Significance
A fraction of the 400 million people infected with dengue annually progresses to severe dengue (SD). Yet, there are currently no biomarkers to predict disease progression. We profiled the landscape of host transcripts and viral RNA in thousands of single blood cells from dengue patients prior to progressing to SD. We discovered cell type-specific immune activation and candidate predictive biomarkers. We also determined preferential virus association with specific cell populations, particularly naive B cells and monocytes. We explored immune activation of bystander cells, clonality and somatic evolution of adaptive immune repertoires, as well as viral genomics. This multifaceted approach could advance understanding of pathogenesis of any viral infection, map an atlas of infected cells, and promote the development of prognostics.
Keywords: dengue, single cell, transcriptomics, biomarkers, virus–host interactions
Abstract
Dengue virus (DENV) infection can result in severe complications. However, the understanding of the molecular correlates of severity is limited, partly due to difficulties in defining the peripheral blood mononuclear cells (PBMCs) that contain DENV RNA in vivo. Accordingly, there are currently no biomarkers predictive of progression to severe dengue (SD). Bulk transcriptomics data are difficult to interpret because blood consists of multiple cell types that may react differently to infection. Here, we applied virus-inclusive single-cell RNA-seq approach (viscRNA-Seq) to profile transcriptomes of thousands of single PBMCs derived early in the course of disease from six dengue patients and four healthy controls and to characterize distinct leukocyte subtypes that harbor viral RNA (vRNA). Multiple IFN response genes, particularly MX2 in naive B cells and CD163 in CD14+ CD16+ monocytes, were up-regulated in a cell-specific manner before progression to SD. The majority of vRNA-containing cells in the blood of two patients who progressed to SD were naive IgM B cells expressing the CD69 and CXCR4 receptors and various antiviral genes, followed by monocytes. Bystander, non-vRNA–containing B cells also demonstrated immune activation, and IgG1 plasmablasts from two patients exhibited clonal expansions. Lastly, assembly of the DENV genome sequence revealed diversity at unexpected sites. This study presents a multifaceted molecular elucidation of natural dengue infection in humans with implications for any tissue and viral infection and proposes candidate biomarkers for prediction of SD.
Dengue virus (DENV) is a major threat to global health, estimated to infect 400 million people annually in over 100 countries (1). The four serotypes of DENV are transmitted by a mosquito vector. A licensed vaccine has shown limited efficacy and increased hospitalizations in children (2, 3), and there are currently no approved antivirals available for dengue treatment. The majority of symptomatic patients present with dengue fever experiencing flu-like symptoms. Five to twenty percent of these patients progress within 4 to 7 d to severe dengue (SD), manifested by bleeding, plasma leakage, shock, organ failure, and sometimes death (3–6). Early administration of supportive care reduces mortality in patients with SD (7); however, there are no accurate means to predict which patients will progress to SD. The currently utilized warning signs to identify dengue patients at risk for progressing to severe disease are based on clinical parameters that appear late in the disease course and are neither sensitive nor specific. This situation promotes ineffective patient triage and resource allocation as well as continued morbidity and mortality (6, 8, 9).
DENV strains are classified in four clades called serotypes 1–4. The greatest risk factor for SD is previous infection with a heterologous DENV serotype, which can cause antibody-dependent enhancement (ADE) during the secondary infection (10–13). Aberrant activation of cross-reactive T cells may also play a role (14). Biomarkers for early detection of SD based on molecular features of the patient’s blood have been proposed. These efforts have focused on two experimental techniques: (i) flow cytometry of fixed blood cell populations (15), and (ii) gene expression in bulk RNA extracted from blood or peripheral blood mononuclear cells (PBMCs) (16–19). Although useful, these studies suffer from several limitations. The majority of these studies identified genes whose altered expression is associated with, but does not precede, the onset of SD and therefore cannot be used as prognostic biomarkers. From a technical standpoint, while flow cytometry has a high throughput, it is constrained to a few protein markers that are selected a priori, making it excellent for separating known, discrete cell populations but less appropriate for screening the complex, dynamic landscape of cell types, subtypes, and states characteristic of immune responses. Transcriptomics performed on bulk cell populations can screen thousands of genes, but its resolution is limited because it cannot capture tissue heterogeneity. Averaging the signal over various cell populations is confounded by changes both in the abundance of cell types and activation states. Coupling fluorescence activated cell sorting (FACS) with single-cell transcriptomics can potentially combine the advantages of both approaches (20). It has also been challenging to identify immune cells containing DENV RNA in humans. Since the composition of the immune system is complex and dynamic, we hypothesized that an unbiased approach would be more suitable than techniques based on a small number of markers to pinpoint the exact subset of cells containing DENV RNA in vivo and identify biomarkers of severity in distinct cell populations.
To facilitate the identification of host cells containing viral RNA (vRNA) and characterize their transcriptional response, we recently developed a virus-inclusive single-cell RNA-seq (viscRNA-seq) platform (21). We previously used this platform to characterize transcriptome dynamics of infection with flaviviruses in cultured human hepatoma (Huh7) cells (21). We simultaneously quantified host-cell mRNA and vRNA abundance at the single-cell level, thereby identifying proviral and antiviral factors that correlated with intracellular viral abundance (21). A similar approach was used to monitor the host response to Zika virus infection in neuronal stem cells (22). Here, we coupled FACS with viscRNA-Seq to identify cells with vRNA from human patients and studied the molecular signatures preceding the development of SD infection. The use of antibodies against surface proteins during FACS enabled enrichment for specific cell populations. Moreover, since viscRNA-Seq requires no genetic manipulation of the cells of interest, this approach enabled high-resolution screening of the whole human transcriptome for changes in gene expression at the single-cell level.
