Abstract
In this work, we develop a fully automatic algorithm named “MCDT” (Migrating Cell Detector and Tracker) for the integrated task of migrating cell detection, segmentation and tracking from in vivo fluorescence time-lapse microscopy imaging data. The interest of detecting and tracking migrating cells arouses from the scientific question in understanding the impact of oligodendrocyte progenitor cells (OPCs) migration in vivo, using advanced microscopy imaging techniques. Current practice of OPC mobility analysis relies on manual labeling, suffering from massive human labor, subjective biases, and weak reproducibility. Existing cell tracking methods have difficulties in analyzing such challenging data due to the extra complexity of in vivo data. Designed for in vivo data, MCDT circumvents the common strong assumption of separable feature distributions between foreground and background. Besides, by focusing on migrating cells (OPCs) only, MCDT relieves the burden of tracking all irrelevant cells correctly, not only accelerating the analysis but also achieving better accuracy in OPCs. Seed based segmentation and tracking by topology-preserved motion estimation endows MCDT with robustness to complex surroundings of the cell under tracking and to occasional inaccurate segmentation in some frames. We tested MCDT on imaging data of transgenic zebrafish larval spinal cord and MCDT showed very promising performance.
Keywords: migrating cell detection, cell tracking, in vivo, time-lapse imaging, oligodendrocyte progenitor cells
1. INTRODUCTION
In the developing central nervous system (CNS), oligodendrocytes, one of the major classes of glial cells, are uniformly and periodically distributed to ensheath axons with myelin membrane, which facilitates rapid conduction of nerve impulses [1]. The migration and differentiation of oligodendrocyte progenitor cells (OPCs) play an important role in the spatial arrangement of oligodendrocytes and their myelinating processes [2]. But the underlying mechanism remains mysterious and is an active research area. In vivo time-lapse fluorescence imaging holds great potential to resolve the puzzle [3]. Despite the advances in in vivo time-lapse fluorescence imaging techniques and plenty of scientific discoveries made using them, current practice of OPC migration analysis using such data relies on manual labeling and tracking, which suffers from not only massive human labor but also subjective biases and weak reproducibility.
As a foundation of automated OPC migration analysis, the integrated problem of OPC detection and tracking is critical but very challenging, especially for in vivo data. Firstly, compared with natural images, fluorescence microscopy images usually have problems such as low SNR, poor staining, unstable and non-rigid cell morphology, and lack of within-object texture patterns. Secondly, in vivo imaging data usually suffers from extra difficulties like global changes of field of view due to tissue motion and growth, spatially clustered cells, weak boundaries between adjacent cells, non-homogeneous intensity distribution within foreground or within background, and inseparable intensity distributions between foreground cells of interest (COI) and background tissues. More importantly, due to the lack of exclusive genetic-labelling marker of OPCs that leaves out non-OPC oligodendrocyte lineage cells, in terms of appearance in images, OPCs can only be distinguished from some non-OPC cells by motion pattern. And, unfortunately, the non-OPC cells are not static, either, but pervasively vibrate locally under the constraints of relative intercellular spatial structure in the tissues (Figure 1). These make detecting and tracking of OPCs distinct from classic cell tracking problems.
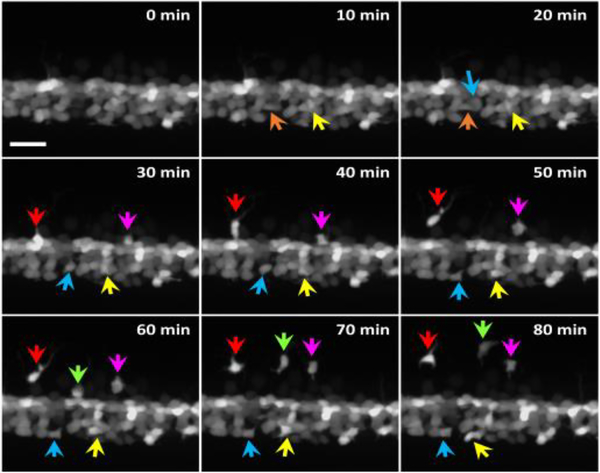
Fig. 1.
