From the pioneering discovery of the first single-gene mutants that controlled circadian behavior by Konopka and Benzer (1), the quest to probe the fundamentally important relationship between gene expression and behavior has been ongoing. The ability to control cellular function and behavior with exquisite precision in vivo through genetic, optogenetic, and pharmacological manipulation already exists, but the search is still afoot for approaches to rapidly and reversibly control gene expression. Genetic code expansion (GCE) has proved to be a powerful tool to control protein function in vitro and inside the cellular milieu (2) and, more recently, in a wide array of vertebrates in vivo (3). In PNAS, Maywood et al. (4) use GCE to create a fast, conditional, and reversible translational switch to reconstitute the molecular circadian clock in the suprachiasmatic nucleus (SCN) and circadian behavior in arrhythmic mice. The rapid kinetics with which protein expression is controlled with this translational switch provides unprecedented temporal control over neuronal function in vivo, restoring complex behavioral responses reversibly on the timescale of about a day. This pioneering approach should be broadly applicable to many other systems, from neurobiology to the periphery, opening the door to a new era of genetic control and biological insight.
In mammals, the SCN acts as the master circadian pacemaker that provides temporal control over nearly every biological process, from the secretion of melatonin to body temperature (5). From its position in the hypothalamus, the SCN receives retinal input that is used to entrain clocks to the environmental light–dark cycle. It also provides molecular cues to synchronize the rhythms of molecular circadian clocks in peripheral tissues to globally coordinate physiological and behavioral rhythms. Therefore, manipulating circadian clocks in the SCN has powerful control over behavioral responses, making it a particularly tractable and interesting initial target for the reversible control of behavior upon reconstitution of a single gene. At the molecular level, circadian rhythms are driven by a set of interlocked transcriptional–translational negative feedback loops. The core loop is established by the transcription factor CLOCK:BMAL1, which activates transcription of Period and Cryptochrome (Cry) genes that feed back to repress their expression and complete the feedback loop (6). At least one cryptochrome must be present to interact directly with CLOCK:BMAL1 (7, 8) and coordinate the assembly of large, multimeric repressive complexes (9) that close the feedback loop.
Traditional approaches via single-gene knockouts have demonstrated that Cry1−/− mice have a short period, while Cry2−/− mice have a long period (10, 11), consistent with the observation that CRY1 is a “stronger” repressor that binds to CLOCK:BMAL1 more tightly (12). Prior work from Hastings and colleagues (13) demonstrated that reconstitution of an arrhythmic SCN clock in a Cry-deficient mouse via adeno-associated virus delivery of a Cry1::EGFP fusion expressed under its own minimal promoter (pCry1) could reconstitute the clock with the expected long period (14). In PNAS, Maywood et al. expand upon this approach by using GCE to reversibly and rapidly control Cry1::EGFP in neurons to advance our understanding of the minimal determinants needed to acutely establish robust circadian rhythms.