Results
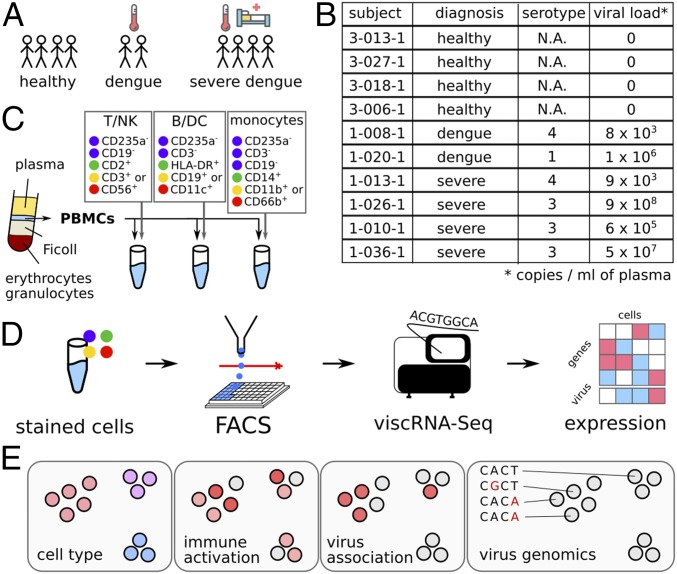
We combined FACS with viscRNA-Seq to profile the host and viral transcriptomes in PBMCs collected early in the course of natural dengue infection in humans. Blood samples were derived from four healthy control subjects and six DENV infected patients: two who experienced an uncomplicated disease course and four who subsequently progressed to SD (Fig. 1 A and B). All subjects were prospectively enrolled in a cohort that we established in Colombia (“Colombia cohort”) (SI Appendix, Tables S1–S3). Disease severity was classified on-site using 2009 World Health Organization criteria upon presentation and discharge (4). Patients were enrolled within 2–5 d of symptoms onset based on clinical presentation compatible with dengue or dengue with warning signs and positive NS1 antigen and/or anti-DENV IgM antibody. Notably, patients who were displaying signs of SD upon presentation were excluded. PBMCs, whole blood and serum samples were obtained upon presentation and at convalescence. qRT-PCR (23) and serological assays confirmed the diagnosis of DENV infection and excluded other arboviral infections (including Zika and chikungunya) (24). IgG level and avidity testing distinguished primary from secondary dengue (SI Appendix, Table S1) (24).
Fig. 1.
FACS-assisted viscRNA-Seq workflow on PBMCs from DENV-infected and healthy control human subjects. (A and B) Blood samples were collected from human subjects enrolled in the Colombia cohort (healthy, dengue, and SD). (C) PBMCs were isolated via Ficoll centrifugation and stained with three antibody panels to distinguish various cell types: T, B, NK, DC, and monocytes. (D) Single cells from each aliquot were sorted and processed by viscRNA-Seq to simultaneously quantify single-cell virus abundance and host transcriptome changes. (E) The information provided for each single cell includes: cell type, immune activation, infection state, and virus population genomics. N.A., nonapplicable.
To sort multiple types of immune cells in patient PBMC samples and enable viscRNA-Seq with high specificity and throughput, we assembled two panels of antibodies against host cell surface markers. The PBMC samples were split into several aliquots, immunostained with one of the antibody panels, and sorted via FACS into T cells, natural killer (NK) cells, B cells, monocytes, and dendritic (DC) cells (Fig. 1C and SI Appendix, Figs. S1–S3 and Tables S4 and S5). The viscRNA-Seq protocol was then followed, and each cell was sequenced at a depth of ∼1 million reads on NextSeq 500 and NovaSeq (Illumina) instruments (Fig. 1D). To measure intracellular DENV RNA abundance, we conducted viscRNA-Seq using the previously reported pan-DENV capture oligonucleotide (21). The information provided by this approach on each individual cell included the cell type, immune activation state, vRNA levels, and sequence of the virus strain (Fig. 1E).