In vivo fluorescence time-lapse imaging on the zebrafish transgenic line Tg(olig2:egfp) using confocal microscopy. The transgene marked a subset of oligodendrocyte-lineage cells including OPCs and motor nerve cells. Different colored arrows indicate different OPCs. The other cells (not migrating) are not of interest. The scale bar equals 15 μm.
Though conceptually the task of detecting and tracking OPCs can be solved by tracking all cells using classic cell tracking tools and then classifying cells into OPCs and non-OPC cells based on the extracted trajectories, we argue that this is not only a waste of computational resource but also prone to more errors than focusing on tracking OPCs only. On the one hand, it is commonly seen in real data that the number of OPCs is much smaller than non-OPC cells. Whatever cell tracking method is used, by actually endowing OPCs and non-OPC cells with the same importance, errors in segmentation and tracking of non-OPC cells can cause unnecessary errors in the tracking of OPCs around such non-OPC cells. On the other hand, as a consequence of the challenges discussed above, such as weak boundaries between adjacent cells and non-homogeneous intracellular intensity, accurate segmentation of a large cluster of mutually contacted cells in in vivo data is much more challenging than segmentation tasks in in vitro data. In spite of the fact that many methods have been developed for cell tracking in the last two decades, most of them were focusing on in vitro data and only a small part of them made efforts for more complex in vivo data [4–5].
In this work, we propose a novel, fully automatic algorithm named “MCDT” (Migrating Cell Detector and Tracker) for the integrated task of OPC (migrating cell) detection, cell segmentation and cell tracking from 2D fluorescence time-lapse imaging data. Principally MCDT can also be applied to similar types of data in which cells have basically homogeneous or smooth intracellular texture. The flowchart is shown in Figure 2. Focusing on migrating cells only, by motion detection we first find “OPC pieces of motion” (known to be within OPCs, and used as initial seeds for segmentation of the complete OPC cell, also referred to as “motion seeds”), then sequentially detect OPC boundaries at one frame starting from the seeds and search for new seeds in the next frame. Given a seed within one OPC at some time point, we design a seed-expansion-based cell segmentation algorithm specifically for complex in vivo fluorescence imaging data, modelling the segmentation problem as a shortest path search problem in graph, which can be solved very efficiently. Finally, after all OPC candidates and their traces are obtained one by one, we design a graphical trace reorganization module to refine the results, utilizing the spatiotemporal relationships among OPC candidates. Our algorithm is tested on both synthetic data and real data of transgenic zebrafish. The experimental results show that MCDT has very promising performance and superiority for OPC problems compared to several popular generic cell tracking algorithms.
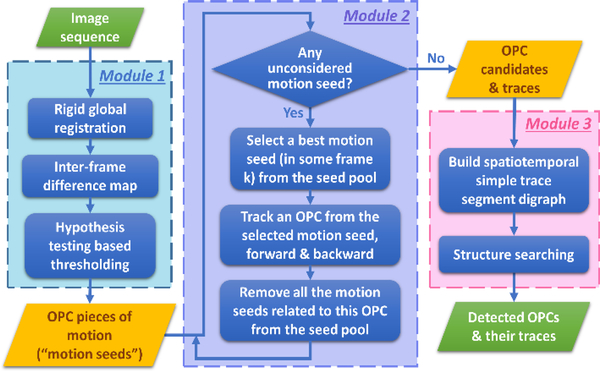
Fig. 2.
Flowchart of MCDT. Three major modules are: 1.) identification of motion seeds; 2.) sequentially tracking OPCs based on motion seeds; 3.) graphical spatiotemporal trace reorganization. Green blocks are overall input and output; blue blocks are procedures; and yellow blocks are critical intermediate products.
2. METHODS
2.1. Overview of the Algorithm
MCDT is designed following several principles. Firstly, we focus on modeling and analyzing migrating cells (OPCs) only. We do not track irrelevant cells. Secondly, considering the property of in vivo fluorescence microscopy imaging data, cell segmentation should not rely on separable distributions of feature between foreground (OPCs) and background. Thirdly, as the basis of tracking between subsequent frames, motion estimation should preserve topological structure among pixels within a cell, but allow totally free spatial relationship changes between cells. Last but not least, OPCs should not be considered independently but, instead, should be considered systematically utilizing spatiotemporal relationships among them.