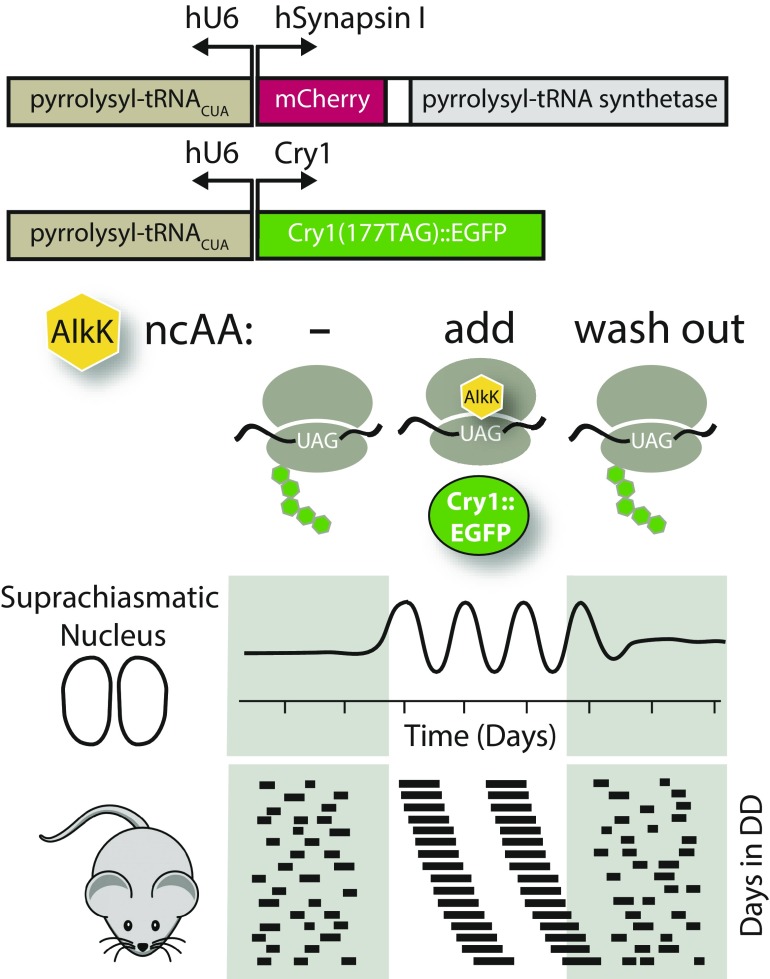
GCE utilizes an engineered aminoacyl-tRNA synthetase and tRNACUA pair that allows the incorporation of a noncanonical amino acid (ncAA) into a protein of interest in response to an amber stop codon (i.e., UAG). Maywood et al. used a pyrrolysyl-tRNA synthetase/pyrrolysyl-tRNACUA pair that templated the insertion of the ncAA alkyne lysine N6-[(2-propynyloxy)carbonyl]-l-lysine (AlkK). Providing AlkK in the culture media for SCN slices ex vivo, or in the drinking water for mice, allowed for the rapid and reversible translation of Cry1::EGFP. Remarkably, this led to reconstitution of the molecular circadian clock and its resultant behavioral control on a timescale of about a day (Fig. 1). Carefully removing AlkK through a series of washouts once again eliminated Cry1::EGFP expression and the loss of molecular and behavioral circadian rhythms.
Fig. 1.
Reversible control of the circadian clock using translational switching of the core clock gene, Cry1. Using the hSynapsin promoter (Synapsin I), expression of Cry1::EGFP was restricted to neurons that expressed the required orthogonal tRNA synthetase (marked by coexpressed mCherry protein) and the tRNACUA under the hU6 promoter. Cry1::Egfp mRNA was controlled by its own minimal promoter (pCry1), which directs its rhythmic transcription. Translational of Cry1::EGFP was strictly conditional on the presence of AlkK, an ncAA that is inserted into the protein upon readout of an amber stop codon (i.e., UAG). Circadian rhythms were monitored before and after AlkK administration with Per2::Luc bioluminescence in SCN explants or behavioral analysis in mice.
One powerful feature of this approach is that increasing the concentration of AlkK allowed for dose-dependent control over the expression of Cry1::EGFP and circadian period, which could be valuable for future studies of analysis of network perturbation by titrating clock protein stoichiometries. Additionally, use of ncAAs like AlkK opens the chemical biology toolkit by allowing for bioorthogonal labeling approaches via click chemistry (15). Here, Maywood et al. use AlkK to label nascent Cry1::EGFP protein with an azido fluorophore for imaging purposes. The ability to rapidly label a specific cellular population with superbright fluorophores, cross-linkers, and other chemical moieties could open the door to new studies of rare and/or transient endogenous protein complexes in situ.