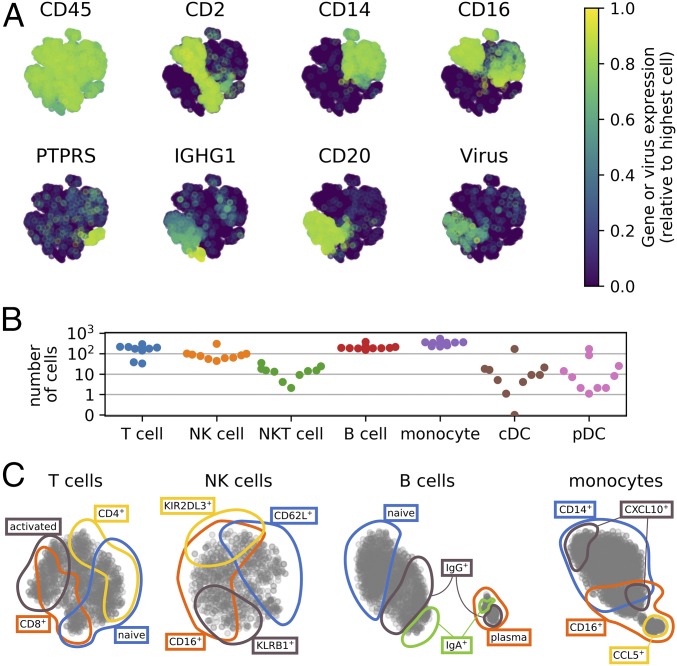
Most human tissues including blood present a skewed composition of cell types. Unbiased cell capture, as routinely done in microfluidic protocols (e.g., ref. 25), produces detailed data on the most abundant cell populations, but fails to represent rare cell populations. To overcome this limitation, we combined FACS with a plate-based protocol to capture immune cells from samples containing less than 1 million cells (because cells are sorted directly into single wells) with high sensitivity (as assessed by CD45 expression), and adequate representation of various cell populations (Fig. 2A) (26, 27). In total, we sequenced over 13,000 cells, of which several hundred showed robust signal for DENV RNA (Fig. 2A). Following quality filtering, tens to hundreds of cells were analyzed for most cell types of each sample, for a total of ∼8,700 cells (Fig. 2B and SI Appendix, Fig. S4 and Table S6). Within each cell type, multiple distinct overlapping immune cell subtypes and cell states were well represented in the dataset (Fig. 2C). In particular, within B cells alone we profiled many naive, IgM/IGD double-positive cells as well as isotype switched cells. Most B cells formed a continuum of differentiation, except for plasmablasts and plasma cells, which formed an additional cluster (Fig. 2C).
Fig. 2.
Overview of the types of PBMCs surveyed. (A) Two-dimensional representation of the cells color coded by the expression level of cell type-specific marker genes or the abundance of virus reads within the cell (>30 virus reads per million reads in samples from two SD patients, p1-026-1 and p1-036-1). (B) Number of cells analyzed for each cell type from each subject, see also SI Appendix, Table S6. (C) tSNE visualizations within T, NK, B cells, and monocytes, highlighting some broad cell subpopulations. The colored lines were drawn manually following inspection of the marker genes for visualization only.
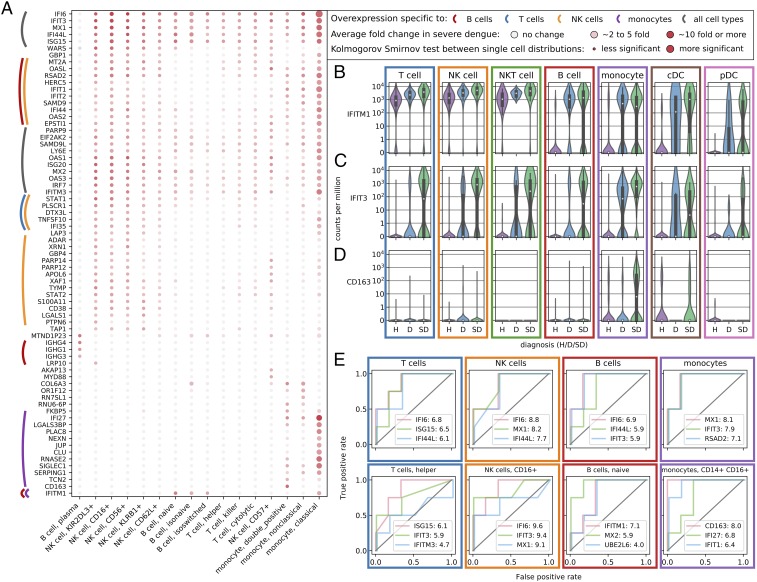
Next, we profiled the host transcriptome responses in the various PBMC populations. Since blood samples were obtained early in the course of dengue infection, this analysis was aimed at revealing alterations in gene expression that preceded the progression to SD. For each cell subtype and gene, we compared the distribution of expression values across the three categories of subjects: healthy control (H), uncomplicated dengue (D), and SD. To identify differentially expressed genes, we used a two-sample Kolmogorov–Smirnov test together with a computation of fold change in the averages across cells. We identified multiple genes whose expression was strongly up-regulated in subjects that subsequently progressed to SD. Many of these genes belonged to the antiviral IFN response, yet they were up-regulated in a cell type-specific manner (Fig. 3A and Dataset S3). Some genes were expressed in multiple cell types but were up-regulated more strongly in specific cells from SD subjects (Fig. 3B); other genes were expressed almost exclusively in SD patients except in a few cell types (Fig. 3C); a few genes were expressed only in one cell type and only in subjects who subsequently developed SD (e.g., CD163 in monocytes, Fig. 3D). These results indicate that distinct cell populations respond differently to the same viral infection, confounding the performance of bulk assays, such as microarrays. Since this heterogeneity is not a hindrance but rather a resource within the single-cell approach, we then explored the predictive potential of gene expression in specific cell types. To do so, we averaged across cells within the same patient and cell population and tested binary classification of severity at increasing thresholds of expression, de facto simulating a pseudobulk assay that could be implemented in the clinic. We identified a number of genes in specific cell populations that showed great statistical power for distinguishing SD before its development from an uncomplicated dengue course, as assessed by receiver operating characteristic (ROC) curves (Fig. 3E). Three notable examples with high ROC performance (area under the curve >95%) are MX2 in naive B lymphocytes and CD163 and IFIT1 in double-positive CD14+/CD16+ monocytes. We also developed a machine learning algorithm to predict whether a single cell originated from a SD patient and found that monocytes yielded the best results in train/test cross-validations, in line with the bulk predictors above (SI Appendix, Fig. S5).
Fig. 3.
