Significance
The cyanobacterium Synechococcus elongatus PCC 7942 is widely used in basic and applied research. However, this model organism appears to have lost, through laboratory domestication, behaviors that are important in a natural environment, such as biofilm formation and phototaxis. We characterized a wild isolate of S. elongatus, UTEX 3055, that forms biofilms and is phototactic and investigated the mechanisms that regulate phototaxis. Our findings suggest a simpler design for phototactic motility in UTEX 3055 than that previously described for the cyanobacterium Synechocystis, because a single 5-GAF–domain photoreceptor senses the direction of illumination by wavelengths that induce both positive and negative responses. This study expands our knowledge of the mechanisms responsible for phototaxis in cyanobacteria and establishes a phototactic model organism.
Keywords: cyanobacteria, photoreceptor, GAF domain, phototaxis, Synechococcus elongatus
Abstract
Many cyanobacteria, which use light as an energy source via photosynthesis, have evolved the ability to guide their movement toward or away from a light source. This process, termed “phototaxis,” enables organisms to localize in optimal light environments for improved growth and fitness. Mechanisms of phototaxis have been studied in the coccoid cyanobacterium Synechocystis sp. strain PCC 6803, but the rod-shaped Synechococcus elongatus PCC 7942, studied for circadian rhythms and metabolic engineering, has no phototactic motility. In this study we report a recent environmental isolate of S. elongatus, the strain UTEX 3055, whose genome is 98.5% identical to that of PCC 7942 but which is motile and phototactic. A six-gene operon encoding chemotaxis-like proteins was confirmed to be involved in phototaxis. Environmental light signals are perceived by a cyanobacteriochrome, PixJSe (Synpcc7942_0858), which carries five GAF domains that are responsive to blue/green light and resemble those of PixJ from Synechocystis. Plate-based phototaxis assays indicate that UTEX 3055 uses PixJSe to sense blue and green light. Mutation of conserved functional cysteine residues in different GAF domains indicates that PixJSe controls both positive and negative phototaxis, in contrast to the multiple proteins that are employed for implementing bidirectional phototaxis in Synechocystis.
Many organisms have evolved the ability to sense and alter their location in response to various beneficial or noxious stimuli. A classic example of this behavior is chemotaxis, in which the organism moves toward nutrients and away from toxins to locate an optimal position in a gradient of stimuli. Light is one of the most important environmental factors that affect life on Earth. For photosynthetic organisms, light is an energy source, but too much light can induce DNA damage and photooxidative stress. Thus, it is sensible to hypothesize that phototaxis is an evolutionarily beneficial behavior, guiding cell movement toward (positive) or away from (negative) a light source to receive optimal light energy. Phototactic behavior has been observed in all domains of life (1). In prokaryotes phototaxis has been characterized in purple bacteria and haloarchaea that swim with flagella or archaella as well as in cyanobacteria, which move over moist surfaces using type IV pili (2, 3).
The unicellular coccoid cyanobacterium Synechocystis sp. strain PCC 6803 (hereafter, “Synechocystis”) has been studied extensively as a model for bacterial phototaxis (4, 5). Cyanobacterial phototactic signaling has been compared with that of chemotaxis in Escherichia coli, the best-studied bacterial taxis behavior (SI Appendix, Fig. S1A). The core signal-processing complex in E. coli consists of a methyl-accepting chemotaxis protein (MCP, chemoreceptor), a kinase (CheA), and an adaptor protein (CheW) (6). Binding of ligands to MCP regulates autophosphorylation of CheA, which can subsequently phosphorylate its response regulator, CheY. Phosphorylated CheY binds to the flagellar motor and changes the rotational direction of flagella, resulting in a reorientation of the cell. Dephosphorylation of CheY is catalyzed by a phosphatase, CheZ. An adaptation system consisting of methyltransferase CheR and methylesterase CheB confers a short-term memory for temporal comparison of ligand concentration, enabling cells to travel up or down the gradient (6–9). The core signal-processing complex is similar in Synechocystis phototaxis (SI Appendix, Fig. S1B), but instead of a chemical-binding domain, the MCP homolog has light-sensing domains at its N terminus (10). No homologs of CheR, CheB, or CheZ are found in the Synechocystis sp. strain PCC 6803 genome, indicating a different and still elusive mechanism of signal transduction in cyanobacterial phototaxis (11).
Cyanobacteria evolved a range of sensory photoreceptors that detect a rainbow of colors. Cyanobacteriochromes (CBCRs) comprise a class of phytochrome-related photoreceptors found only in cyanobacteria that have been reported to sense wavelengths that span the entire visible spectrum (12–15). CBCRs use GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA) domains for photoreception and a C-terminal domain, such as a histidine kinase or an MCP domain, for signal output (12, 16). PixJ1 is a CBCR that mediates positive phototaxis in Synechocystis, and its inactivation results in negative phototaxis (11, 17). PixJ1 contains two GAF domains, but only the second carries the conserved Cys residue that covalently binds the bilin chromophore phycoviolobilin (PVB) (10). Purified PixJ1-GAF2 (PixJg2) switches between two conformational states that absorb either blue or green wavelengths (blue/green photocycle) when exposed to light, as does the GAF domain from the Thermosynechococcus elongatus photoreceptor TePixJ (18, 19).