The algorithm framework can be summarized as three modules as shown in Figure 2: 1.) identification of motion seeds; 2.) sequentially tracking OPCs seeded from moving OPC pieces; 3.) graphical spatiotemporal trace reorganization.
2.2. Identification of OPC motion seeds
The most critical and perhaps the only difference between OPCs and non-OPC cells lies in their motion patterns: in spite of local vibrations, non-OPC cells remain at a local position relative to other non-OPC cells and background tissue structures; while OPCs migrate away from their initial locations. Specifically, we define OPCs as cells that migrate more than an average diameter of cells. Here we first detect some potential OPC pieces that are expected to be contained in OPCs when they are moving rapidly. In this work, they are referred to as “OPC motion seeds” and will be utilized as initiation for follow-up OPC tracking process.
Firstly we find regions where objects (cells) are moving. One move of a cell, in the perspective of images, means that the cell (or a part of it) is no longer where it was in a proceeding frame. Thus we can detect the original position of the cell by finding regions with obvious intensity change between frames. To distinguish pixels with statistically significant intensity change due to motion from pixels with pervasive intensity change due to noises or due to global illumination change, we do hypothesis test based thresholding to the intensity difference map between two frames. The null distribution of intensity change is learned from the frames under discussion, by fitting a gamma distribution to the empirical distribution of intensity change. Secondly, from all the detected regions of motion, we select the regions of OPCs (“OPC motion seeds”) by determining whether the regions are comparable in size to a complete cell. In addition, to avoid confounding motions due to global deformation of the tissue, before all these operations we register the tissue in all frames as preprocessing. Although the tissue deformation is essentially non-rigid, too flexible registration will falsely compensate true cell motions. So a rigid global registration is applied as an approximate.
2.3. Sequential tracking of OPCs from motion seeds
Given a pool of OPC motion seeds, we select one with the largest area as the initial seed for tracking. The seed is within a certain OPC in some frame, and large area suggests the OPC moves fast in that frame. Then we start to track the corresponding OPC temporally forward and backward, separately. Since multiple OPC motion seeds can be within the same OPC but at different time points, after the tracking of one OPC is done, all OPC motion seeds covered by this OPC are removed from the pool. Then a new initial seed will be selected from the remaining pool of OPC pieces of motion, from which the tracking procedure for a new OPC will start. This sequential process ensures that all detected OPC pieces of motion are considered and covered by tracked OPC candidates. Even if errors occur and the algorithm loses track of a certain OPC before it disappears or the video ends, the lost parts are likely to be found back starting from another OPC motion seeds. This endows our algorithm with robustness and strong detection power.
Beginning from a selected OPC motion seed, we track a single OPC using a “segmentation – motion estimation – segmentation” iterative strategy. Given a seed in frame k, denoted as Ik, the OPC’s boundary in Ik is detected using our single cell segmentation method (see Section 2.3.1). Once the cell boundary in Ik is obtained, we estimate the motion of the OPC from Ik to Ik+1. With all pixels within a cell assumed to move together, an extreme case of preserved topological structure among intracellular pixels, the motion estimation can be seen as a template matching problem. The detected OPC region in Ik is used as the template, and the matching score is a function of center position of the matched region in Ik+1. The optimal center position in Ik+1 is expected to be with the local maximum of matching score that is spatially the closest to original position in Ik. Then a small disk of radius 1 around the estimated center position in Ik+1 is considered as the new seed.
2.3.1. Seed based cell segmentation
Now that a seed region within an OPC in some frame is known but the whole region of the cell is unknown, we segment this cell of interest (COI) containing the seed from its complex neighborhood. COI and its adjacent cells often have very similar appearance (e.g. intensity, gradient, morphology), lacking of differentially distributed features between objects and background (Figure 1). Instead, it is mainly the relatively sharp intensity contrast on cell boundaries that indicates the separation of adjacent cells. Besides, due to the discussed limitations of image quality, it is often the case that parts of the cell boundaries are too weak to tell by using only the intensity contrast, but they can be inferred from other visible parts of the contours using spatial continuity and directions of linelike structures. Additionally, if we have two candidate COI contours with exactly the same average intensity contrast, we prefer the one that is closer to the seed center, because intensity changes smoothly within cells and high-contrast pixels either locate on the COI’s contour or on the contours of other cells (outside the COI).
