journal of neurochemistry. 1999;72(4):1529–1533. doi: 10.1046/j.1471-4159.1999.721529.x. [DOI] [PubMed] [Google Scholar]
- 47.Stanwood GD, Parlaman JP, Levitt P. Genetic or pharmacological inactivation of the dopamine D1 receptor differentially alters the expression of regulator of G-protein signalling (Rgs) transcripts. The European journal of neuroscience. 2006;24(3):806–818. doi: 10.1111/j.1460-9568.2006.04970.x. [DOI] [PubMed] [Google Scholar]
- 48.Schwendt M, Gold SJ, McGinty JF. Acute amphetamine down-regulates RGS4 mRNA and protein expression in rat forebrain: distinct roles of D1 and D2 dopamine receptors. Journal of neurochemistry. 2006;96(6):1606–1615. doi: 10.1111/j.1471-4159.2006.03669.x. [DOI] [PubMed] [Google Scholar]
- 49.Taymans JM, Kia HK, Claes R, Cruz C, Leysen J, Langlois X. Dopamine receptor-mediated regulation of RGS2 and RGS4 mRNA differentially depends on ascending dopamine projections and time. The European journal of neuroscience. 2004;19(8):2249–2260. doi: 10.1111/j.0953-816X.2004.03336.x. [DOI] [PubMed] [Google Scholar]
- 50.Geurts M, Hermans E, Maloteaux JM. Opposite modulation of regulators of G protein signalling-2 RGS2 and RGS4 expression by dopamine receptors in the rat striatum. Neuroscience letters. 2002;333(2):146–150. doi: 10.1016/s0304-3940(02)01004-2. [DOI] [PubMed] [Google Scholar]
- 51.Taymans JM, Leysen JE, Langlois X. Striatal gene expression of RGS2 and RGS4 is specifically mediated by dopamine D1 and D2 receptors: clues for RGS2 and RGS4 functions. Journal of neurochemistry. 2003;84(5):1118–1127. doi: 10.1046/j.1471-4159.2003.01610.x. [DOI] [PubMed] [Google Scholar]
- 52.Schwendt M, Sigmon SA, McGinty JF. RGS4 overexpression in the rat dorsal striatum modulates mGluR5- and amphetamine-mediated behavior and signaling. Psychopharmacology. 2012;221(4):621–635. doi: 10.1007/s00213-011-2606-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwendt M, McGinty JF. Regulator of G-protein signaling 4 interacts with metabotropic glutamate receptor subtype 5 in rat striatum: relevance to amphetamine behavioral sensitization. The Journal of pharmacology and experimental therapeutics. 2007;323(2):650–657. doi: 10.1124/jpet.107.128561. [DOI] [PubMed] [Google Scholar]
- 54.Dripps IJ, Wang Q, Neubig RR, Rice KC, Traynor JR, Jutkiewicz EM. The role of regulator of G protein signaling 4 in delta-opioid receptor-mediated behaviors. Psychopharmacology. 2016;234(1):29–39. doi: 10.1007/s00213-016-4432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park SW, Shen X, Tien LT, Roman R, Ma T. Methamphetamine-induced changes in the striatal dopamine pathway in mu-opioid receptor knockout mice. J Biomed Sci. 2011;18:83. doi: 10.1186/1423-0127-18-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen W, Plotkin JL, Francardo V, Ko WK, Xie Z, Li Q, et al. M4 Muscarinic Receptor Signaling Ameliorates Striatal Plasticity Deficits in Models of L-DOPA-Induced Dyskinesia. Neuron. 2015;88(4):762–773. doi: 10.1016/j.neuron.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding J, Guzman JN, Tkatch T, Chen S, Goldberg JA, Ebert PJ, et al. RGS4-dependent attenuation of M4 autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat Neurosci. 2006;9(6):832–842. doi: 10.1038/nn1700. [DOI] [PubMed] [Google Scholar]
- 58.Krashes MJ, Kravitz AV. Optogenetic and chemogenetic insights into the food addiction hypothesis. Front Behav Neurosci. 2014;8:57. doi: 10.3389/fnbeh.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