Other photoreceptors that regulate positive or negative phototaxis have been reported in Synechocystis. PixD, a sensor that uses flavin adenine dinucleotide (FAD) as a cofactor in a domain called “BLUF” (sensors of blue light using FAD), regulates the direction of phototaxis through binding to its interaction partner PixE, which inhibits positive phototaxis in its monomeric state (20, 21). The photoreceptor UirS/PixA is a UV-A sensor that, together with its response regulator UirR/NixB and a PatA-like protein called “LsiR/NixC,” is involved in switching between positive and negative phototaxis (22). Moreover, UirS/UirR-regulated LsiR is required for negative phototaxis (23). The photoreceptor Cph2 is involved in the inhibition of phototaxis in the presence of blue light. Upon blue-light detection by its third GAF domain, Cph2 catalyzes c-di-GMP formation through a C-terminal GGDEF domain, which results in the inhibition of pili-based motility (SI Appendix, Fig. S1B) (24–26). It is still not clear whether any of the receptors described above is genuinely required for detecting light direction, because disruption of any of them results in a reversal of the direction of movement or general inhibition of motility, but not random movement, under directional light (3). A recent study demonstrated that Synechocystis cells sense a light source by acting as a microlens that focuses incoming light onto the membrane at the opposite side of the cell. Cells undergoing positive phototaxis move away from the focused-light spot and therefore move toward the light source (27). This model implies that the spherical shape is important for the mechanism of phototaxis for coccoid species and does not explain how phototaxis would occur in nonspherical species.
Although widely used for studies of circadian rhythms (28), light-regulated gene expression (29, 30), and metabolic engineering (31), the rod-shaped, unicellular model species Synechococcus elongatus PCC 7942 (hereafter, “PCC 7942”) does not exhibit some environmentally relevant and often interrelated behavioral responses, such as biofilm formation and phototaxis. It has been shown that the ability to form biofilms is encoded in the PCC 7942 genome but is locked in a permanently repressed state unless mutations in the type II secretion or type IV pilus assembly system are made that prevent the production or secretion of a repressing agent (32, 33). Phototaxis in the thermophilic rod-shaped T. elongatus has been reported (34), but the paucity of genetic tools for that organism has limited investigation. Here, we report a recent wild isolate of S. elongatus, the strain UTEX 3055, that readily forms biofilms under laboratory conditions and shows strong phototactic behavior under directional light exposure. The mechanism of phototaxis in this cyanobacterium relies on a single 5-GAF domain containing an MCP-like photoreceptor that localizes at the cell poles. Despite structural and spectroscopic similarities to the blue/green photoreceptor PixJ of other cyanobacteria, this receptor alone is responsible for both negative and positive directional orientation. Like Synechocystis, UTEX 3055 focuses light with its rod-shaped cell, roughly opposite to the direction of incidence.
Results
A Wild S. elongatus Isolate Shows Photo-Induced Migration and Biofilm Formation.
To evaluate whether PCC 7942, which has been a domesticated laboratory strain for more than four decades (35), was once capable of phototaxis or other environmentally relevant behaviors, we isolated a wild strain of S. elongatus. Cyanobacterial cells were enriched from rock-attached scum in Waller Creek at the University of Texas, Austin, where the species had been isolated previously (36, 37). Cells of the wild isolate have a morphology, as observed by light microscope, very similar to that of PCC 7942 (SI Appendix, Fig. S2). Sequencing and assembly of the genome of this wild isolate (hereafter, “UTEX 3055”) revealed a 98.46% average nucleotide identity (ANI) with PCC 7942. Based on their identical 16S rRNA sequences and an ANI >95%, we concluded that they are the same species. UTEX 3055 has chromosomal insertions and deletions relative to PCC 7942, a large plasmid homologous to pANL of PCC 7942 but with expansions, and a 24-kb plasmid (GenBank accession nos.: CP033061, CP033062, and CP033063). UTEX 3055 also has an inversion in the chromosome relative to PCC 7942 that has been previously described for another domesticated laboratory strain, S. elongatus PCC 6301 (38). UTEX 3055 has 38,774 SNPs compared with PCC 7942, of which 9,446 predict nonsynonymous amino acid substitutions (SI Appendix, Table S1). Because PCC 7942 is easily manipulated genetically and because it is the premier model organism for bacterial circadian rhythms, we first examined whether the genetic tools used for PCC 7942 would work in the wild isolate and whether UTEX 3055 exhibits similar circadian rhythms. The UTEX 3055 genome includes the entire set of S. elongatus clock genes (SI Appendix, Table S2). When transformed with the same luciferase-reporter vector used for PCC 7942 circadian studies and with the same transformation protocol developed for PCC 7942, UTEX 3055 produced rhythmic patterns of bioluminescence, demonstrating both natural competence and the presence of a functional circadian clock system (SI Appendix, Fig. S3).
