Significance
This report provides striking evidence that expression of a chimeric RNA mimicking a fusion RNA can drive the formation of gene fusions in mammalian cells. However, it is the antisense rather than sense chimeric RNAs that effectively drive gene fusion. The discovery that the cellular AZI1 RNA, not AZI1 protein, can act as an “initiator” RNA to induce TMPRSS2–ERG gene fusion indicates that this mechanism may have important biological relevance to oncogenesis. RNA-mediated gene fusion, a mechanism that relies on sequence-specific interactions, can account for the “specificity” of genes that were selected to undergo gene fusion. The results could also have fundamental implications in mammalian genome stability, as well as gene-editing technology via mechanisms native to mammalian cells.
Keywords: gene fusion, chimeric RNA, prostate cancer, R-loop, noncoding RNA
Abstract
One of the hallmarks of cancer is the formation of oncogenic fusion genes as a result of chromosomal translocations. Fusion genes are presumed to form before fusion RNA expression. However, studies have reported the presence of fusion RNAs in individuals who were negative for chromosomal translocations. These observations give rise to “the cart before the horse” hypothesis, in which the genesis of a fusion RNA precedes the fusion gene. The fusion RNA then guides the genomic rearrangements that ultimately result in a gene fusion. However, RNA-mediated genomic rearrangements in mammalian cells have never been demonstrated. Here we provide evidence that expression of a chimeric RNA drives formation of a specified gene fusion via genomic rearrangement in mammalian cells. The process is: (i) specified by the sequence of chimeric RNA involved, (ii) facilitated by physiological hormone levels, (iii) permissible regardless of intrachromosomal (TMPRSS2–ERG) or interchromosomal (TMPRSS2–ETV1) fusion, and (iv) can occur in normal cells before malignant transformation. We demonstrate that, contrary to “the cart before the horse” model, it is the antisense rather than sense chimeric RNAs that effectively drive gene fusion, and that this disparity can be explained by transcriptional conflict. Furthermore, we identified an endogenous RNA AZI1 that functions as the “initiator” RNA to induce TMPRSS2–ERG fusion. RNA-driven gene fusion demonstrated in this report provides important insight in early disease mechanisms, and could have fundamental implications in the biology of mammalian genome stability, as well as gene-editing technology via mechanisms native to mammalian cells.
Fusion genes are among the most cancer-specific molecular signatures known to date. They are important for understanding cancer mechanisms and developing useful clinical biomarkers and anticancer therapies (1). Fusion gene formation as a result of chromosomal translocations is presumed to occur before fusion RNA expression. However, several studies have reported the presence of fusion transcripts in individuals without detectable fusion genes at the genomic DNA level (2, 3). For example, the AML1–ETO fusion transcript, associated with a subtype of acute myeloid leukemia, was present in patients who were negative for chromosomal translocations (2). Other fusion RNAs—such as BCR–ABL, MLL–AF4, TEL–AML1, PML–RARα, and NPM–ALK—were reported in healthy individuals (3). Although the discrepancy between the presence of fusion transcripts and the absence of fusion genes could result from detection limitations of the methodologies employed, fusion transcripts in normal cells could also arise from RNA transsplicing in the absence of chromosomal translocations (4). Indeed, JAZF1–JJAZ1 fusion transcripts are expressed in normal human endometrial tissue and an endometrial cell line in the absence of chromosomal translocation (5). Furthermore, transsplicing between JAZF1 and JJAZ1 was demonstrated to occur in vitro using cellular extracts, resulting in a fusion RNA similar to that transcribed from the JAZF1–JJAZ1 fusion gene in endometrial stromal sarcomas (5). These observations raise the possibility that cellular fusion RNAs created by transsplicing act as guide RNAs to mediate genomic rearrangements. A precedent for RNA-mediated genomic rearrangements is found in lower organisms, such as ciliates (6, 7). Rowley and Blumenthal (8) coined this as “the cart before the horse” hypothesis, in that “RNA before DNA” defies the normal order of the central dogma of biology: DNA → RNA → protein (9). Despite important implications in biology and human cancer, RNA-mediated genomic rearrangement in mammalian cells has not been directly demonstrated. In this report, we provide evidence that expression of a specific chimeric RNA can lead to specified gene fusion in mammalian cells.
Results
To test whether the expression of a fusion RNA in mammalian cells can lead to a specific gene fusion, the TMPRSS2–ERG fusion (10, 11), identified in ∼50% of prostate cancers, was selected as a model. Both the TMPRSS2 and ERG genes are located on chromosome 21 separated only by 3 Mb, an intrachromosomal configuration prone to rearrangements. To recapitulate TMPRSS2–ERG fusion gene formation, we used the LNCaP prostate cancer cell line that lacks the TMPRSS2–ERG fusion (11, 12). Furthermore, treating LNCaP cells with androgen induces the chromosomal proximity between the TMPRSS2 and ERG genes (13–15), which was thought to enhance the possibility of gene fusion. To test “the cart before the horse” hypothesis (4, 8), we transiently expressed a short fusion RNA consisting of two exons, TMPRSS2 exon-1 joined to ERG exon-4, which is a short fragment of a full-length TMPRSS2–ERG fusion RNA that is most common in prostate cancer (Fig. 1A, Upper) (11). This short fusion RNA mirrors the presumptive transspliced fusion RNA product that is generated only in the sense orientation because the correct splice sites are absent in the antisense orientation. However, because the “antisense” sequence should, in theory, contain the same template information for guiding genomic rearrangements, we tested both the sense and antisense short fusion RNA. Each was individually expressed using either a CMV or a U6 promoter (Fig. 1A, Upper) and designated as “input RNA” to distinguish them from the “endogenous” full-length fusion RNA transcribed from the genome.
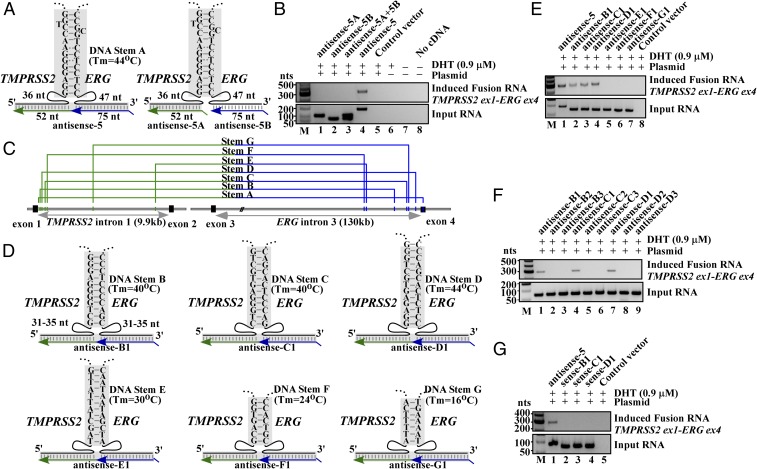
Fig. 1.
Exogenously expressed input chimeric RNAs induce the expression of endogenous fusion transcripts. (A, Upper) Schematics of the designed input RNAs containing complete TMPRSS2 exon-1 (78 nt, uc002yzj.3) and ERG exon-4 (218 nt, uc021wjd.1), expressed in the sense or antisense orientation from the CMV or U6 promoters. pA, poly-A signal; ts, transcriptional stop “TTTTTT” for U6 promoter. (Lower) RT-PCR detection of induced fusion transcript (Upper gel) and input RNA (Lower gel). Antisense short fusion RNAs (lanes 3 and 4), but not sense short fusion RNAs (lanes 1 and 2), induced a band of fusion transcript. For negative controls, transfection with a parental plasmid expressing mCherry sequence (lane 5), DHT treatment without plasmid transfection (lane 6), cells without transfection and DHT treatment (lane 7), and PCR without cDNA (lane 8) all resulted in the absence of endogenous fusion transcripts. M, DNA markers. (B) Length and positional effect of antisense input RNAs. (Upper) Antisense input RNAs with 75 nt (blue) targeting ERG exon-4 and varying lengths (82, 67, 52, and 33 nt, green) targeting TMPRSS2. Dashed line links ERG and TMPRSS2 sequence in the input RNA and contains no sequence. A 20-nt mutation (black line) was introduced to the input RNAs to discern input RNAs from the induced fusion transcript (SI Appendix, Fig. S1B). (Lower) RT-PCR detection of induced fusion transcript (Top), input RNA (Middle), or detection of induced fusion transcript using a different primer pair (Bottom). (C) Expression of the corresponding sense input RNAs (Lower) all failed to induce the fusion transcript (Upper, lanes 1–4). (D) Induction by antisense-5 occurred at physiologically relevant DHT concentrations as low as 20 nM. Three-rounds of nested PCR were performed to reveal the lowest amount of DHT required. (E) Antisense-5 led to clear induction while antisense-3 and -7 did not, indicating that it is not the length of input RNAs but targeted regions that is critical.