Differential expression across disease severity and cell types shows hallmarks of progression to SD. (A) Genes that are overexpressed in subjects before progressing to SD versus all other subjects across cell types and subtypes. Color (white to red) indicates the average log-fold change; size of the dot indicates lower P value in a distribution statistical comparison (two sample Kolmogorov–Smirnov). (B) Many inflammatory genes such as IFITM1 are expressed ubiquitously during both mild and SD infection. (C) Other genes such as IFIT3 are specifically expressed before the development of SD in various types of lymphocytes. (D) A number of genes show double specificity for both SD and a single cell type, for instance CD163 in monocytes. (E) Averaging across cells within specific cell types and subtypes indicates promising candidate predictors of SD as assessed by ROC curves at increasing discriminatory thresholds for gene expression versus disease severity. The numbers after the gene name indicate log-twofold changes of average expression in patients progressing to SD versus other dengue patients, indicating an overexpression of these genes by a 100-fold or more in our cohort. D, dengue; H, healthy subject.
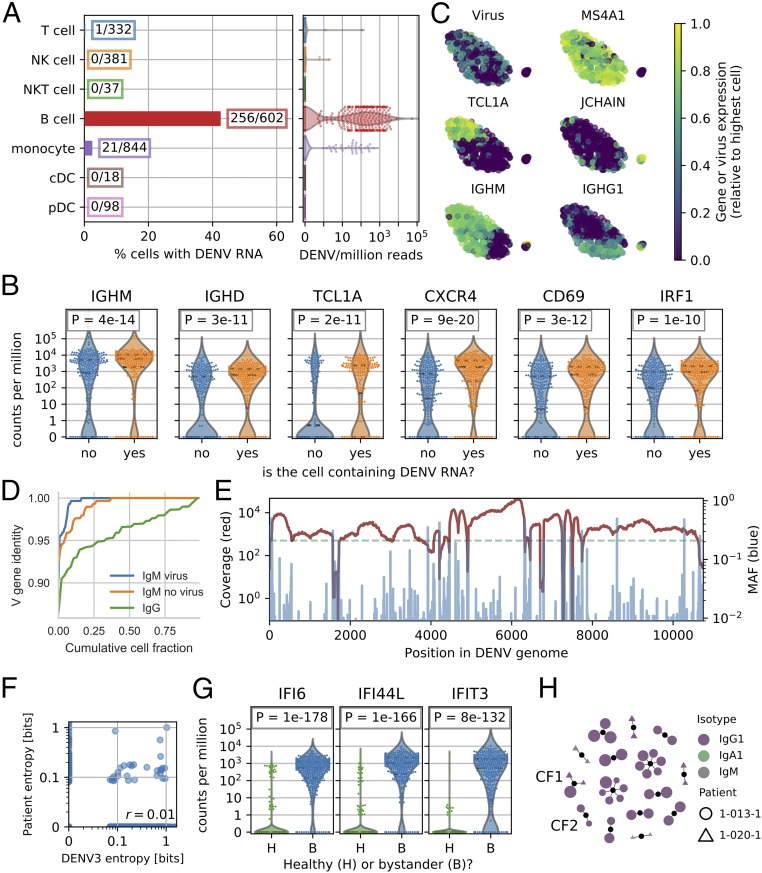
To define the cell subtypes that contain DENV RNA in our PBMC samples, we then focused on cells with vRNA reads. We detected viral reads in two samples only (of six dengue confirmed samples analyzed), both of which were derived from subjects who had high viral loads in their serum and who subsequently progressed to SD (samples 1-026-1 and 1-036-1, SI Appendix, Table S2). In both samples, around 3% of monocytes contained vRNA, in agreement with previous observations by flow cytometry (28). A weak up-regulation of CD4 and other genes, especially immune pathways, was observed in these vRNA-containing monocytes (SI Appendix, Fig. S6 and Table S7). The majority of cells with vRNA were B lymphocytes, representing 45% of the total B cell population from those patients (Fig. 4A). No viral reads were detected in other types of leukocytes or in control samples, except for low levels of cross-talk (SI Appendix, Fig. S7). These findings are in line with a previous report based on bulk qPCR assays (29). The fraction of uniquely mapped reads corresponding to vRNA in both B cells and monocytes was heterogeneous but generally 1% or less, corresponding to several hundred reads per cell but much lower than we measured in cultured Huh7 cells (21).
Fig. 4.
Naive B cells are the main cell type containing DENV RNA in two SD patients. (A) Fraction of cells containing vRNA across cell types from the two subjects and relative amount of virus RNA from each cell. (B) vRNA-containing B cells from the same subjects show a higher expression of specific surface receptors (CXCR4, CD69) and immune activation genes (IRF1, FCRL1). (C) tSNE visualization of the B cells from the two subjects. The expression level of DENV RNA and MS4A1 (CD20), TCL1A, JCHAIN, IGHM, and IGHG1 are highlighted. (D) Fractional identity of heavy chain V loci to their germ-line counterparts in vRNA-containing IgM, bystander IgM, and IgG B cells from the subjects 1-026-1 and 1-036-1. (E) Coverage (red) and minor allele frequency (MAF, blue) along the DENV genome in the viral reads from all cells from patient sample 1-026-1 show the genetic diversity of the virus population. (F) Site-specific Shannon entropy of a cross-sectional DENV serotype 3 alignment does not correlate with entropy from the viral reads of patient sample 1-026-1. Only sites with a coverage of 200 or more reads are considered (dashed green line in E). (G) B cells that do not contain DENV (bystanders) but are derived from subjects with vRNA-containing cells (B, blue) show a clear IFN response compared with B cells derived from healthy controls (H, green). (H) Graph of heavy chain CDR3 antibody clonality showing clonal expansion of IgG1 plasmablasts in patients 1-013-1 and 1-020-1 (no viral reads were detected in these two patients). Each dot is a unique antibody sequence; larger size corresponds to more somatic hypermutation. Clonal families CF1 and CF2 referred to in the text are labeled.