Despite sharing high nucleotide identity, UTEX 3055 shows some interesting phenotypes that are absent in the laboratory strain. Wild-type PCC 7942 exhibits an entirely planktonic phenotype when grown in liquid medium and remains suspended indefinitely in the absence of agitation. Only when specific mutations are introduced does PCC 7942 tend to settle and form biofilms (32). In contrast, wild-type UTEX 3055 cells flocculate and form biofilms on the wall of a culture flask under the same conditions (Fig. 1A). Moreover, UTEX 3055 exhibits strong taxis toward a lateral directional light source, whereas PCC 7942 does not (Fig. 1B). As observed in Synechocystis (39, 40), UTEX 3055 forms finger-like projections directed toward the light source on soft agarose plates (Fig. 1C and Movies S1–S3). Because bacterial movement can be monitored by microscopy (SI Appendix, Fig. S4), we tested whether individual UTEX 3055 cells respond to a light-intensity gradient projected onto the surface by placing a dark filter on one side of the light field. The light gradient that was generated parallel to the plane of the surface by a perpendicular light source resulted in migration that is significantly different from the biased movement under lateral directional light (Fig. 1 D and E and compare Movie S4 with Movie S5) but still exhibits directionality compared with a uniform circular distribution (Hodges–Ajne test). The results suggest that cells may sense light direction rather than a gradient of intensity during phototaxis, but more refined experiments are needed to exclude the effect of light scattering caused by the agarose medium. In contrast, PCC 7942 cells were minimally motile and had a random direction of movement under directional light when observed in the microscope (Movie S6), which is consistent with the lack of phototaxis on agarose plates (Fig. 1B).
Fig. 1.
Biofilm formation and phototaxis phenotypes in S. elongatus UTEX 3055. (A) Biofilm formation in S. elongatus UTEX 3055 and PCC 7942. The purple color after crystal violet staining indicates the formation of biofilm by UTEX 3055. (B) Phototaxis assays with S. elongatus UTEX 3055 and PCC 7942. Cells were spotted onto BG-11 soft agarose pads and provided with directional white fluorescent light illumination (28 µmol photons⋅m−2⋅s−1), indicated by the light bulb symbol. (C) Formation of finger-like projections by UTEX 3055. Cells were grown under directional light for 10 h on BG-11 soft agarose pads before imaging. (D) End-to-end displacement of cells moving in a projected light gradient from 0 to 20 µmol photons⋅m−2⋅s−1 and showing no significant directional bias (Left) or moving directionally when illuminated from an LED light source (broadband, peak 470 nm; Schott LLS) at a 45° angle with intensity of 50 µmol photons⋅m−2⋅s−1 (Right). Sample size (n) and end-to-end mean resultant length (r) values are indicated. A circular Kuiper test showed the r values from the two measurements are significantly different (P < 0.001). (E) Frame-to-frame change of mean resultant length (r) (overall movie average) from a Rayleigh test over time from the two plots in D; r = 0 indicates perfect nondirectionality, and r = 1 indicates maximal clustering in one direction. The apparent oscillation pattern may represent noise in the measurements.
Examination of the distribution of cell speed after transferring UTEX 3055 from a 12-h light:12-h dark environment to a constant-light environment showed no time-of-day variation (SI Appendix, Fig. S5), indicating that UTEX 3055 motility is not under circadian control.
Genetic Identification of a Phototaxis Operon.
The ability to manipulate UTEX 3055 genetically with tools and techniques developed for PCC 7942 enables the dissection of the molecular mechanism of UTEX 3055 phototaxis. In other organisms phototaxis-signaling components are encoded by chemotaxis-like (Che) genes that are often encoded in operons. Two Che operons, named “tax1” and “tax2” to follow the nomenclature used in Synechocystis, were found in both the PCC 7942 and UTEX 3055 genomes (Fig. 2A). Both operons encode homologs of an MCP, CheA and CheW, whereas cheY is present only in tax1. One gene in the tax1 operon (Synpcc7942_0858/UTEX3055_0948) is predicted to encode a protein with N-terminal GAF domains and a C-terminal MCP domain, similar to the Synechocystis protein PixJ1 that mediates positive phototaxis (Fig. 2A) (10). However, the version of this gene in UTEX 3055, also present in PCC 7942, encodes five contiguous GAF domains that all have the potential to bind a chromophore, whereas Synechocystis PixJ1 carries a single bilin-binding GAF domain (10). Thus, it is reasonable to speculate that the Synpcc7942_0858 protein (hereafter, “PixJSe”) functions as a photoreceptor that mediates phototaxis in UTEX 3055. To test this hypothesis, a pixJ disruption mutant was created by transforming UTEX 3055 with a mutagenic cosmid that carries a transposon insertion in pixJ from the PCC 7942 uni-gene set (UGS) library (41, 42). Indeed, inactivation of pixJ resulted in loss of phototaxis in UTEX 3055 (Fig. 2B). Disruption of other genes in the tax1 operon, such as Synpcc7942_0859 (CheA-like; hereafter, “PixLSe”), Synpcc0855 (CheY-like; PixGSe) or Synpcc0856 (CheY-like; PixHSe), also led to nonphototactic phenotypes (Fig. 2B). However, disruption of Synpcc7942_0857 or Synpcc0860 (both are CheW-like; hereafter “PixISe-1” and “PixISe-2”) individually did not affect phototaxis, nor did individual disruptions of the MCP-like (Synpcc7942_1015), CheA-like (Synpcc7942_1014), or CheW-like (Synpcc7942_1016) genes in the tax2 operon (Fig. 2B). Proficiency for phototaxis in the mutants that are disrupted upstream of pixJ argues against polar effects contributing to the pixJ phenotype. To verify that the nonphototactic phenotype is caused by mutations in tax1 genes, a shuttle vector that carries the corresponding intact UTEX 3055 ORF was recombined into the chromosome of respective mutants at neutral site I (NS1) (43). Introduction of pixJ, pixL, pixG, or pixH restored the respective mutant’s phototaxis; however, complementation by pixJ and pixH was observed only at specific expression levels (Fig. 2C and SI Appendix, Fig. S6 B and C). The extent of phototaxis was rarely restored to wild-type levels by expression of a gene from an ectopic site, but because the mutants designated as nonphototactic showed no biased movement in more than 10 assays, even modest restoration of biased movement was scored as complementation.