We transiently transfected LNCaP cells with either plasmid and treated the cells with dihydrotestosterone (DHT, a metabolite of testosterone) for 3 d. If the expression of an input RNA leads to a TMPRSS2–ERG gene fusion, it is expected that the endogenous full-length fusion RNAs would be transcribed from the newly induced fusion gene. Specific RT-PCR assays were designed to distinguish between endogenous full-length fusion RNAs and the input RNAs exogenously expressed from the plasmids (see SI Appendix, Fig. S1A for primer designs). As shown in Fig. 1A, expression of the sense short fusion RNA resembling the transspliced product, either by the CMV or U6 promoter (Fig. 1A, Lower, lanes 1 and 2, respectively), led to no detection of an induced endogenous fusion transcript. Expression of a longer version of sense fusion RNA consisting of four exons (TMPRSS2 exon-1 joined to ERG exon-4/5/6) also failed to induce an endogenous fusion transcript (SI Appendix, Fig. S2). In contrast, expression of antisense short fusion RNAs induced a band of 721 bp (Fig. 1A, Lower, lanes 3 and 4). Sanger sequencing revealed that the induced band contains TMPRSS2 exon-1 fused to ERG exons-4/5/6/7 (SI Appendix, Fig. S3), and that the exons are joined by annotated splice sites, which would be expected of mature endogenous fusion mRNA derived from the TMPRSS2–ERG fusion gene. Because the precise annotated splice junctions strongly indicate that the fusion transcripts are generated and processed through cellular mechanisms, it rules out the possibility that the observed fusion transcripts are the results of RT-PCR artifacts produced by template switching. Furthermore, these induced fusion transcripts cannot possibly arise from the sequence of input RNAs as the expression plasmids contain only TMPRSS2 exon-1 and ERG exon-4 without the ERG exon-5/6/7 sequence. Notably, the induction was more pronounced when the antisense input RNA was driven by the U6 promoter (Fig. 1A, Lower, antisense-2, lane 4) compared with the CMV promoter (Fig. 1A, Lower, antisense-1, lane 3), presumably because exogenous input RNAs transcribed by U6 accumulate in the nucleus. These differences (antisense vs. sense, U6 vs. CMV) are not caused by differing amounts of input RNA because all input RNAs were expressed at relatively equal levels (Fig. 1A, Lower). Transfection with a parental plasmid containing mCherry sequence (Fig. 1A, Lower, lane 5), DHT treatment without plasmid transfection (Fig. 1A, Lower, lane 6), and PCR without cDNA served as RT-PCR controls (Fig. 1A, Lower, lane 8), all resulted in the absence of endogenous fusion transcripts. In addition, all experiments were performed independently at least four times and the results were identical. Taken together, the data suggest that expression of an input chimeric RNA can lead to the induction of a specified endogenous fusion transcript in human cells. Surprisingly, the antisense, rather than the sense version of input RNA, exhibits the capacity of induction.
Antisense input RNAs described above contain 218 nt against the entire ERG exon-4 and 78 nt against the entire TMPRSS2 exon-1 (Fig. 1A), suggesting that 78 nt is sufficient to specify a parental gene for a fusion event. Furthermore, because the effective input RNAs are of the antisense orientation, the data imply that the input RNAs may not require an RNA junction resembling that of the TMPRSS2–ERG fusion transcript generated by splicing in the sense orientation. To further analyze the sequence requirement, we used the U6 promoter to express a series of antisense input RNAs with 75 nt complementary to ERG exon-4 joined to various segments (33, 52, 67, 82 nt) that are complementary to TMPRSS2 near the exon-1/intron-1 boundary (Fig. 1B). A parallel set of sense input RNAs were also tested as controls (Fig. 1C). As shown in Fig. 1B, Lower, all antisense RNAs, with the exception of antisense-3, induced fusion transcripts even though their target regions span the exon/intron boundary. The level of induction peaked for antisense-5, which contains 52 nt designed to anneal with TMPRSS2, suggesting that this length might be optimal to engage a parental gene for fusion event. The results were confirmed using a different, but more efficient, primer pair (Fig. 1B, Lower; primer design in SI Appendix, Fig. S1B) followed by Sanger sequencing of the induced band (SI Appendix, Fig. S4). In contrast to the antisense input RNA, all corresponding sense input RNAs failed to induce endogenous fusion transcripts (Fig. 1C, Lower). This was true even when the sense input RNA was intentionally expressed at a much higher level than the antisense RNA (SI Appendix, Fig. S5). The plasmids expressing sense RNA contain the same DNA sequences as the plasmids expressing antisense RNA except that the promoter is placed in the opposite direction (Fig. 1A). Therefore, the inability of sense plasmids to induce fusion transcripts argues against the possibility that it is the DNA sequences in the plasmids that induce fusion transcripts. Additional experiments using plasmids with a severed U6 promoter (SI Appendix, Fig. S6), to eliminate input RNA expression, confirmed that it is the antisense input RNAs expressed from plasmids—not the DNA sequence of plasmids—that induce the observed TMPRSS2–ERG fusion transcripts.
As shown in Fig. 1D, the amount of endogenous fusion transcript induced by antisense-5 (the most effective antisense input RNA) appears to correlate with the concentration of DHT used, presumably because the hormone induces the chromosomal proximity between the TMPRSS2 and ERG genes (13–15). Antisense-5 was effective at DHT concentrations as low as 20 nM, as revealed by sensitive nested PCR (Fig. 1D, lane 3), indicating that fusion events induced by input RNA can occur under physiologically relevant androgen conditions (16). As a control, DHT treatment alone up to 2 µM failed to induce fusion (SI Appendix, Fig. S7). Titration of DHT showed that the induction by antisense-5 reaches the 50% maximal level (EC50) at 0.9 µM DHT (SI Appendix, Fig. S7). Under this standard EC50 condition, we estimated that the percentage of LNCaP cells induced by antisense-5 to express the TMPRSS2–ERG fusion transcript is ∼1 in 103 or 104 cells (see assay in SI Appendix, Fig. S8). Together, these results demonstrated that the induction of fusion events by input RNA can occur at physiologically relevant hormone levels, but does not represent a high-frequency event.
Although induced fusions are infrequent, all antisense RNAs described in Fig. 1B successfully induced endogenous fusion RNA except antisense-3, which is only 30-nt longer than antisense-5 in the arm targeting TMPRSS2 intron-1 (Fig. 1B, Upper). To test whether its inability to induce fusion transcripts was due to input RNA length or the specific target sequence in TMPRSS2 intron-1, we made a hybrid antisense (antisense-7) that shifted the 52-nt recognition window of antisense-5 to target the TMPRSS2 intron-1 region covered by antisense-3 (Fig. 1E). This alteration resulted in the loss of induction (Fig. 1E, lane 1 vs. lane 3), implying that the inability of antisense-3 to induce is not reflective of input RNA length. Rather, its targeting arm may interfere with a motif important for the fusion process. BLAST alignment of the genomic DNA sequence revealed an imperfect stem (named stem A) potentially formed by the sense genomic TMPRSS2 sequence complementary to the sense genomic ERG sequence (Fig. 2A, Left). We reasoned that this genomic DNA stem (Tm = ∼44 °C) could stabilize a three-way junction that involves an RNA/DNA duplex formed by the antisense-5 RNA and its targeted genomic DNA in a sequence-specific manner. If correct, then the formation of this putative three-way junction would be disrupted by antisense-3 because its recognition sequence invades the genomic DNA stem. Consistent with the idea that induction requires bringing TMPRSS2 and ERG gene in close proximity, expression of antisense-5 as two separate halves (Fig. 2A, Right, antisense-5A and -5B) severed the link between the TMPRSS2 (52 nt) and ERG (75 nt) sequences in the input RNA, resulting in the loss of induction (Fig. 2B, lanes 1–3).