To determine whether a specific subpopulation of B cells contained DENV RNA, we identified the most up-regulated genes in the vRNA-containing population versus other B cells from the same patients. DENV RNA was enriched in B cells expressing IgM/IgD as well as other markers of naive B lymphocytes, such as the transcription factor TCL1A (see SI Appendix, Fig. S8 for more markers) (30). The surface receptors CD69, FCRL1, and CXCR4 that signal B cell activation and tissue-specific homing, and IRF1 that encodes an IFN-related protein, were also up-regulated (Fig. 4B). We computed a two-dimensional embedding of the B cells via t-Distributed Stochastic Neighbor Embedding (tSNE) from the two PBMC samples with detectable viral reads and measured no viral reads associated with cells belonging to the plasma cells (Fig. 4C and SI Appendix, Fig. S8) (31). We then assembled the whole B cell receptor (BCR) locus de novo and found that IgM B cells containing vRNA tended to have less hypermutations than other IgM B cells from the same subjects (Fig. 4D). In contrast, V/J usage in heavy and light chains was not apparently different between vRNA-containing and bystander (non-vRNA containing) B cells in the same subjects (SI Appendix, Fig. S9).
VRNA-containing cells may harbor vRNA either on their surface or intracellularly (SI Appendix, Fig. S10). Notably, treatment of B cells derived from dengue patients with protease E was previously shown to remove virus particles attached on the cell surface, while maintaining about half of the total viral content inside the cells, suggesting that the detected vRNA likely represents both surface bound and intracellular virus particle populations (32). However, the intracellular vRNA may be actively replicating or not. To attempt to distinguish between these possibilities, we studied the expression of a number of genes that are known to participate in viral replication and that we have previously shown to be altered in DENV-infected Huh7 cells in correlation with viral abundance (21). The expression of these genes in the vRNA-containing B cells was not altered, suggesting that the virus may not be actively replicating. Our attempts to define this by single molecule fluorescence in situ hybridization (smFISH) for detection of both positive-strand and negative-strand (indicative of actively replicating virus) DENV RNA were, however, limited by the inability to infect naive B cells derived from a healthy blood donor with DENV. Whereas both DENV RNA strands were strongly detected in DENV-infected Huh7 control cells, only positive strand DENV RNA was detected in blood donor monocytes, and neither strands were detected in blood donor B cells (SI Appendix, Fig. S11).
In addition to counting the DENV reads, we mapped them in an iterative manner and recovered ∼300,000 viral reads from patient 1-026-1 and ∼2,000 reads from patient 1-036-1. We obtained high coverage across the whole DENV genome and a third of the genome, respectively. The intrapatient population genomics showed a wide range of conservation levels, as determined by minor allele frequencies (Fig. 4E and SI Appendix, Fig. S12A). Site-specific Shannon entropy restricted to positions with 200 or more virus reads did not correlate with cross-sectional entropy in DENV serotype 3 (Fig. 4F and SI Appendix, Fig. S12B).
Hundreds of non-vRNA–containing B cells were recovered from samples containing vRNA-containing cells. We computed differential gene expression between these bystanders and B cells from healthy controls and identified a strong antiviral response via IFN stimulated genes IFI6, IFI44L, and IFIT3 (Fig. 4G). Moreover, we considered whether the diversity of the immune repertoire [BCR and T cell receptors (TCR)] could play a role in virus-cell association. Whereas assembled BCRs from patients with detected vRNA-containing B cells scattered into small clones, the BCR repertoire of patients 1-013-1 and 1-020-1, who had no vRNA-containing B cells, contained large clonal families comprised of multiple plasmablasts sharing similar antibody heavy chains, indicating a rapid and large clonal expansion in the B cell compartment (Fig. 4H). The fact that such plasmablast expansions were captured simply as part of these patients’ circulating B cell populations was surprising given the vast diversity of possible BCR rearrangements (33). This could be indicative of a more extensive plasmablast response and concurrent rise in neutralizing antibody titers known to occur in response to acute dengue infection (34). One clonal family (CF1, Fig. 4H and Dataset S1) had members belonging to both patients, while another (CF2) featured two plasmablasts with nearly identical antibody heavy chains, but distinct light chains, which supports the idea of heavy chain convergence in response to dengue (35). Since no DENV RNA reads were detected in these patient samples (in contrast to samples 1-026-1 and 1-036-1), we hypothesized that this oligoclonal plasmablast population reduces binding of DENV by the host B cells. However, serum neutralization studies revealed that a sample derived from only one of the two patients (1-013-1) potently neutralized DENV (SI Appendix, Fig. S13). Within the T cell compartment, we found that clustering by TCRβ/δ CDR3s produced clonal families that were largely private to an individual, while clustering according to TCRα/γ CDR3s revealed known invariant T cell subsets, including invariant NK T cells and mucosal associated invariant T cells, as well as public γ chain CDR3 sequences (SI Appendix, Fig. S14) (36).