Fig. 2.
Chemotaxis-like operon tax1 is responsible for UTEX 3055 phototaxis. (A) Gene organization of chemotaxis-like operons in PCC 7942. Short black bars over genes indicate SNPs in UTEX 3055 (tax1 ORF: UTEX3055_0945-0950). The domain organization of MCP-like proteins is depicted below each operon. Gray rectangles represent a transmembrane domain. A, CheA; HAMP, domain found in histidine kinases, adenylyl cyclase, methyl-accepting proteins, and phosphatases; HP, hypothetical protein; MA: methyl-accepting chemotaxis-like domains; W, CheW; Y, CheY. (B) Phototaxis phenotypes of tax1 and tax2 mutants. A solid triangle indicates a Tn5-insertion mutant, and an open triangle indicates a deletion mutant. (C) Complementation of tax1 phototaxis mutants. Phototaxis phenotypes of pixG, pixH, pixJ, and pixL mutants expressing the respective gene integrated at NS1, as indicated after a slash and with a plus sign. Wild-type UTEX 3055 strains expressing an SpSm cassette and lacking pixJ served as positive and negative controls, respectively. See SI Appendix, Fig. S6A for the experimental setup.
Because wild-type UTEX 3055 cells switch from directional movement under directional light to nondirectional movement in the dark (SI Appendix, Fig. S7), we tested whether motility that is unrelated to direction is also affected in the phototaxis mutants by assessing the speed of movement of the pixJ mutant while applying a dark pulse. The results showed that the mutant cells are motile and move in random directions under both light and dark conditions (SI Appendix, Fig. S7). This experiment demonstrates that tax1 does not control type-IV pilus biogenesis and function in S. elongatus. Throughout this study we found that motile but nonphototactic mutants yield varying colony phenotypes in phototaxis assays. Sometimes all the cells appeared to be trapped at the inoculation site, forming a spot with a smooth edge; alternatively, some cells appeared to burst out in a few random or in all directions and sometimes formed twisted tree-like branches (SI Appendix, Fig. S8). We speculate that this phenotypic variability relates to differences in agarose surface conditions. Because disruption of pixJ or pixL causes loss of phototaxis in UTEX 3055, we asked whether the nonphototactic phenotype of PCC 7942 is due to impairment of key proteins encoded in the tax1 operon. However, we found that both pixJ and pixL from PCC 7942 restore phototaxis when expressed in their respective UTEX 3055 mutants (SI Appendix, Fig. S9A), indicating that loss of phototaxis in PCC 7942 is caused by factors other than genetic differences in the photoreceptor or kinase genes.
PixJSe Is a Blue/Green-Sensing DXCF Subfamily CBCR Photoreceptor.
PixJSe harbors five contiguous GAF domains at its N terminus. Alignment and clustering of the sequences of these domains with GAF domains from other CBCR and phytochrome proteins demonstrates that the GAFs of PixJSe are most similar to each other, followed by a high degree of similarity with the previously described DXCF/CBCR/GAF domains PixJg2 from Synechocystis and the single GAF from TePixJ of T. elongatus, all of which contain two conserved Cys residues (SI Appendix, Fig. S10 A and B). The DXCF subclass of CBCRs detects blue/green light via its PVB chromophore (12); PVB is derived by chemical isomerization of the precursor phycocyanobilin (PCB) following its covalent attachment to the protein via thioether linkages to the two conserved Cys residues (44). The first Cys is essential for binding the chromophore, and the second Cys is essential for detecting light at shorter wavelengths (12, 45). Light excitation triggers photoisomerization of the chromophore’s C15, C16 double bond, which leads to changes in chromophore–protein interaction that control signal transmission to the output domain. To determine whether any of the PixJSe GAF domains also binds a chromophore, we assayed for fluorescence of covalently bound bilin chromophores in SDS/PAGE gels under UV light in the presence of zinc acetate (46). Protein gel electrophoresis of a wild-type UTEX 3055 lysate showed multiple fluorescent bands, indicating the presence of chromophore-bound proteins, including the abundant light-harvesting phycobiliproteins and minor species (SI Appendix, Fig. S10C). We identified one of these minor bands as PixJSe based on the protein’s predicted size of 153 kDa, the absence of this band in the pixJ-knockout strain, and the band’s reappearance upon complementation (SI Appendix, Fig. S10C). Consistent with complementation data demonstrating that PixJSe from PCC 7942 is functional (SI Appendix, Fig. S9A), a PCC 7942 lysate also produces a fluorescent PixJ band in this assay (SI Appendix, Fig. S9B). These results demonstrate that PixJSe is a chromophore-bound protein that has the potential for light sensing.