Fig. 2.
Formation of a three-way junction may facilitate fusion induction. (A, Left) Schematics of three-way junction that could be formed between genomic DNA (black) and antisense-5 input RNA (green/blue). The sense genomic strands of both TMPRSS2 and ERG genes are on the minus strand of chromosome 21, separated by 3 Mb. Short lines in shaded regions represent base pairings. Imperfect DNA stem A includes a high-energy G-T and A-C wobble pair known to have Watson–Crick-like geometry in a DNA double helix (30, 31). A spacer region of 36 nt and 47 nt separate stem A from the regions targeted by antisense-5 input RNA. (Right) Expressing antisense-5 as two separate halves (antisense-5A and -5B) that severed the link between TMPRSS2 (52 nt) and ERG (75 nt) RNA sequence. (B) RT-PCR assays of fusion transcripts showed that the severed input RNAs resulted in the loss of fusion transcript induction (lanes 1, 2, 3, vs. lane 4). (C) Locations of putative stems A to G identified by BLAST analyses. The DNA stem B to G are located in the introns. Genomic coordinates are listed in SI Appendix, Fig. S9. (D) The putative three-way junction formed between the indicated genomic DNA stem B to G (black) and designed antisense input RNA (green/blue). These antisense RNAs target introns and contain no exon sequence. (E) Targeting genomic DNA stem B, C, and D that exhibit higher DNA stem stability (Tm = 40 °C, 40 °C, and 44 °C, respectively) by antisense RNAs induced fusion transcripts (lanes 2–4). In contrast, targeting less stable stem E, F, and G (Tm = 30 °C, 24 °C, and 16 °C, respectively) failed to induce fusion transcripts (lanes 5–7). (F) Antisense input RNAs designed to invade each of the respective genomic DNA stem B, C, and D (antisense-B2, B3, C2, C3, D2, and D3) resulted in the loss of induction. (G) Corresponding sense input RNAs targeting stem B, C, and D failed to induce fusion transcripts (lanes 2–4).
To test whether the proposed three-way junction formation could facilitate fusion induction, we used BLAST alignment to identify several intron locations where the sense genomic TMPRSS2 sequence can pair with the sense genomic ERG sequence to form a DNA stem (stems B to G in Fig. 2 C and D; genomic coordinates in SI Appendix, Fig. S9; sequences flanking the stems in SI Appendix, Fig. S10). Matching antisense input RNAs (termed antisense-B1 to -G1) were then designed to facilitate the formation of a three-way junction with the possible intron stems (Fig. 2D) that would mirror the three-way junction formed by antisense-5 on stem A, as postulated in Fig. 2A. Because these input RNAs target the introns (Fig. 2C) and contain no exon sequence, any observed induction of endogenous fusion transcripts composed of exons cannot arise from the sequence of input RNAs or plasmids used for expression. As shown in Fig. 2E, targeting genomic DNA stems B, C, and D that exhibit higher DNA stem stability (Tm = 40 °C, 40 °C, and 44 °C, respectively) by the corresponding antisense input RNAs clearly induced fusion transcripts (Fig. 2E, lanes 2–4). In contrast, targeting less stable stems E, F, and G (Tm = 30 °C, 24 °C, and 16 °C, respectively) failed to induce fusion transcripts (Fig. 2E, lanes 5–7). To disrupt the three-way junction involving stems B, C, and D, six additional antisense RNAs (antisense-B2, -B3, -C2, -C3, -D2, and -D3) were designed with one side of their recognition sequence altered to invade each of the respective genomic DNA stems on the TMPRSS2 side or the ERG side (SI Appendix, Fig. S11). These modifications were chosen to mirror the interference on stem A by antisense-3. Similar antisense RNAs were also designed to invade stem A (SI Appendix, Fig. S12). In all cases, invasion of the genomic DNA stems by the modified input RNAs resulted in the significant loss of induction (Fig. 2F and SI Appendix, Fig. S12). While these results by no means necessitate that a three-way junction is required for fusion transcript induction, they nevertheless suggest that such transiently stabilized structures may “facilitate” the process and could have important implications in developing gene-editing technologies via mechanisms native to mammalian cells. Consistent with earlier observations, the corresponding sense version of the effective antisense input RNAs (sense-B1, -C1, -D1) all failed to induce fusion transcripts (Fig. 2G, lanes 2–4).
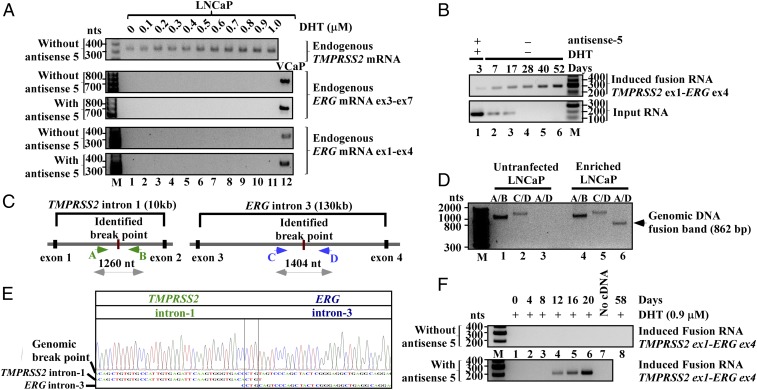
The fact that antisense input RNAs, but not their sense counterparts, induce fusion transcripts, raises the possibility that the former act as a docking station to mediate transsplicing between endogenous sense TMPRSS2 and ERG pre-mRNAs. Because the antisense, but not the sense input RNAs, are complimentary to both sense TMPRSS2 and ERG pre-mRNAs, they can base pair with both parental pre-mRNAs, thus resulting in spliced fusion transcripts without the requirement of genomic rearrangement. However, the following experimental results indicate that this is unlikely. First, although TMPRSS2 is expressed in LNCaP cells (Fig. 3A, Top), endogenous ERG mRNA is not detected in LNCaP cells (11) in the presence or absence of DHT or before and after transfection of antisense-5 (Fig. 3A, Middle and Bottom with different primer pairs). In fact, parental ERG mRNA was not detected in the presence of high-level DHT (Fig. 3A), or even using three rounds of nested RT-PCR using various primer sets (SI Appendix, Fig. S13). Therefore, before and during induction, no or an insufficient number of parental ERG mRNAs are available in LNCaP cells as raw material for transsplicing to account for the level of induced fusion transcript. Second, after initial transient transfection and DHT treatment for 3 d, we continued to propagate and enrich the induced LNCaP population for 52 d in the absence of DHT (experimental procedures described in SI Appendix, Fig. S14). As shown in the Lower panel of Fig. 3B, antisense-5 RNA transiently expressed by plasmids was degraded and completely absent beyond day 17. In contrast, the induced fusion transcript was continuously expressed and enriched up to day 52 in the absence of antisense input RNA and DHT (Fig. 3B, Upper), indicating the persistent nature of the induced fusion product. Taken together, these results strongly suggest that the induced expression of the TMPRSS2–ERG fusion transcript is the consequence of gene fusion at the DNA level, which has a permanent nature. This is in contrast to the result of induced transsplicing at the RNA level mediated by antisense input RNA, which is transient and requires the continuous presence of input RNAs.
Fig. 3.