Discussion
We recently developed viscRNA-Seq, a scalable approach to quantify host and nonpolyadenylated vRNAs from the same cell (21). In the current study, we apply viscRNA-Seq to in vivo samples and show that it can be used to effectively profile the landscape of host transcripts and vRNA in thousands of single immune cells during natural dengue infection of human subjects. The human samples studied here posed additional challenges, beyond those presented in cultured cells. First, the exact sequence of the viral strain infecting the patients was unknown. We therefore designed an oligonucleotide for virus capture in a conserved region of the viral genome (21). Second, the target cell types of DENV in vivo are incompletely characterized, mandating assembly of antibody panels for FACS that maximize the probability of capturing the vRNA-containing cell population(s) (Fig. 1C). Third, cell viability and integrity of the RNA after freezing, shipping, and thawing the PBMC samples was much lower than in cultured cells, especially for certain cell types. Although we could not recover all types of blood cells (e.g., granulocytes, see SI Appendix, Fig. S3) we increased the throughput and PCR preamplification to ensure a sufficient number of high-quality cells from many cell types. These modifications have successfully addressed the above challenges, as indicated by our findings. Because the viscRNA-seq approach can be extended to any virus of interest and is compatible with surface markers for FACS, we expect it to be readily extensible to dissociated solid tissues, for instance to characterize viral reservoirs of various viral infections. A similar approach was recently applied to influenza infection in mouse lungs (37) and to in vitro Zika virus infection of neuronal stem cells (22).
There are currently no clinically usable biomarkers to predict the development of severe complications associated with DENV infection, which include bleeding, shock, vascular leakage, and organ failure (4). Previous work on molecular biomarkers for dengue severity has focused on flow cytometry, which has a high throughput but requires an a priori choice of a limited number of protein markers (15), and bulk transcriptomics, which can quantify the expression of all genes in parallel but is confounded by the superimposition of cell types (e.g., B and T lymphocytes) and cell state (activation of SD specific genes) (16, 17). The blood samples analyzed in this study, obtained before the progression to SD and combined with the single cell resolution and ability to sample a wide range of cell types and activation states via viscRNA-seq, provided a unique opportunity to discover candidate biomarkers of progression to severity. Our data suggest the expression of MX2 within naive B cells and of CD163 and IFIT1 within CD14+ CD16+ monocytes is greatly up-regulated before the development of SD (Fig. 3E). It has been previously reported that MX2 is one of only four IFN-induced genes induced in an IRF3 and IRF7 independent manner in DENV-infected mice (38) and that CD163 in macrophages and CD14+ CD16+ monocytes contributes to the pathogenesis of SD (39, 40). Given the small number of subjects analyzed in this study and the female predominance, the predictive power of these candidate biomarkers warrants further validation in larger cohorts with a balanced number of females and males. Nevertheless, these findings underscore the utility of the viscRNA-Seq approach to identify candidate prognostic biomarkers for dengue.
Cell lines such as Huh7 and primary cells such as monocyte-derived DC are commonly used to study DENV host–cell interactions. However, elucidating the immune target cells of DENV in vivo has proven challenging. This study performs high-dimensional profiling of vRNA-containing cells in vivo. Twenty-one vRNA-containing monocytes were identified in samples derived before the progression to SD from two patients whose viral load was high and demonstrated a weak up-regulation of CD4 and other genes (SI Appendix, Fig. S6). While only positive-strand vRNA was detected at a high signal in monocytes derived from healthy donors, it remains to be determined whether this reflects nonreplicating virus or limited detection resulting from the low (<1:100) ratio of negative to positive strand vRNA (29, 32).
The majority of vRNA-containing cells in these patient samples were, however, B cells, in line with previous reports on bulk samples (29, 41). Distribution-level statistics and dimensionality reduction indicated that the most abundant vRNA-containing B cells were IgM positive, naive B cells expressing the surface markers CD69 and CXCR4, although other B cells of other isotypes were also represented. In contrast, plasmablasts and plasma cells had no viral reads (Fig. 4 A–D). B cells are known to be involved in SD pathogenesis by producing antibodies that mediate ADE (12–14). Discovering that naive B cells possessing diverse BCR sequences, rather than specific isotype switched, sequence-restricted memory B cells, typically contain DENV RNA was therefore surprising. Based on the previous work demonstrating that DENV vRNA is comparably distributed on the surface and intracellularly in B cells from dengue patients (32), we predict that the detected vRNA was derived from both of these viral populations. The expression of genes that are known to be altered in DENV infection was not altered in B cells derived from the SD patients, supporting that these cells may not harbor an actively replicating virus. Our attempt to further probe this with FISH, however, was limited since we could not infect blood donor B cells with DENV. More work is thus needed to conclusively define whether the virus harbored in these B cells is actively replicating or not. Regardless of the replicative status, vRNA-containing B cells may serve in shuttling DENV within the human body.
Bystander B cells from patient samples with detectable vRNA-containing cells had elevated levels of immune genes, particularly of the IFN response (Fig. 4G). Additionally, B cells from one SD and one non-SD patient (1-013-1 and 1-020-1) showed an interesting clonal structure in terms of antibody sequences. Specifically, multiple antibody sequences of heavy and light chains from several cells from these patients clustered into a few, presumably very large, clonal families of mostly heavily hypermutated IgG1 plasmablasts. No DENV RNA reads were detected in these patient samples, yet only one of the two samples (1-013-1) potently neutralized DENV. It is thus unclear whether this oligoclonal plasmablast population reduces binding of DENV by the host B cells. Further work is needed to understand the role of vRNA-containing and bystander B cells, determine whether DENV affects hypermutation dynamics upon binding naive B cells, and decipher whether the antibodies from these specific clonal families are protective against DENV.