To determine the absorption spectrum of PixJSe, we heterologously expressed a single GAF domain, GAF2, to avoid the difficulties of purifying a large membrane protein. The well-studied cyanobacterial phytochrome Cph1 was also purified as a positive control (47). Recombinant C-terminally tagged GAF2 (PixJSeGAF2-His) was expressed in PCB-producing E. coli and purified by nickel-affinity chromatography (SI Appendix, Fig. S10D). The presence of a covalently bound chromophore in both proteins was confirmed by zinc-induced fluorescence on SDS/PAGE (SI Appendix, Fig. S10E). The UV-VIS absorption and difference spectra of PixJSeGAF2-His show two major peaks at 429 nm and 529 nm (Fig. 3 A and B), corresponding to blue-absorbing (Pb) and green-absorbing (Pg) states, respectively. The Pb state of PixJSeGAF2-His appeared yellow after green-light exposure, which was efficiently converted to magenta after blue-light irradiation (Fig. 3A, Inset). A dark-reversion experiment showed that, after blue-light irradiation, the Pg state is slowly converted to the Pb state in the dark (Fig. 3C). In contrast, the green-light–induced Pb state was stable in the dark (Fig. 3D). These experiments demonstrate that PixJSeGAF2-His photoconverts between the Pb and Pg states, where Pb is the dark-adapted state and Pg is the photoproduct, similar to the previously studied blue/green sensors PixJg2, TePixJg, and cce_4193g2 (10, 18, 48). Based on the sequence conservation of the five GAF domains in PixJSe, we hypothesize that the characterized photoresponse of GAF2 is common for all five domains.
Fig. 3.
Blue/green light sensing by PixJSeGAF2 and blue/green-induced motility in UTEX 3055. (A) Absorbance spectra of PixJSeGAF2 after illumination with blue or green light (color-coded lines). Green-light–exposed protein showed a peak of blue-light absorption (Pb); blue-light–exposed protein showed a peak of green-light absorption (Pg). Absorption of buffer without protein, measured as a negative control (red curve), showed no absorption at tested wavelengths. (Inset) Exposure to blue light caused the protein to become visibly magenta (Pg, absorbs green light). Subsequent exposure of the same protein to green light changed the protein color to yellow (Pb, absorbs blue light); this process is reversible. (B) Difference spectrum of purified PixJSeGAF2 produced by subtracting the green spectrum from the blue spectrum in A. (C and D) Dark reversion of PixJSeGAF2 after exposure to blue (C) or green (D) light, measured at the indicated time points. (E) Phototactic movement of UTEX 3055 cells exposed to blue and green light. Cells were placed at four distances from the LED bulbs (represented by colored triangles below the images) to create a range of light fluences (indicated by the triangle at right). See SI Appendix, Fig. S11A for detailed experimental setup and lighting conditions.
Based on the blue/green photocycle of PixJSeGAF2-His, we speculated that the in vivo photosensory behavior of PixJSe involves sensing blue and green light during phototactic movement. To test this hypothesis, phototaxis assays were performed on wild-type UTEX 3055 under lateral directional illumination from different colors of lights. Blue light alone resulted in bidirectional migration of UTEX 3055 cells, with high intensities causing repulsion and low intensities causing attraction (Fig. 3E). In contrast, green-light exposure did not elicit directed cell movement, regardless of light intensity. However, the combination of blue and green light induced positive phototaxis in a manner that resembles that of white-light exposure. Lateral directional red-light illumination of wild-type UTEX 3055 significantly stimulated cell growth but did not induce any phototactic movement, and the same effect was observed for a red-and-green combination (SI Appendix, Fig. S11). Red and blue light together stimulated migration similar to that of blue light alone but with better cell growth and enhanced positive migration (SI Appendix, Fig. S11A). As a negative control, pixJ-knockout mutants did not respond to either blue or blue-and-green light (SI Appendix, Fig. S11B). These results are consistent with the blue/green cycles of PixJSe and further demonstrate the role of this photoreceptor in determining the direction of motility in response to blue and green illumination.
GAF Domain Signaling Is Important for Regulating the Direction of Cell Movement.
To better understand the role of the multiple GAF domains (GAF1–5) in PixJSe, we made Cys → Ala mutations in the first Cys residue, which is essential for chromophore binding (10, 49), in various GAF domains, singly and in combination. Most mutants that retain at least one bilin-binding GAF domain exhibited positive phototaxis to white light, like the wild type (SI Appendix, Fig. S12). These include single mutations in GAF1, GAF3, or GAF5 and almost all other double, triple, or quadruple mutants tested (SI Appendix, Fig. S12). Except for the quintuple GAF1–5 mutant, all mutants produced fluorescent bands upon SDS/PAGE zinc staining, demonstrating that PixJSe mutant proteins with at least one functional GAF domain continue to bind chromophores (SI Appendix, Fig. S13). The quintuple mutation of all GAF domains resulted in a nonphototactic phenotype similar to that of the pixJ-knockout mutant (Fig. 4 and SI Appendix, Fig. S12). Mutation of the GAF4 domain alone or both GAF4 and GAF5 resulted in negative phototaxis under light conditions where the wild type shows positive phototaxis (Fig. 4). Mutation of GAF domains other than GAF5 in addition to GAF4 restored positive phototaxis, indicating that integration of signals among the domains is important for signaling. Thus, PixJSe-mediated phototaxis requires at least one chromophore-bound GAF domain; the presence of multiple GAF domains enables switching between positive and negative directions of movement; and GAF4 is specifically important for positive phototaxis.