Induced TMPRSS2–ERG fusion is the result of genomic arrangements. (A, Top) RT-PCR shows that LNCaP cells express TMPRSS2 mRNA, which is up-regulated by DHT. Primers used are specific to TMPRSS2 exon-2 and exon-4. (Middle and Bottom) ERG mRNA, however, was not detected in LNCaP cells under a wide range of DHT in the presence or absence of antisense-5 (lanes 1–11). RT-PCR assays were performed using two independent primer pairs that selectively amplify exon-3 to -7 (Middle), or exon-1 to -4 (Bottom) of ERG mRNA. Both primer pairs amplified ERG mRNA in VCaP cells (lane 12), but will not amplify the induced TMPRSS2–ERG fusion transcript which has ERG exon-4 to -12. (B) RT-PCR shows the transient nature of input RNA that was degraded by day 17 (Lower), and the persistent nature of the induced fusion transcript (Upper) up to 52 d postinitial treatment in the enriched LNCaP population (see SI Appendix, Fig. S14 for enrichment procedure). (C) Schematics of identified genomic breakpoints and the primer A, B, C, and D used to amplify the breakpoints. (D) The unrearranged wild-type TMPRSS2 and ERG alleles were revealed by primer pair A/B (∼1,404 bp) and C/D (∼1,260 bp), respectively (lanes 1, 2, 4, and 5). The genomic fusion band of 862-bp amplified by fusion-specific primer pair A/D was present only in the enriched LNCaP population (lane 6) and absent in untransfected LNCaP cells (lane 3). (E) Sanger sequencing of the fusion band showed a 500-bp segment of TMPRSS2 intron-1 fused to 362 bp of ERG intron-3 defined by primer A/D. The genomic breakpoint contains a “CTG” microhomology (boxed). The full-length Sanger sequence is shown in SI Appendix, Fig. S16. (F) Prolonged expression of antisense-5 for 12 d induced the TMPRSS2–ERG fusion transcript in PNT1A cells as detected by three-round nested PCR.
To provide definite evidence of gene fusion via genomic rearrangement, we used genomic PCR to identify the genomic breakpoint induced by antisense-5 in the enriched LNCaP population (primer designs in Fig. 3C and SI Appendix, Fig. S15A). As shown in Fig. 3D, the unrearranged wild-type TMPRSS2 and ERG alleles were amplified by gene-specific primer pair A/B and C/D both in untransfected cells (Fig. 3D, lanes 1 and 2) and enriched LNCaP cells (Fig. 3D, lanes 4 and 5). In contrast, a genomic fusion band of ∼862 bp amplified by fusion-specific primer pair A/D was present only in the enriched LNCaP population (Fig. 3D, lane 6) and absent in untransfected LNCaP cells (lane 3). Sanger sequencing of the excised fusion band (Fig. 3D, lane 6) revealed the exact genomic breakpoint located within TMPRSS2 intron-1 (chr21:41502038, GRCh38/hg38) and ERG intron-3 (chr21:38501207, GRCh38/hg38) (Fig. 3E; full-length Sanger sequence shown in SI Appendix, Fig. S16). Intriguingly, within TMPRSS2 intron-1 the induced breakpoint lies within an Alu, a transposable element known to contribute to genomic arrangements (17). In ERG intron-3, the breakpoint resides in a hot spot clustered with genomic breakpoints previously identified in prostate cancer patients (SI Appendix, Fig. S15B) (18). There is no obvious sequence homology between TMPRSS2 and ERG at the genomic breakpoint except for a three nucleotide “CTG” microhomology (Fig. 3E and SI Appendix, Fig. S16), suggesting that this gene fusion may be mediated by nonhomologous break-repair mechanisms (19, 20).
To test whether antisense input RNA can cause TMPRSS2–ERG fusion in nonmalignant cells before cancerous transformation, we performed experiments using immortalized normal prostate epithelium cells (PNT1A) that express very low levels of androgen receptors (21). As shown in the Lower panel of Fig. 3F, prolonged expression of antisense-5 for 12 d induced fusion transcripts (Sanger sequencing confirmation in SI Appendix, Fig. S17). This induction was not due to prolonged exposure to DHT because continuous treatment of 0.9 µM DHT alone for up to 2 mo resulted in no detectable fusion transcripts in PNT1A cells (Fig. 3F, lane 8). Thus, our results indicate that the induction of TMPRSS2–ERG fusion by antisense input RNA can occur in normal prostate epithelial cells before malignant transformation and is not restricted to the pathological cellular context of malignant cells.
To test whether an input RNA can specify a pair of genes to undergo fusion other than TMPRSS2–ERG in a sequence-specific manner, we designed a series of input RNAs to induce TMPRSS2–ETV1, an interchromosomal fusion gene found in ∼1% of prostate cancers (11, 22). Eight antisense RNAs (SI Appendix, Fig. S18) were designed to target different chosen regions in the introns where three-way junctions potentially can be forged between the genomic DNA and input RNAs (SI Appendix, Figs. S18 and S19). Again, because these input RNAs target introns and contain no exon sequence, it rules out the possibility that induced endogenous fusion transcripts composed of exons arise from the sequence of input RNAs or the plasmids. As shown in Fig. 4A, targeting TETV stem 1, which has the highest genomic DNA stem stability (Tm = 72 °C) among this group, led to clear induction of the TMPRSS2–ETV1 fusion transcript (Fig. 4A, lane 1). Sanger sequencing validated that the induced transcript contains TMPRSS2 exon-1 joined with ETV1 exon-3 (uc003ssw.4) by annotated splice sites (SI Appendix, Fig. S20). Similar to earlier observations, targeting with sense versions of input RNAs (Fig. 4B, lane 1 vs. lane 2), or using antisense input RNAs designed to form three-way junctions with lower genomic DNA stem stabilities (Fig. 4A, lanes 2–8 and SI Appendix, Fig. S18), resulted in no detectable induction. Furthermore, the input RNA designed to target TMPRSS2 and ETV1 induced TMPRSS2–ETV1 but not TMPRSS2–ERG fusion (Fig. 4C, lane 2). Conversely, antisense-5 targeting TMPRSS2 and ERG induced TMPRSS2–ERG but not TMPRSS2–ETV1 fusion (Fig. 4C, lane 1), indicating that fusion formation is specified by the sequence of input RNA and not secondary effects, such as global genomic instability.
Fig. 4.
RNA-mediated interchromosomal gene fusion between TMPRSS2 and ETV1. (A) RT-PCR shows that only the antisense RNA targeting stem TETV-1 (with the highest stem Tm = 72 °C) led to induced fusion transcript (lane 1). Antisense RNAs targeting stems with lower genomic DNA stem stabilities (TETV-2 to -8, lanes 2–8) resulted in no detectable induction. (B) The corresponding sense input RNA targeting the same TETV-1 stem failed to induce fusion transcript (lane 1 vs. lane 2). (C) Gene fusion is specified by the sequence of input RNA used. The antisense TETV-1 induced TMPRSS2–ETV1 fusion but not TMPRSS2–ERG fusion (lane 2). Conversely, antisense-5 induced TMPRSS2–ERG fusion but not TMPRSS2–ETV1 fusion (lane 1). (D) RT-PCR shows the transient nature of input RNA which was present at day 3 but not day 47 postinitial treatment (Lower, lane 1 vs. lane 2), and the persistent nature of the induced fusion transcript (Upper) up to 47 d postinitial treatment in the enriched LNCaP population. (E, Upper) Schematics of three identified genomic breakpoints marked as x, y, and z, and the primers used to amplify the breakpoints. (Lower) The unrearranged wild-type TMPRSS2 allele was revealed by primer pair E/F (990 bp; lanes 1 and 4) and G/H (956 bp; lanes 7 and 10), and the unrearranged wild-type ETV1 allele by primer pair M/N (1,024 bp; lanes 2, 5, 8, and 11). The genomic fusion band x (1,150 bp) and y (1,044 bp) amplified by fusion-specific primer pair E/N, and fusion band z (1,043 bp) amplified by primer pair G/N, were present only in the induced and enriched LNCaP population but absent in untransfected LNCaP cells (lane 6 vs. lane 3, and lane 9 vs. lane 12). (F) Sanger sequencing of the x, y, and z fusion band identified the exact genomic breakpoints. Region of microhomology at the breakpoints are boxed by solid lines, and indels by dash lines. The full-length Sanger sequences are shown in SI Appendix, Figs. S22–S24. (G) FISH probes against TMPRSS2 gene (red) on chromosome 21 and against ETV1 gene (green) on chromosome 7 were used to confirm the gene fusion. Examples of FISH signal in an untransfected cell (i) and a cell carrying induced TMPRSS2–ETV1 gene fusion (ii) are shown. Arrow points to the colocalized FISH signals indicative of TMPRSS2–ETV1 gene fusion, which is shown at a higher magnification in the inlet. About 0.9% of the enriched population (30 of 3,301 cells) was positive for TMPRSS2–ETV1 fusion gene based on the colocalized FISH signals. In contrast, none of the cells from the untransfected population (0 of 620 cells) showed colocalized FISH signals.