From the viral reads of two patients, 1-026-1 and 1-036-1, we assembled the entire or a third of the DENV genome, respectively (Fig. 4E and SI Appendix, Fig. S12A). Although the viral capture oligonucleotide corresponds to the 3′ untranslated region (UTR) of DENV, we do not detect a strong 3′ bias in the DENV genome coverage, supporting that most vRNA is of genomic origin. Nevertheless, it is possible that a small fraction of the viral reads originates from subgenomic flavivirus RNA (sfRNA), previously reported in B cells (42). We observed some high-variability genomic sites (Fig. 4E). Previous work on other RNA viruses, particularly HIV-1, has shown that due to error-prone viral polymerases and fast generation times, intrapatient genomic viral diversity can represent a subsampled snapshot of the global diversity of the same virus in multiple infected individuals, implying a universal landscape of fitness costs (43, 44). DENV behaves quite differently, as globally variable sites do not correspond to variable sites within our patients (Fig. 4F). An optimized approach with higher sensitivity and sample selection (PBMCs or solid tissues) that maximizes the number of viral reads will facilitate a deeper understanding of the genomic diversity of viruses inhabiting the human body at the single-cell level.
In this study, we leveraged the viscRNA-Seq approach to explore many different facets of virus infection in uncomplicated dengue and SD in humans at the single-cell level. This multifaceted profiling included investigation of transcriptional up-regulation in specific subpopulations as a predictor of disease severity. Further validation in larger cohorts is warranted to determine the effectiveness of the identified candidate biomarkers as potential prognostic tools. Cell purification (e.g., by magnetic beads) followed by a rapid bulk expression assay (e.g., qPCR) is one option to translate such findings into a near-care, sample-to-answer system assay to be used for predicting progression of SD upon patient presentation. We also explored preferential association of virus with certain host cells, immune activation of bystander cells, clonality and somatic evolution of the adaptive immune repertoire, and intrapatient viral genomics. This technological convergence, combined with a high level of experimental and computational automation, underscores the utility of viscRNA-Seq as a powerful tool to rapidly gain a broad knowledge of emerging infectious diseases from just a few tissue samples.
Methods
Blood samples were collected from individuals presenting to the Fundación Valle del Lili in Cali (Colombia) between 2016 and 2017 with symptoms compatible with dengue. Patients that already showed severe symptoms at presentation were not considered. All work with human subjects was approved by the Stanford University Administrative Panel on Human Subjects in Medical Research (Protocol #35460) and the Fundación Valle del Lili Ethics committee in biomedical research (Cali/Colombia). All subjects, their parents, or legal guardians provided written informed consent, and subjects between 6 to 17 years of age and older provided assent. PBMCs were extracted using SepMate tubes (Stemcell Technologies), frozen, stored, and shipped in liquid nitrogen. FACS was performed on a Sony SH800 using fluorescently labeled antibodies to enrich for various immune cell types. The viscRNA-Seq protocol was followed and the libraries were sequenced on Illumina NextSeq 500 or NovaSeq. The sequencing reads were mapped and genes counted as reported before (21). Data analysis was performed using singlet (https://github.com/iosonofabio/singlet) and custom Python scripts. Detailed methods and protocols are available as SI Appendix.
Supplementary Material
Acknowledgments
We thank the reviewers whose suggestions greatly improved the manuscript and to the patients who participated in this study and to their families. This work was supported by seed grants from the Stanford Bio-X Interdisciplinary Initiatives Seed Grants Program, the Stanford Translational Research and Applied Medicine program, the Stanford SPARK program, Stanford Child Health Research Institute, and Stanford Institute for Immunity, Transplantation, and Infection (to S.E.). This work was also supported by NIH Grant 5U19AI057229-15, the Chan Zuckerberg Biohub, and California Institute for Regenerative Medicine Grant GC1R-06673 (to S.R.Q.). F.Z. was supported by a long-term European Molecular Biology Organization Fellowship ALTF 269–2016. M.L.R. was supported by the Stanford Advanced Residency Training at Stanford Fellowship Program.
Footnotes
Conflict of interest statement: F.Z., M.L.R., D.C., L.G., S.R.Q., and S.E. have filed US Provisional Application No. 62/715,628 related to this manuscript. S.R.Q. and A.K.S. are coauthors on a 2017 review article.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE116672), and gene expression counts and metadata were deposited in FigShare under doi 10.6084/m9.figshare.7149134.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813819115/-/DCSupplemental.
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Kouri G. Dengue and dengue hemorrhagic fever in the Americas: Lessons and challenges. J Clin Virol. 2003;27:1–13. doi: 10.1016/s1386-6532(03)00010-6. [DOI] [PubMed] [Google Scholar]
- 3.Khursheed M, et al. A comparison of WHO guidelines issued in 1997 and 2009 for dengue fever–Single centre experience. J Pak Med Assoc. 2013;63:670–674. [PubMed] [Google Scholar]
- 4.World Health Organization . Dengue Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. World Health Organization Press; Geneva: 2009. [PubMed] [Google Scholar]
- 5.World Health Organization 2018 Dengue and Severe Dengue. Available at www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue. Accessed June 25, 2018.