Fig. 4.
Amino acid substitution to prevent bilin binding, in GAF4 alone or in combination with GAF5, reverses the direction of phototactic movement of UTEX 3055. PixJSe variants were expressed from NS1 in the pixJ-deletion mutant as represented by sketches of the five GAFs (g1–g5). Intact GAF domains with bound chromophores (blue shapes) are shown in yellow, and mutated GAF domains (Cys → Ala) are shown in gray. The direction of the light and the resulting movement are indicated.
Bipolar Localization of Photoreceptor PixJSe.
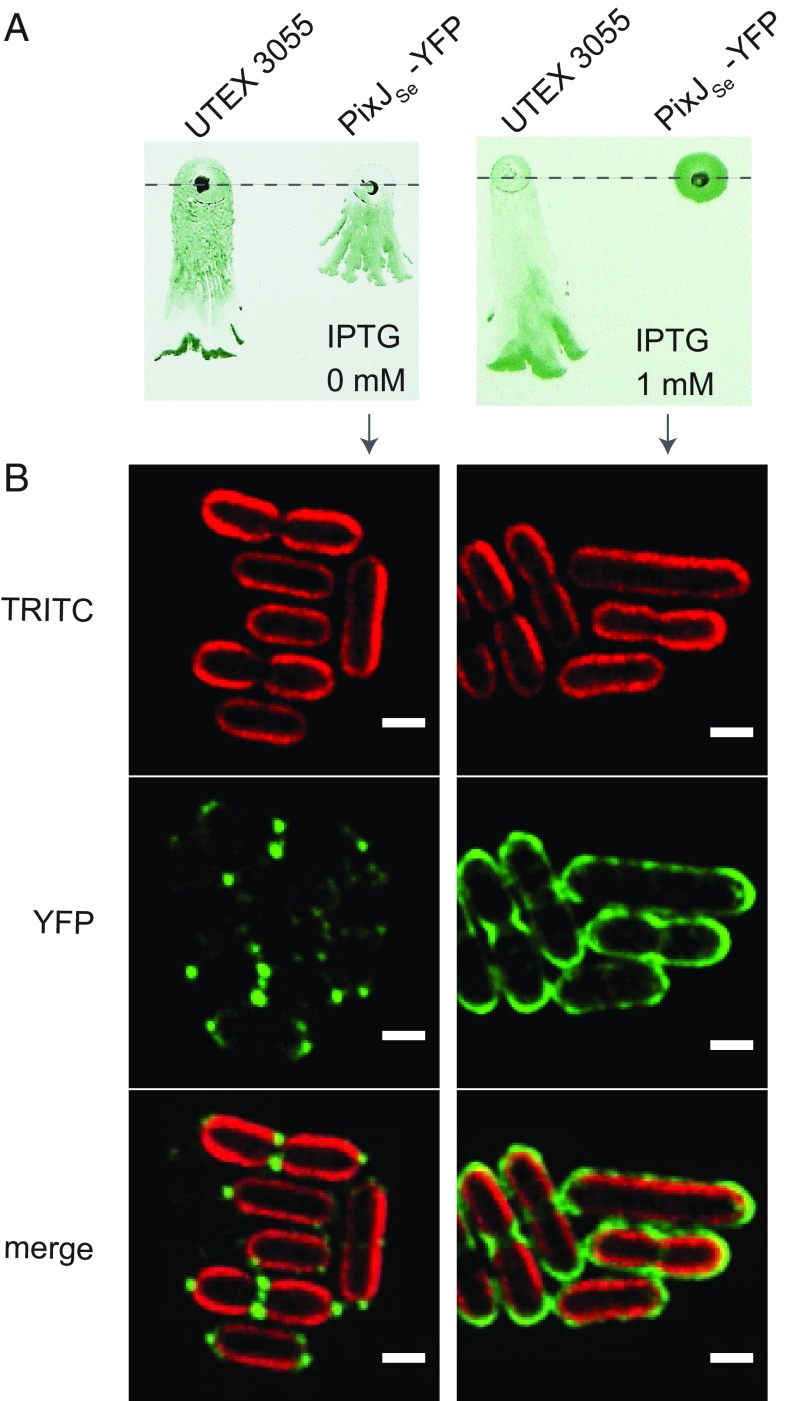
To explore the function of PixJSe further, we investigated whether this photoreceptor accumulates at a specific location within the cell. YFP was fused to the C terminus of PixJSe with a GSGGG linker and introduced into the UTEX 3055 pixJ mutant. An immunoblot showed the expression of full-length fused protein (SI Appendix, Fig. S14A), while zinc staining confirmed the presence of covalently bound chromophore (SI Appendix, Fig. S14B). YFP-tagged PixJSe appears to be fully functional in the cell because PixJSe-YFP restored the pixJ mutant’s phototaxis at uninduced (leaky) expression levels (Fig. 5A, Left). Notably, tight fluorescent clusters were observed on membranes primarily at cell poles (Fig. 5B, Left), similar to those seen for chemoreceptor complexes in E. coli (8). Note that this polar localization is not caused by the intrinsic localization of YFP, because previous studies showed that unfused YFP distributes homogeneously throughout the cytoplasm (50). However, overexpression of PixJSe-YFP caused a nonphototactic phenotype (Fig. 5A, Right), possibly due to the disruption of polar localization or stoichiometry of the signaling complex (Fig. 5B, Right). PixJSe-YFP also showed bipolar localization when heterologously expressed in E. coli (SI Appendix, Fig. S14C), indicating that polar localization either may use an evolutionarily conserved mechanism or is an inherent property of the protein.