To verify that TMPRSS2–ETV1 serves as a second example of induced fusion that is indeed the consequence of genomic translocation, we propagated and enriched the induced LNCaP population for 47 d after the initial transfection of input RNA and DHT treatment (experimental procedures same as described for TMPRSS2–ERG enrichment in SI Appendix, Fig. S14). The transiently expressed antisense input RNA had been degraded and was absent by day 47 (Fig. 4D, lane 1 vs. lane 2). The induced TMPRSS2–ETV1 fusion transcript, however, was continuously expressed beyond day 47 (Fig. 4D, lane 2). Once again, this observation indicated that the sustained expression of an induced fusion gene does not require the continuous presence of input RNA. Moreover, genomic PCR assays identified three distinct genomic breakpoints between the TMPRSS2 and ETV1 genes (labeled as x, y, and z in Fig. 4E) that were present only in the enriched LNCaP population but absent in untransfected LNCaP cells (Fig. 4E, lane 3 vs. lane 6, lane 9 vs. lane 12). Similar to earlier observations, no obvious sequence homology between TMPRSS2 and ETV1 was observed at the genomic breakpoints except for a few nucleotides of microhomology (Fig. 4F and SI Appendix, Figs. S22–S24), indicating that the gene fusion is mediated by nonhomologous break-repair mechanisms (19, 20).
Unlike TMPRSS2 and ERG that are located near each other on the same chromosome, TMPRSS2 and ETV1 are located on different chromosomes. Thus, gene fusion as a result of chromosomal translocation could be confirmed unequivocally by evidence of chromosomal colocalization of the latter pair. Using probes specific to TMPRSS2 and ETV1, we performed FISH followed by deconvolution microscopic imaging of 3,301 cells from the enriched LNCaP cell population and 620 cells from the control untransfected LNCaP population. Analyses of constructed 3D images showed that ∼0.9% of the enriched population (30 of 3,301 cells) were positive for colocalization of the TMPRSS2 and ETV1 gene in the cellular nucleus (Fig. 4G; examples of constructed 3D images are shown in Movies S1 and S2). In contrast, none of the cells from the untransfected population showed colocalized FISH signals as determined by the same 3D image criteria (Fisher’s exact test, P < 0.01). Together, the evidence of chromosomal colocalization, the identified genomic breakpoints by genomic PCR at single base resolution (Fig. 4 E and F), and the observation that the sustained expression of induced fusion does not require the continuous presence of input RNA (Fig. 4D), strongly indicate that the induced expression of the TMPRSS2–ETV1 fusion transcript represents the consequence of gene fusion caused by chromosomal translocation.
The mechanism central to our hypothesis is that the input chimeric RNA acts as a guide RNA to mediate genome rearrangement by annealing to TMPRSS2 or ERG genes. Resolving such an RNA/DNA duplex by DNA break/repair mechanisms yield the final gene fusion through recombination in regions prone to DNA breaks. Accordingly, overexpression of RNaseH in cells, which degrades the RNA in an RNA/DNA duplex, should reduce the probability of fusion gene formation. To test whether the RNA/DNA duplex is indeed required for an RNA-mediated fusion gene, we cotransfected input chimeric RNA expression plasmid together with a second plasmid that expresses wild-type RNaseH (23), which degrades the RNA in the RNA/DNA duplex. As a control, an inactive mutant RNaseH (D10R E48R mutant) (23) that lacks the ability to degrade RNA was used for head-to-head comparisons. As shown in Fig. 5A, induction of the TMPRSS2–ERG fusion gene by antisense chimeric RNA was significantly reduced in the presence of wild-type RNaseH vs. the mutant RNaseH (Fig. 5A, lane 2 vs. lane 1). Similarly, induction of the TMPRSS2–ETV1 fusion gene was also significantly reduced in the presence of wild-type vs. the mutant RNaseH (Fig. 5A, lane 6 vs. lane 5). These results indicate that the induction of gene fusions requires the formation of an RNA/DNA hybrid. Consistent with previous observations, sense input RNAs failed to induce fusion regardless of the expression of RNaseH (Fig. 5A, lanes 3, 4, 7, and 8).
Fig. 5.
(A) RNA-mediated gene fusion requires RNA/DNA hybrid formation. Plasmids expressing input RNAs (antisense-5 for TMPRSS2–ERG, and antisense TETV-1 for TMPRSS2–ETV1) were cotransfected in ratio of 2:3 with plasmids expressing either wild-type or mutant RNaseH. The corresponding sense input RNAs were also cotransfected with wild-type or mutant RNaseH as the controls. (Top and Middle) Induction of fusion gene by antisense input RNA was significantly reduced in the presence of wild-type RNaseH vs. the mutant RNaseH (TMPRSS2–ERG: lane 2 vs. lane 1; TMPRSS2–ETV1: lane 6 vs. lane 5). Sense input RNAs failed to induce fusion regardless of the expression of RNaseH (lanes 3, 4, 7, and 8). (Bottom) RT-PCR showed that equal amount of wild-type or mutant RNase H was expressed. (B) The disparity between antisense versus sense input RNA is due to transcriptional conflict. The input RNAs were expressed by U6 (a pol-III promoter) while α-amanitin was used to inhibit pol-II transcription of the parental genes for various time periods (0, 2, 6, 12, and 24 h). α-amanitin was then rinsed off so that the newly induced fusion gene can express the fusion RNA. The induced fusion RNA was assayed by RT-PCR at day 3. Antisense-5 and sense-5 are for inducing TMPRSS2–ERG (Upper); antisense TETV-1 and sense TETV-1 are for inducing TMPRSS2–ETV1 (Lower). The sense input RNAs that previously failed to induce fusion, began to induce TMPRSS2–ERG (lanes 9 and 10) after 12 h of α-amanitin treatment, and TMPRSS2–ETV1 fusion (lane 20) after 24 h of α-amanitin treatment, respectively. GAPDH is used as internal loading control.
One important observation emerging from our study is that sense input RNAs consistently fail to induce gene fusion. This disparity was observed throughout our study despite that the sense and the antisense input RNA should form similarly stable DNA/RNA hybrids by annealing to the opposing strand of genomic DNA of the same site. This raises the possibility that the observed disparity could be a consequence of the transcriptional activity of targeted parental genes. The sense input RNAs forming DNA/RNA hybrids with the antisense strands of TMPRSS2 genomic DNA, which is the template strand used by RNA polymerase-II for RNA synthesis, could be frequently “bumped” off by RNA polymerase-II, thus unable to transiently stabilize the DNA/RNA hybrids. To test whether the parental gene transcriptional activity is responsible for the ineffectiveness of sense input RNAs, we used α-amanitin, a specific inhibitor of RNA polymerase-II, to shut down the endogenous TMPRSS2 and ETV1 gene transcription, as well as residual ERG transcription. In parallel, we used the U6 promoter, a polymerase-III promoter insensitive to α-amanitin, to continuously express the input chimeric RNAs. This setting essentially removes transcriptional conflict as a confounding factor, and allows the comparison of fusion induction by sense vs. antisense input RNA under an equivalent condition. As shown in Fig. 5B, antisense-5 specifically induced TMPRSS2–ERG fusion (Fig. 5B, Top, lanes 1–5), but not TMPRSS2–ETV1 fusion (Fig. 5B, Middle, lanes 1–5), regardless of the duration of α-amanitin treatment. Similarly, antisense TETV-1 specifically induced TMPRSS2–ETV1 fusion (Fig. 5B, Middle, lanes 11–15) but not TMPRSS2-ERG fusion (Fig. 5B, Top, lanes 11–15) under the same conditions. The results indicate that α-amanitin does not alter the specificity of the respective antisense input RNAs. Nonetheless, the toxicity exhibited by α-amanitin was readily observed during longer treatment periods. However, the toxicity decreased, rather than increased, the fusion induction (compare Fig. 5B, Top, lane 1 vs. lane 5, for TMPRSS2–ERG induction, and lane 11 vs. lane 15, Middle, for TMPRSS2–ETV1 induction), presumably because that gene fusion is more likely to occur in a healthy cell. Strikingly, the sense-5 input RNA that previously failed to induce TMPRSS2–ERG (Fig. 5B, Top, lane 6) began to induce TMPRSS2–ERG, but not TMPRSS2–ETV1, after 12 h of α-amanitin treatment (Fig. 5B, Top, lanes 9 and 10). Conversely, the sense TETV-1 input RNA that previously failed to induce TMPRSS2–ETV1 began to induce TMPRSS2–ETV1, but not TMPRSS2–ERG, after 24 h of α-amanitin treatment (Fig. 5B, Middle, lane 20). Thus, this latent induction by sense input RNAs is specified by the RNA sequence, not a property of unspecific toxicity. Additional controls using a parental plasmid vector lacking the sense input RNA sequences, DHT treatment without plasmid transfection, and PCR reactions without cDNA, all failed to induce fusion under the same α-amanitin treatment (SI Appendix, Fig. S25). The results are consistent with the notion that the sense versus antisense disparity is largely due to the transcriptional activity of parental genes.