Thus we search for a closed curve surrounding the seed that is most likely to be COI’s contour. To do this, from the image window containing the COI we build a directed graph D where each non-seed pixel corresponds to a vertex. Two vertices are connected in both directions if they are 8-connecting neighbor pixels in the image. Therefore a closed curve in the image is mapped to a cycle in the digraph D. We design an arc weight score w(e) such that the more likely an arc e ∈ E(D) is on some cell boundaries, the smaller the weight w(e) is. In this way, the problem of finding the best closed curve can be transformed into a problem of finding shortest cycle in the weighted digraph. In order to meet the constraint of “surrounding the seed”, we utilize the fact that if the center of the seed is regarded as the origin of a relative coordinate system, any closed curve surrounding the seed must go across the positive horizontal axis. Specifically, we split the vertex of any positive-horizontal-axis pixel into two vertices: a “tail” vertex that only connects to pixels in the first quadrant, and a “head” vertex that only connects to pixels in the fourth quadrant. Therefore the best closed curve that goes across a positive-horizontal-axis pixel is just the shortest path in the digraph from the pixel’s “tail” vertex to its “head”. This can be solved in (|E| + |V| log|V|) time using Dijkstra’s algorithm [6–7]. After comparing the best curves respectively found starting at all positive-horizontal-axis pixels, we finally get the overall best solution.
The arc weight score w(e) is defined as:
| (1) |
where λ is a parameter balancing the appearance in image data and inference from neighborhood. We set λ = 10 in all our experiments. s1(e) is the average intensity contrast score of the two end vertices of the edge e, enhanced by applying scale-auto-selected line filter [8] to the gradient map of original image. s2(e) = 0.5*(1 + cos θ) evaluates how consistent the following two directions are: 1) the average direction of line structure around the two end pixels (got from the mentioned line filters) and 2) the arc e’s physical direction (i.e. point from the arc tail vertex’s pixel to the arc head vertex’s pixel in the image). θ is the angle between these two directions. Using similar idea, s3(e) = 0.5*(1 + cos ψ) assesses whether the gradient vector at e is pointing to the seed center, and ψ is the angle between gradient vector and the connecting vector from e (position on the image) to the seed center. indicates how unlikely (e), the average intensity on e, is from the empirical distribution of the seed area.
2.4. Graphical spatiotemporal trace reorganization
The OPC candidates generated from the first two modules may have errors due to under-segmentation, over-segmentation, intrinsic ambiguity between adjacent cells, and inaccuracy in motion estimation, etc. As a consequence, two OPC candidates may totally or partially overlap in space at some time point, where we call them “spatiotemporally overlapping” with each other. Perfect OPC tracking results should not contain spatiotemporal overlapping OPCs. It is also possible that one OPC is captured by different OPC candidates in different time periods, which is not desirable, either. In this module, we utilize spatiotemporal relationships among the detected OPC candidates to infer true OPC traces, resolving the errors by operations such as merging and splitting.
In our refinement module, the basic unit of consideration and operation is a “simple trace segment”, that is, a consecutive part (in time) of an OPC candidate trace during which no ambiguity occurs. Ambiguity arises when multiple OPC candidate traces converge into one trace (by overlapping in some frame), or a trace diverges into several. Our goal therefore is to perform a set of merging/splitting operations on simple trace segments such that eventually each OPC trace is exactly one isolated simple trace segment without any ambiguity. Besides, selecting among all possible sets of operations towards this end, we prefer simpler operation set, namely, smaller number of operations on shorter trace segments. This is because the more modifications to the originally detected OPC candidates, the higher risk we introduce artificial errors.