Fig. 5.
Polar localization of photoreceptor PixJSe in UTEX 3055. (A) Phototaxis of UTEX 3055 that expresses PixJSe-YFP on soft agarose plates without (Left) or with (Right) 1 mM IPTG. (B) Fluorescent images indicating cellular localization of PixJSe-YFP in cells grown without or with 1 mM IPTG. (Top) TRITC autofluorescence from photosynthetic pigments. (Middle) YFP channel. (Bottom) Merge of TRITC and YFP channels. (Scale bars: 2 µm.)
Lensing Effect of Rod-Shaped S. elongatus Cells.
A compact complex is consistent with the hypothesis that PixJSe acts in conjunction with a cellular lens. The round cells of the cyanobacterium Synechocystis act as microlenses that focus incoming light onto the membrane at the back of the cell, enabling cells to sense the light direction (27). We tested whether such a lensing mechanism also operates in rod-shaped S. elongatus cells. Both PCC 7942 and UTEX 3055 exhibited a strong lensing effect under directional illumination (Fig. 6A, SI Appendix, Fig. S15, and Movie S7). Although the focusing effect was greatly reduced at an orientation of 45° and 90° to the light direction, this result shows that a rod-shaped cell is also capable of lensing, focusing incoming light onto the opposite side of the cell. Thus, the optical properties of the cell resemble those known from Synechocystis. Mathematical finite difference time domain (FDTD) simulations with an assumed uniform refractive index of 1.4 (51) of the cell immersed in water reproduced this lensing effect (Fig. 6B). Although a uniform refractive index of the cells is only an approximation, it seems that the microscopically observed lensing effects can be modeled sufficiently by this simulation approach. Notably, lensing was not affected in cells that lack PixJSe (Movie S8). As shown for Synechocystis, the cells did not respond to the projection of a light gradient, indicating that the stimulus is a directional light source rather than a spatial change in light intensity (Fig. 1D).
Fig. 6.
Lensing effect in rod-shaped S. elongatus. (A) S. elongatus UTEX 3055 cell imaged under oblique illumination from the left at the indicated cell-body orientation relative to the light direction. (Scale bars: 3 µm.) (B) FDTD simulation of light (460 nm) passing through rod-shaped UTEX 3055 cell at the orientation shown in A, using the actual cell size measured in SI Appendix, Fig. S2. The color indicates relative light intensity.
Discussion
UTEX 3055 Is a Useful Model Cyanobacterium for Studying Environmentally Relevant Behaviors.
The discovery of robust biofilm formation and phototaxis in an isolate of S. elongatus that can be studied using the extensive genetic tools developed for PCC 7942 provides an opportunity to understand environmentally relevant cyanobacterial behaviors. The functionality of pixJ and pixL genes of PCC 7942, when expressed in UTEX 3055 mutants, argues strongly that original isolates of PCC 7942 were also phototactic before laboratory propagation. Similarly, although wild-type PCC 7942 does not form biofilms under laboratory growth conditions, a number of mutations can enable biofilm formation by triggering the expression of proteins that contribute to the biofilm phenotype (32, 33). Taken together, the data suggest that domestication over four decades may have selected for either the loss or repression of these phenotypes. These losses have limited the use of PCC 7942 for studying complex behaviors that are both environmentally and perhaps commercially important. UTEX 3055 preserves the circadian clock system characterized in PCC 7942, is naturally competent, and performs homologous recombination in a manner similar to PCC 7942. In passaging the strain, we have been mindful of our culturing techniques to prevent the loss of these traits that are shared with PCC 7942 as well as the wild traits of biofilm formation and phototaxis (Methods). Because of the high sequence identity between the strains, a wide array of genetic tools developed for PCC 7942 can be directly applied to UTEX 3055, establishing the wild strain as a model organism.
Phototaxis in S. elongatus Is Encoded by a Single Phototaxis Operon.