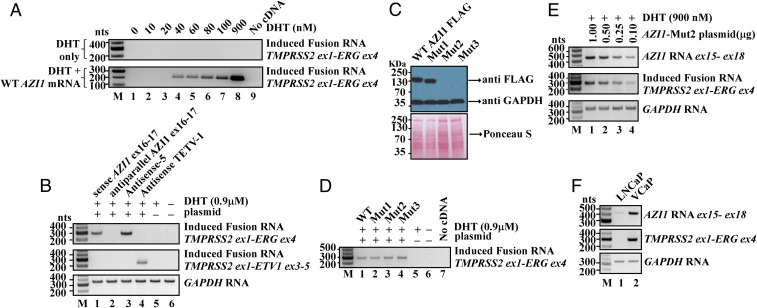
With the plausibility of RNA-mediated gene fusion established, we then sought evidence that specific endogenous cellular RNAs can act as the “initiator” to induce TMPRSS2–ERG fusion, which is found in ∼50% of prostate cancers. To identify candidate cellular initiator RNAs, we analyzed an mRNA-sequencing (mRNA-seq) database consisting of prostate tumors and matched benign tissues (24). However, bioinformatics analyses of this database failed to uncover any endogenous antisense chimeric RNAs in which the TMPRSS2 sequence was joined to any ERG sequence by discernable 5′ and 3′ splice sites in the antisense orientation. This suggests that if endogenous initiator RNAs do exist, they might arise from unrelated genomic sources that coincidentally resemble an imperfect chimeric RNA antisense to both TMPRSS2 and ERG. To identify such cellular initiator RNAs, we took the sequence of TMPRSS2 intron-1 and ERG intron-3 and used them as the bait templates to BLAST search for cellular RNAs with partial sequence complementarity and antisense to the TMPRSS2 and ERG intron sequences. The analysis utilized thermodynamic calculations of RNA/DNA hybrid stability, and permitted both Watson–Crick base pairing (G-C, A-U) as well as G-U wobble base pairing that commonly present in RNA/DNA hybrids. The analysis identified that AZI1 mRNA (also known as CEP131) (25, 26) could form high-affinity RNA/DNA hybrids with TMPRSS2 and ERG introns. To test whether the expression of AZI1 indeed induces TMPRSS2–ERG fusion, we cloned the full-length AZI1 mRNA (3,619 nt, uc002jzn.1) and overexpressed it in LNCaP cells by transient transfection. As shown in Fig. 6A, expressing AZI1 mRNA induced the TMPRSS2–ERG fusion transcript in LNCaP cells. The induction occurred at physiological DHT concentrations as low as 40 nM (Fig. 6A, lane 4, Lower). As a control, DHT treatment alone up to 0.9 µM failed to induce fusion (Fig. 6A, Upper). Furthermore, expression of exon16-17 of AZI1, a short 220-nt segment containing an imperfect sequence antisense to TMPRSS2 and ERG, was sufficient to induce TMPRSS2–ERG fusion (Fig. 6B, Top, lane 1), suggesting that the induction is mediated by an RNA sequence that resides in exon16–17. Consistent with previous observations that sense input RNAs are ineffective for the fusion process, the expression of exon16–17 in the antiparallel orientation also failed to induce TMPRSS2–ERG fusion (Fig. 6B, Top, lane 2). Moreover, the expression of exon16–17 of AZI1 specifically induced TMPRSS2–ERG fusion, but not TMPRSS2–ETV1 fusion (Fig. 6B, lane 1, Top vs. Middle). Thus, the induction by exon16–17 of AZI1 RNA is specified by the RNA sequence, not a property of unspecific global genomic instability.
Fig. 6.
Endogenous AZI1 RNA acts as the initiator RNA to induce TMPRSS2–ERG gene fusion. (A) Expression of AZI1 RNA in LNCaP cells for 3 d led to induced TMPRSS2–ERG fusion gene. The induction occurred at physiological DHT concentrations as low as 40 nM (lane 4, Lower). As a control, DHT treatment alone up to 0.9 µM failed to induce fusion (Upper). Three-rounds of nested PCR were performed to reveal the lowest amount of DHT that permits AZI1-mediated fusion induction. (B) Expression of AZI1 exon16-17, but not its antiparallel sequence, led to TMPRSS2–ERG gene fusion (lane 1 vs. lane 2). AZI1 exon16-17 induced TMPRSS2–ERG but not TMPRSS2–ETV1 (lane 1, Top vs. Middle). Antisense-5 (lane 3) and antisense TETV-1 RNA (lane 4) were used as the positive controls. GAPDH was the loading control. (C) Western blotting showed that while wild-type (WT) and Mut1 (first in-frame ATG mutated to TAA) produced a full-length and a shorter AZI1 protein, respectively, Mut2 (first three in-frame ATGs mutated), and Mut3 (coding frame was altered by a “G” insertion after the first ATG) completely eliminated the production of AZI1 protein in LNCaP cells. Anti-GAPDH antibody and Ponceau S staining revealed the total loaded protein. (D) All four AZI1 constructs (WT, Mut1, Mut2, Mut3) retained the ability to induce TMPRSS2–ERG gene fusion, indicating that AZI1 RNA but not protein is required. (E) Increasing the expression of AZI1 RNA (Top) resulted in increased levels of induced TMPRSS2–ERG fusion (Middle). GAPDH was the loading control. (F) LNCaP cells, which contain no TMPRSS2–ERG fusion gene, express very low level of endogenous AZI1 RNA (lane 1). In contrast, the VCaP cells, a prostate cancer cell line that harbors the TMPRSS2–ERG fusion gene, display a highly elevated level of endogenous AZI1 RNA (lane 2).