We model this problem as structure searching in a “spatiotemporal simple trace segment digraph” Dtrc. We set up a vertex in Dtrc for each simple trace segment. Two vertices are linked in temporal order if they contain a common OPC candidate in two consecutive frames. The “operation cost” of any vertex is the time duration of the simple trace segment. The structure searching is done using a greedy iterative algorithm. In each round, the smallest-cost vertex in current Dtrc is selected, then an operation is chosen from the operation pool according to this vertex’s condition. For example, if this vertex is a hub of multiple precursors and multiple successors, we spatially split it into several pieces and assign the pieces to different precursors and successors by considering spatial closeness. Or if this vertex has exactly the same precursors and successors as another vertex, we merge them, taking union of their spatial regions and filling out the region in between them. In this process, the structure of Dtrc is iteratively updated until converged.
3. RESULTS
3.1. Data
We performed time-lapse imaging on the transgenic zebrafish line Tg(olig2:egfp) using spinning disk confocal microscopy at 40x magnification with step size of 1μm. The spatial resolution is 0.315μm*0.315μm. A maximum projection is done to get 2D+t videos. The transgene expresses enhanced green fluorescent protein (EGFP), marking a subset of oligodendrocyte lineage cells including OPCs and motor nerve cells. The zebrafish larvae were imaged from 48 hours post-fertilization (hpf) to examine spinal cord OPC behaviors. Most of the marked cells that appear bright in the images are motor nerve cells which pile up together in the ventral spinal cord, while OPCs are the only motile cells (Figure 1). The dataset includes 25 videos of 100 frames each, with spatial resolution of 1024×1024 pixels. Different image sequences may have different temporal resolution, with time intervals 1, 2, 5, or 10 minutes.
3.2. Overall performance
Considering the complexity of real in vivo imaging data, synthetic data can hardly capture the challenges in real data. Thus we evaluate the performance of MCDT by manually labeling and tracking OPCs and then comparing the results of MCDT with manual labels (typically 3~30 OPCs in one image sequence sample).
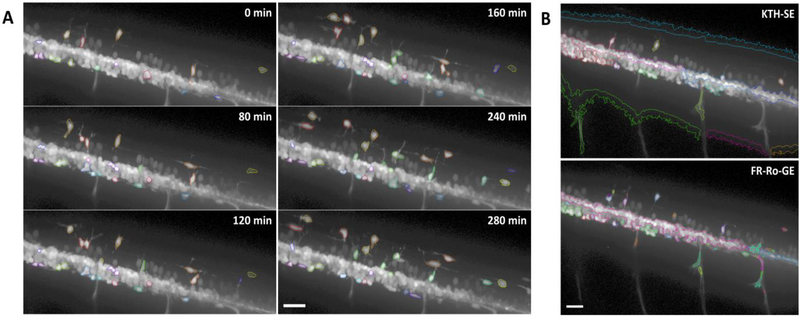
Figure 3A shows an example of MCDT’s results. Most OPCs except for two are successfully detected and correctly tracked. Particularly, OPCs surrounded by motor nerve cells are also detected and distinguished from vibrating neighbor cells. Seed-based segmentation shows its power to extract COIs from adjacent clustered cells. The results also show MCDT’s strength in robustness to occasional errors in segmentation. In some frames, the cell boundary detection is extremely difficult for some OPCs, since they are too weak to tell even for human experts. MCDT also reported inaccurate contours there, but the tracking process was still going well and cell contours were detected correctly in following frames. This robustness is owed to the tolerance of seed updating process to the variation of contours. No matter how the contour looks like in the last frame, as long as it roughly encloses the cell, MCDT’s motion estimation module will find a new seed locating within the cell in current frame, and seed-based segmentation will get a new contour that is not contaminated from the last one.
Fig. 3.
Exemplary results of MCDT and comparison with existing methods. (A) Detection and tracking results of MCDT on one image sequence. Each detected OPC is circled using one color, consistently over time. The scale bar equals 25 μm. (B) Results on an exemplary frame, generated by two existing methods, KTH-SE and FR-Ro-GE, respectively. The scale bar equals 25 μm.