GCE offers several improvements over conventional methods to manipulate protein expression. Current strategies generally target gene transcription, making use of well-studied constitutive or inducible promoters to control expression of the protein of interest. However, in many cases, changes in gene expression are permanent, such as with the use of Cre-loxP and Flp-FRT systems. Other systems, like those based on tetracycline (Tet)-controlled transcriptional control (Tet-on/off), can reversibly regulate protein expression upon addition of doxycycline. However, leaky control of gene expression and the use of orthogonal constitutive promoters can disrupt circadian rhythms, because many clock genes need to be expressed with temporal precision that is encoded by their feedback loops (14, 16). This study therefore establishes an elegant tool that should be applicable to other systems in which transcription can remain under tight regulation, further expanding the current genetic toolkit for studying and controlling dynamic processes.
Through the use of the neuron-specific hSynapsin promoter, this work demonstrates that, despite arrhythmic behavior in the absence of Cry expression, the circadian network is apparently primed and ready to run upon reconstitution with the CRY protein. Given the complex network compensation that occurs upon permanent gene knockouts in the circadian network (17), the reversibility of this approach could be useful in parsing out compensation by paralogs and bringing a deeper understanding of this complicated and dynamic process. Furthermore, recent work has highlighted an exciting role for astrocyte–neuron communication in establishing circadian networks in the SCN (18, 19). Using cell type-specific promoters, GCE now opens the door to further studying the roles of intercellular communication in vivo.
In the future, it will be interesting to see whether GCE strategies could be employed to reversibly manipulate posttranslational modifications of clock proteins. Most core components of the clock are regulated by posttranslational modifications at different points throughout the circadian cycle (20). Recent advances in the use of GCE approaches to encode site-specific phosphorylation in mammalian cells (21) tease the possibility of probing clock function at high temporal precision, thereby eliminating potential artifacts from expressing genes with constitutive mimetic substitutions. The future looks bright: As our toolkit for the genetic code expands, so will our insight into biology.
Acknowledgments
Our research is supported by Grants R01 GM107069 and R01 GM121507 from the National Institute of General Medical Sciences of the National Institutes of Health, as well as by Grant IOS 1656647 from the National Science Foundation.
Footnotes
The authors declare no conflict of interest.
See companion article on page E12388.
References
- 1.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 3.Brown W, Liu J, Deiters A. Genetic code expansion in animals. ACS Chem Biol. 2018;13:2375–2386. doi: 10.1021/acschembio.8b00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maywood ES, et al. Translational switching of Cry1 protein expression confers reversible control of circadian behavior in arrhythmic Cry-deficient mice. Proc Natl Acad Sci USA. 2018;115:E12388–E12397. doi: 10.1073/pnas.1811438115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: Interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb Perspect Biol. 2017;9:a027706. doi: 10.1101/cshperspect.a027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael AK, et al. Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc Natl Acad Sci USA. 2017;114:1560–1565. doi: 10.1073/pnas.1615310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, et al. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol. 2015;22:476–484. doi: 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aryal RP, et al. Macromolecular assemblies of the mammalian circadian clock. Mol Cell. 2017;67:770–782.e6. doi: 10.1016/j.molcel.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 11.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosensweig C, et al. An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat Commun. 2018;9:1138. doi: 10.1038/s41467-018-03503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards MD, Brancaccio M, Chesham JE, Maywood ES, Hastings MH. Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling. Proc Natl Acad Sci USA. 2016;113:2732–2737. doi: 10.1073/pnas.1519044113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Lang K, Chin JW. Bioorthogonal reactions for labeling proteins. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baggs JE, et al. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH. Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron. 2017;93:1420–1435.e5. doi: 10.1016/j.neuron.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tso CF, et al. Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr Biol. 2017;27:1055–1061. doi: 10.1016/j.cub.2017.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano A, Fu YH, Ptáček LJ. The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol. 2016;23:1053–1060. doi: 10.1038/nsmb.3326. [DOI] [PubMed] [Google Scholar]
- 21.Beránek V, et al. Genetically encoded protein phosphorylation in mammalian cells. Cell Chem Biol. 2018;25:1067–1074.e5. doi: 10.1016/j.chembiol.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]