Because AZI1 is a known protein-coding gene, the expression of cloned full-length AZI1 cDNA will produce both the RNA and the protein. To answer the question of whether it is AZI1 RNA or protein that induces TMPRSS2–ERG fusion, we made three additional mutant constructs: Mut1 with the start codon ATG mutated; Mut2 with the first three in-frame ATGs mutated; and Mut3 with the coding frame of AZI1 shifted by a “G” insertion after the start codon. Western blotting showed that while wild-type and Mut1 produced a full-length and a shorter AZI1 protein, respectively, Mut2 and Mut3 completely eliminated the production of AZI1 protein (Fig. 6C). Nonetheless, all four AZI1 constructs (wild-type, Mut1, Mut2, Mut3) retained the ability to induce TMPRSS2–ERG gene fusion (Fig. 6D). The results demonstrated that the induction of TMPRSS2–ERG gene fusion is mediated by the AZI1 RNA, not AZI1 protein, and that AZI1 RNA possesses an aberrant noncoding function that leads to TMPRSS2–ERG gene fusion. To investigate whether the steady-state AZI1 RNA expression level (which reflects the equilibrium of RNA synthesis and degradation) is correlated with the level of fusion induction, we expressed the full-length AZI1 RNA (but not the protein) at different levels by transfecting various amount of AZI1-Mut2 plasmid in LNCaP cells. As shown in Fig. 6E, increasing the expression of AZI1 RNA (Fig. 6E, Top) resulted in increased levels of induced TMPRSS2–ERG fusion (Fig. 6E, Middle). Similarly, increasing the expression of antisense-5 input RNA also resulted in increased levels of induced TMPRSS2–ERG fusion (SI Appendix, Fig. S5, Middle). Consistent with this correlation, the parental LNCaP cells, which contain no TMPRSS2–ERG fusion gene, express a very low level of endogenous AZI1 RNA. In contrast, the VCaP cells, a prostate cancer cell line that harbors the TMPRSS2–ERG fusion gene, display a highly elevated level of endogenous AZI1 RNA (Fig. 6F, lane 1 vs. lane 2). These results suggest that a higher expression level of AZI1 RNA is required to mediate the fusion of TMPRSS2 and ERG gene.
Discussion
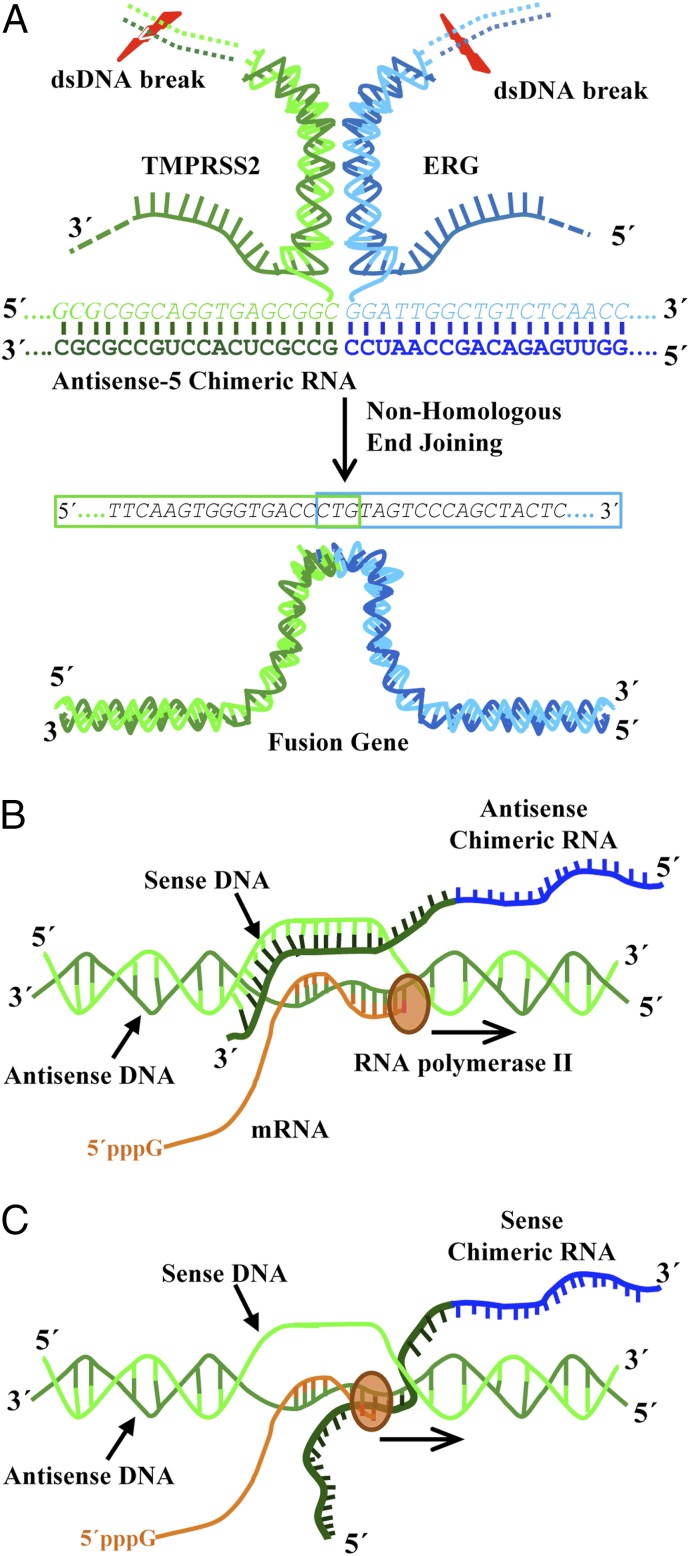
This report provides the striking evidence that expression of a chimeric RNA can drive the formation of gene fusions in mammalian cells. Hence, we propose that “the cart before the horse” hypothesis concerning fusion gene causation is mechanistically plausible. However, our data support a model (Fig. 7 and SI Appendix, Fig. S26) where the initiator RNA with chimeric sequence invades chromosomal DNA to stabilize a transient RNA/DNA duplex [reminiscent of an R-loop (27–29)] using DNA sequences located in two distant genes. Resolution of such an RNA/DNA duplex by DNA repair mechanisms might yield the final gene fusion through recombination in regions prone to DNA breaks. Such events were rare in the initial population of transfected cells (1 in 103 or 104 cells occurred within 3 d). However, the experiments using PNT1A cells showed that the necessary machinery is clearly present in normal prostate epithelial cells before malignant transformation. If the resulting gene fusion provides a survival advantage, a single affected cell among billions of cells in a normal prostate tissue may be conditioned to proliferate abnormally and eventually accumulate additional mutations and contribute to cancer formation. Studying such mechanism and the involved initiator RNAs might provide novel insights into early disease mechanisms, as well as the discovery of new preventive and therapeutic strategies to combat cancer.
Fig. 7.
A model of RNA-mediated gene fusion in mammalian cells. (A) Antisense-5 chimeric sequence invades chromosomal DNA of TMPRSS2 and ERG to stabilize a transient RNA/DNA hybrid reminiscent of an R-loop. Resolution of such an RNA/DNA hybrid by DNA break/repair mechanisms yields the final gene fusion through recombination in regions prone to DNA breaks. For simplicity, only part of the antisense-5 RNA sequence base-pairing with TMPRSS2 and ERG genes is shown. The three-way junction formation proposed in Fig. 2A is also omitted. (B and C) The disparity between antisense and sense chimeric RNA is explained by transcriptional conflict produced by the transcriptional activity of parental genes. (B) The antisense chimeric RNAs are able to form transiently stable DNA/RNA hybrids with sense strands of genomic DNA. (C) In contrast, the sense chimeric RNAs forming DNA/RNA hybrids with antisense strands of genomic DNA (the template strand used for transcription) are likely be “bumped” off by RNA polymerase and unable to stabilize the structures required for initiating genomic arrangements.
Contrary to the previous “cart before the horse” model (4, 8), our results do not support the postulation that a sense fusion mRNA derived from transsplicing between two pre-mRNAs effectively mediates gene fusion. As our experiments have demonstrated, expressing sense input RNAs mirroring the transspliced mRNA at high levels failed to induce fusion in LNCaP cells (Fig. 1A and SI Appendix, Fig. S2). Of 10 antisense RNAs that were demonstrated to be capable of inducing fusion (Figs. 1 A and B, 2E, and 4A), all of their corresponding sense RNAs failed to induce fusion (Figs. 1 A and C, 2G, and 4B). This was true even when the sense input RNA was deliberately expressed at a much higher level than the antisense RNA (SI Appendix, Fig. S5). This remarkable disparity between antisense and sense occurred even though the sense RNAs could, in theory, anneal to the same genomic sites targeted by their antisense counterparts and form similar DNA/RNA hybrids when paired with the antisense strand of genomic DNA. As demonstrated in Fig. 5, transcriptional shut down effectively diminishes this disparity and enables sense input RNAs to induce gene fusions. Therefore, the observed disparity can be readily explained by transcriptional conflict. Because the TMPRSS2 promoter is highly active in LNCaP cells, sense chimeric RNAs forming DNA/RNA hybrids with antisense strands of genomic DNA (the template strand used for transcription) would be frequently “bumped” off and unable to stabilize the transient structures required for initiating genomic arrangements (see illustration in Fig. 7 B and C). While our results do not support the postulation that sense fusion mRNA can effectively mediate gene fusion, it is worth noting that our results do not strictly preclude such a possibility that sense fusion mRNA derived from transsplicing can mediate gene fusion in other cellular contexts, especially under the conditions when the parental gene transcriptions are inactivated temporarily or inactivated in a cyclic manner.