Quantitative performance evaluation was done using metrics in three aspects: 1) OPC detection; 2) segmentation; and 3) tracking. In terms of OPC detection, or OPC/non-OPC labeling, MCDT achieved average recall 92.73%, and true negative rate 94.85%. Segmentation performance was evaluated using Jaccard similarity index (SEG) used in the ISBI Cell Tracking Challenge [5]. MCDT’s average SEG score is 89.47%, and most segmentation errors lie in the long external processes. Somas were mostly accurately segmented. For tracking performance we consider the tracked frame rate of each OPC, defined as the ratio of number of frames where an OPC is correctly hit by MCDT (regardless of boundary accuracy) to the total number of frames that this OPC exist in the video. In the experiments, MCDT got an average tracked frame rate of 93.19%.
3.3. MCDT solves difficulties of generic cell tracking algorithms
To check if our concerns are founded about the difficulties that generic cell tracking algorithms will encounter in OPC detection and tracking problem, we applied two published cell tracking algorithms to our data. KTH-SE [9] and FR-Ro-GE [10] are two among the best methods in the past and ongoing ISBI Cell Tracking Challenges [5]. We use the software provided by the authors. Parameters were tuned to the best we can get, starting from the default settings.
Figure 3B shows the results generated by the two peer methods, respectively. Both of them successfully detected and tracked several OPCs that are isolated from others. However, they failed in separating clustered cells, including OPCs around the motor nerve cell cluster. They also missed a lot of dim cells. Additionally, both KTH-SE and FR-Ro-GE falsely labeled some background regions as cells. One key reason of such errors is the violation of an assumption: the intensity distribution of foreground should be separable from that of background, which is one of the most basic assumptions of KTH-SE and FR-Ro-GE, as well as many other cell tracking algorithms designed for in vitro data. Indeed, separating contacting, adjacent cells of similar appearance is much harder than segmenting cells from darker background (typical in in vitro data). On the contrary, as we can see in Figure 3A, MCDT successfully separating many OPCs from their adjacent cells and had the power to detect and segment OPCs with dim intensity.
4. CONCLUSIONS
We have developed an automatic algorithm, MCDT, for the integrated task of migrating cell detection, segmentation and tracking from in vivo microscopy image series. In experiments MCDT shows very promising performance in both detection and tracking. Though designed for in vivo data, in principle MCDT can also be applied to in vitro fluorescence microscopy data. In addition, the framework of MCDT allows for alternative implementations of each block, with the key principles and strengths unchanged.
ACKNOWLEDGEMENTS
Research reported in this publication was supported by NIH R01MH110504 (G.Y.).
REFERENCES
- [1].Hines JH, et al. , “Neuronal activity biases axon selection for myelination in vivo.” Nature neuroscience, vol. 18, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kirby BB, et al. , “In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development.” Nature neuroscience, vol. 9, no. 12, pp. 1506, 2006. [DOI] [PubMed] [Google Scholar]
- [3].Czopka T, “Insights into mechanisms of central nervous system myelination using zebrafish.” Glia, vol. 64, no. 3, pp. 333–349, 2016. [DOI] [PubMed] [Google Scholar]
- [4].Meijering E, et al. , “Tracking in cell and developmental biology.” In Seminars in cell & developmental biology, vol. 20, no. 8, pp. 894–902. Academic Press, 2009. [DOI] [PubMed] [Google Scholar]
- [5].Maška M, et al. , “A benchmark for comparison of cell tracking algorithms.” Bioinformatics, vol. 30, no. 11, pp. 1609–1617, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dijkstra EW, “A note on two problems in connexion with graphs.” Numerische mathematik, vol. 1, no. 1, pp. 269–271, 1959. [Google Scholar]
- [7].Fredman ML, and Tarjan RE “Fibonacci heaps and their uses in improved network optimization algorithms.” Journal of the ACM (JACM), vol. 34, no. 3, pp. 596–615, 1987. [Google Scholar]
- [8].Sato Y, et al. , “Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images.” Medical image analysis, vol. 2, no. 2, pp. 143–168, 1998. [DOI] [PubMed] [Google Scholar]
- [9].Magnusson KE, and Jaldén J, “A batch algorithm using iterative application of the Viterbi algorithm to track cells and construct cell lineages” in ISBI, pp. 382–385. IEEE, 2012. [Google Scholar]
- [10].Bensch R, and Ronneberger O “Cell segmentation and tracking in phase contrast images using graph cut with asymmetric boundary costs” in ISBI, pp. 1220–1223. IEEE, 2015. [Google Scholar]