- 6.Thein TL, Leo Y-S, Lee VJ, Sun Y, Lye DC. Validation of probability equation and decision tree in predicting subsequent dengue hemorrhagic fever in adult dengue inpatients in Singapore. Am J Trop Med Hyg. 2011;85:942–945. doi: 10.4269/ajtmh.2011.11-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization; UNICEF; UNDP; World Bank; WHO Special Programme for Research and Training in Tropical Diseases . Handbook for Clinical Management of Dengue. World Health Organization Press; Geneva: 2012. [Google Scholar]
- 8.Hadinegoro SRS. The revised WHO dengue case classification: Does the system need to be modified? Paediatr Int Child Health. 2012;32:33–38. doi: 10.1179/2046904712Z.00000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nujum ZT, et al. Comparative performance of the probable case definitions of dengue by WHO (2009) and the WHO-SEAR expert group (2011) Pathog Glob Health. 2014;108:103–110. doi: 10.1179/2047773214Y.0000000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halstead SB, Chow JS, Marchette NJ. Immunological enhancement of dengue virus replication. Nat New Biol. 1973;243:24–26. [PubMed] [Google Scholar]
- 11.Halstead SB, O’Rourke EJ. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J Exp Med. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzelnick LC, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang TT, et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehorn J, Simmons CP. The pathogenesis of dengue. Vaccine. 2011;29:7221–7228. doi: 10.1016/j.vaccine.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Durbin AP, et al. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376:429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ubol S, et al. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J Infect Dis. 2008;197:1459–1467. doi: 10.1086/587699. [DOI] [PubMed] [Google Scholar]
- 17.Fink J, et al. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl Trop Dis. 2007;1:e86. doi: 10.1371/journal.pntd.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sessions OM, et al. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS Negl Trop Dis. 2013;7:e2107. doi: 10.1371/journal.pntd.0002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolayeva I, et al. A blood RNA signature detecting severe disease in young dengue patients at hospital arrival. J Infect Dis. 2018;217:1690–1698. doi: 10.1093/infdis/jiy086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darmanis S, et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanini F, Pu S-Y, Bekerman E, Einav S, Quake SR. Single-cell transcriptional dynamics of flavivirus infection. eLife. 2018;7:e32942. doi: 10.7554/eLife.32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman MJ, et al. An immunocompetent mouse model of Zika virus infection. Cell Host Microbe. 2018;23:672–685.e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waggoner JJ, et al. Single-reaction, multiplex, real-time rt-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Negl Trop Dis. 2013;7:e2116. doi: 10.1371/journal.pntd.0002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B, et al. Diagnosis of Zika virus infection on a nanotechnology platform. Nat Med. 2017;23:548–550. doi: 10.1038/nm.4302. [DOI] [PubMed] [Google Scholar]
- 25.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanini F. 2018 lshknn (Stanford University). Available at https://github.com/iosonofabio/lshknn. Accessed November 19, 2018.
- 27.Carnevali P. 2018 ExpressionMatrix2 (Chan Zuckerberg Initiative). Available at https://github.com/chanzuckerberg/ExpressionMatrix2. Accessed November 19, 2018.
- 28.Wong KL, et al. Susceptibility and response of human blood monocyte subsets to primary dengue virus infection. PLoS One. 2012;7:e36435. doi: 10.1371/journal.pone.0036435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srikiatkhachorn A, et al. Dengue viral RNA levels in peripheral blood mononuclear cells are associated with disease severity and preexisting dengue immune status. PLoS One. 2012;7:e51335. doi: 10.1371/journal.pone.0051335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Said JW, et al. TCL1 oncogene expression in B cell subsets from lymphoid hyperplasia and distinct classes of B cell lymphoma. Lab Invest. 2001;81:555–564. doi: 10.1038/labinvest.3780264. [DOI] [PubMed] [Google Scholar]
- 31.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 32.King AD, et al. B cells are the principal circulating mononuclear cells infected by dengue virus. Southeast Asian J Trop Med Public Health. 1999;30:718–728. [PubMed] [Google Scholar]
- 33.Georgiou G, et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appanna R, et al. Plasmablasts during acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine. 2016;12:178–188. doi: 10.1016/j.ebiom.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parameswaran P, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravens S, et al. Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat Immunol. 2017;18:393–401. doi: 10.1038/ni.3686. [DOI] [PubMed] [Google Scholar]
- 37.Steuerman Y, et al. Dissection of influenza infection in vivo by single-cell RNA sequencing. Cell Syst. 2018;6:679–691.e4. doi: 10.1016/j.cels.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H-W, et al. The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. J Immunol. 2013;191:4194–4201. doi: 10.4049/jimmunol.1300799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwissa M, et al. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ab-Rahman HA, Rahim H, AbuBakar S, Wong P-F. Macrophage activation syndrome-associated markers in severe dengue. Int J Med Sci. 2016;13:179–186. doi: 10.7150/ijms.13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baclig MO, et al. Flow cytometric analysis of dengue virus-infected cells in peripheral blood. Southeast Asian J Trop Med Public Health. 2010;41:1352–1358. [PubMed] [Google Scholar]
- 42.Manokaran G, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zanini F, et al. Population genomics of intrapatient HIV-1 evolution. eLife. 2015;4:e11282. doi: 10.7554/eLife.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanini F, Puller V, Brodin J, Albert J, Neher RA. In vivo mutation rates and the landscape of fitness costs of HIV-1. Virus Evol. 2017;3:vex003. doi: 10.1093/ve/vex003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.