Our results also do not support the hypotheses that antisense input RNAs, acting as a docking station, mediate transsplicing by base-pairing with both endogenous sense parental pre-mRNAs, or by bringing the parental genes in close proximity, thus facilitating transsplicing of parental pre-mRNAs transcribed from two genomic loci. Both mechanisms would require the continuous presence of antisense input RNAs to sustain the expression of induced fusion transcripts. However, we showed that the induced fusion expression has a permanent nature and requires no continuous presence of input RNAs (Fig. 3B for TMPRSS2–ERG, and Fig. 4D for TMPRSS2–ETV1). Furthermore, in the case of TMPRSS2–ERG there is no detectable ERG parental RNA as raw material in LNCaP cells (Fig. 3A) as would be expected for the transsplicing models. In addition, sense input RNAs, which cannot act as docking stations to base pair with parental sense pre-mRNAs, are able to induce the fusion transcripts after a period of transcriptional shut down. In contrast, the transient nature of input chimeric RNAs vs. the permanent nature of induced fusions, the evidence of genomic breakpoints identified by genomic PCR, and chromosomal colocalization provided by FISH, all strongly support that the induced expression of fusion transcript is largely the consequence of gene fusion resulting from chromosomal translocation. While we cannot completely rule out that a minuscule level of transsplicing might occur, our data indicate that if such transsplicing does occur, it contributes insignificantly to the observed fusion transcript induction in our experimental conditions.
Prior works have shown that strong genotoxic stress, such as γ-radiation under high levels of DHT (14, 15), can generate double-stranded DNA breaks and eventually lead to infrequent TMPRSS2–ERG fusions in LNCaP cells. However, such mechanisms of general genotoxicity fail to account for the “specificity” of gene fusions found in prostate cancer. The mechanism of RNA-mediated gene fusion, a mechanism that relies on sequence-specific interactions, can account for the specificity of gene fusion partners that were selected to undergo fusion under physiological conditions. The discovery that the endogenous cellular AZI1 RNA, not AZI1 protein, can act as an initiator RNA to induce TMPRSS2–ERG fusion at physiological DHT levels indicates that this mechanism may have important biological relevance to oncogenesis. Future investigation of the aberrant noncoding function of AZI1 RNA and the underlying mechanisms that lead to gene fusion might provide important insights into early disease mechanisms. In summary, this report demonstrates RNA-mediated gene fusion in mammalian cells. The results could potentially address the issue of specificity concerning fusion gene formation in cancer, and may have fundamental implications in the biology of mammalian genome stability, as well as gene-editing technology via mechanisms native to mammalian cells.
Materials and Methods
RNA was extracted from cells using a RiboPure Kit (Ambion). Reverse transcription was performed using SuperScript III (Invitrogen). The PCR primer designs and sequences, the reagents and conditions used for Western blotting, and the maintenance of the human prostate cancer cell lines are detailed in SI Appendix, Materials and Methods. Additional results described in the main text are also detailed in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Richard Sifers, Michael Ittmann, Richard Kelley, and Tom Cooper for critical suggestions; the Michael Ittmann laboratory for providing the cell lines; and Radhika Dandekar, Fabio Stossi, and Michael Mancini of the Integrated Microscopy Core at Baylor College of Medicine for assistance. The assistance of the Integrated Microscopy Core at Baylor College of Medicine had funding from the NIH (Grants DK56338 and CA125123), the Cancer Prevention Research Institute of Texas (CPRIT; Grant RP150578), the Duncan Cancer Center, and the Dunn Gulf Coast Consortium for Chemical Genomics. S.K.G. has been supported by CPRIT training Grant RP160283. L.Y. was supported by a Duncan Cancer Center Pilot grant, CPRIT Grant HIHRRA RP160795, and NIH Grant R01EB013584.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. F.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814704115/-/DCSupplemental.
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Langabeer SE, et al. MRC Adult Leukaemia Working Party Incidence of AML1/ETO fusion transcripts in patients entered into the MRC AML trials. Br J Haematol. 1997;99:925–928. doi: 10.1046/j.1365-2141.1997.4663270.x. [DOI] [PubMed] [Google Scholar]
- 3.Janz S, Potter M, Rabkin CS. Lymphoma- and leukemia-associated chromosomal translocations in healthy individuals. Genes Chromosomes Cancer. 2003;36:211–223. doi: 10.1002/gcc.10178. [DOI] [PubMed] [Google Scholar]
- 4.Zaphiropoulos PG. Trans-splicing in higher eukaryotes: Implications for cancer development? Front Genet. 2011;2:92. doi: 10.3389/fgene.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 6.Nowacki M, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang W, Landweber LF. RNA-mediated genome rearrangement: Hypotheses and evidence. Bioessays. 2013;35:84–87. doi: 10.1002/bies.201200140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowley JD, Blumenthal T. Medicine. The cart before the horse. Science. 2008;321:1302–1304. doi: 10.1126/science.1163791. [DOI] [PubMed] [Google Scholar]
- 9.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 10.Perner S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 11.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 12.Horoszewicz JS, et al. The LNCaP cell line—A new model for studies on human prostatic carcinoma. Prog Clin Biol Res. 1980;37:115–132. [PubMed] [Google Scholar]
- 13.Bastus NC, et al. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 2010;70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyce MJ, Baisley KJ, Clark EV, Warrington SJ. Are published normal ranges of serum testosterone too high? Results of a cross-sectional survey of serum testosterone and luteinizing hormone in healthy men. BJU Int. 2004;94:881–885. doi: 10.1111/j.1464-410X.2004.05051.x. [DOI] [PubMed] [Google Scholar]
- 17.Rüdiger NS, Gregersen N, Kielland-Brandt MC. One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res. 1995;23:256–260. doi: 10.1093/nar/23.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weier C, et al. Nucleotide resolution analysis of TMPRSS2 and ERG rearrangements in prostate cancer. J Pathol. 2013;230:174–183. doi: 10.1002/path.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coll-Bastus N, Mao X, Young BD, Sheer D, Lu YJ. DNA replication-dependent induction of gene proximity by androgen. Hum Mol Genet. 2015;24:963–971. doi: 10.1093/hmg/ddu508. [DOI] [PubMed] [Google Scholar]
- 22.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britton S, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kannan K, et al. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci USA. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoto H, et al. Isolation of a novel cDNA that encodes a protein localized to the pre-acrosome region of spermatids. Eur J Biochem. 1995;234:8–15. doi: 10.1111/j.1432-1033.1995.008_c.x. [DOI] [PubMed] [Google Scholar]
- 26.Aoto H, Miyake Y, Nakamura M, Tajima S. Genomic organization of the mouse AZ1 gene that encodes the protein localized to preacrosomes of spermatids. Genomics. 1997;40:138–141. doi: 10.1006/geno.1996.4546. [DOI] [PubMed] [Google Scholar]
- 27.Belotserkovskii BP, Tornaletti S, D’Souza AD, Hanawalt PC. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair (Amst) 2018;71:69–81. doi: 10.1016/j.dnarep.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanz LA, et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol Cell. 2016;63:167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keskin H, Meers C, Storici F. Transcript RNA supports precise repair of its own DNA gene. RNA Biol. 2016;13:157–165. doi: 10.1080/15476286.2015.1116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 31.Kimsey I, Al-Hashimi HM. Increasing occurrences and functional roles for high energy purine-pyrimidine base-pairs in nucleic acids. Curr Opin Struct Biol. 2014;24:72–80. doi: 10.1016/j.sbi.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.