Mutagenic disruption of phototaxis pathway genes in the tax1 operon leads to loss of directional light-dependent migration in UTEX 3055. However, these phototaxis mutants and cells of PCC 7942 are still motile and migrate in random directions independent of the location of a directional light source (SI Appendix, Fig. S7 and Movie S6). These results indicate that the tax1 operon is responsible only for phototactic signaling. Mutation in tax2 genes did not affect phototaxis, and the stimuli to which this operon may be related are unknown. Notably, neither operon is required for cell motility, which is most likely mediated by peritrichous type-IV pili based on the moving behavior of UTEX 3055 on the agarose surface (Movie S7) and the observed distribution of pili in PCC 7942 (52). We suspect that the motility of phototaxis mutants causes the observed variation in colony shapes on the semisolid agarose surface in different phototaxis assays, frequently observed as out-grown bursts (Figs. 2B and 4 and SI Appendix, Fig. S8). These bursts occur in random directions with respect to the orientation of lateral illumination, suggesting these are not phototactic migrations caused by second-site suppressor mutations. A reconstruction experiment showed that a minimum of 1 in 100 cells must be phototactic to form functional moving structures toward a light source. This ratio is unlikely to be reached by a spontaneous second-site suppression mutant that emerges during the course of the assay. Synechocystis contains a similar phototaxis operon in its genome, but disruption of the genes in the homologous operons of Synechocystis results in either reversal of phototactic direction or a nonmotile phenotype due to the loss of type-IV pili (11, 17). This comparison suggests that Synechocystis integrates stimuli through competing directional sensing or signaling pathways, one for positive phototaxis and others for negative phototaxis, whereas the single tax1 operon in UTEX 3055 is solely responsible for directional migration. The PixJSe and PixLSe homologs encoded in the genome of PCC 7942 are functional, indicating that the loss of directional movement results from other defects, such as decreased motility (Movie S6) or exopolysaccharide secretion or in downstream signaling to relay the directionality of light.
Bidirectional Phototaxis Is Regulated by Integration of Light Stimuli in a Single Photoreceptor.
S. elongatus UTEX 3055 shows bidirectional responses to lateral directional blue-light stimulation, with strong intensities repelling and weak intensities attracting the cells. Repulsion by high blue-light intensities is reasonable because blue wavelengths can cause cellular damage. However, insufficient light is also detrimental to the photosynthetic organism, consistent with positive phototaxis under weak blue-light intensities. The results show that S. elongatus seeks optimal light conditions by adjusting its location to tune the intensity of blue wavelengths contained in the light source. S. elongatus inhabits aquatic environments, where the components and intensity of light vary throughout the diel cycle and in a depth-dependent manner. In such a complex environment, cells in a microbial consortium use phototaxis to microadjust their locations in a biofilm to enhance exposure to sunlight for photosynthesis while avoiding damage caused by too much light. We hypothesize that the intensity of blue light relative to other colors, which are attenuated by depth in the water column, acts as a signal to determine the direction of phototactic movement. Short-wavelength light-induced bidirectional phototaxis has also been observed in Synechocystis, probably mediated by the blue-light–sensing photoreceptors PixJ1, Cph2, and PixD/PixE (10, 21, 24, 25) and a UV-A–responsive UirS/UirR-controlled LsiR expression system (23). The presence of multiple sensing systems indicates the importance of blue/UV sensing, which may be a common strategy used by cyanobacteria to find optimal light conditions. Similar bidirectional regulation strategies are observed in other taxis systems, such as pH taxis and thermotaxis (53, 54). Green light alone did not induce any phototactic movement in UTEX 3055, but blue and green light presented together stimulated strong positive phototaxis, similar to that of white light. This result correlates well with the blue/green absorbance of purified PixJSeGAF2. Notably, Synechocystis shows positive phototaxis to green-light stimulation (23, 55). Such different responses to blue and green light in Synechocystis and S. elongatus may reflect differences in their natural habitats and/or the use of multiple photoreceptors to guide taxis in Synechocystis.
The most striking difference among the photoreceptors PixJSe, PixJ1 of Synechocystis, and TePixJ of T. elongatus is the presence of five DXCF/GAF domains at the N terminus of the UTEX 3055 protein as compared with only one DXCF/GAF domain in PixJ1 and TePixJ. An additional functional complexity is evident in the S. elongatus protein, in which removal of chromophore attachment through Cys mutations in GAF4 or GAF4/GAF5 resulted in negative phototaxis, indicating that bidirectional phototaxis can be achieved through the integration of signals detected by a multi-GAF–containing photoreceptor. However, we cannot exclude the presence of other photoreceptors that contribute to phototaxis. No other combination of GAF4 and other GAF-domain mutations resulted in negative phototaxis, suggesting that GAF4-mediated conformational changes play a critical role in reversing the output signal from more N-terminal GAF domains. It should be noted that we have not presented direct evidence to show that the other four GAF domains (GAF1, -3, -4, and -5) have the same photocycle as GAF2; however, given the sequence similarity of these domains to each other, to TePixJ, and to PixJg2 (SI Appendix, Fig. S10B), it is likely that all the PixJse GAF domains have blue/green photocycles and bind PVB. This result also demonstrates that the presence of multiple GAF domains in PixJSe is neither a matter of simple redundancy nor solely for the purpose of signal addition and amplification. Instead, the multiple GAF domains appear to play a regulatory role in controlling the direction of movement. Numerous multiple-GAF CBCRs are encoded in genomes throughout the cyanobacterial clade. In filamentous Nostoc punctiforme, photoreceptor NpF1883 contains three chromophore-bound GAF domains (NpF1883g2/3/4) that all show similar blue/teal photocycles (14). An MCP-like photoreceptor in Nostoc punctiforme, PtxD, contains six chromophore-bound GAF domains, all with different photocycles (15). Loss of PtxD has been reported to cause loss of phototaxis of motile filaments called “hormogonia” (56), a phenotype similar to that observed for PixJSe in this study. These examples suggest that multi-GAF photoreceptors present a common mechanism for regulating the direction of phototaxis.
How Do Rod-Shaped Cells Determine the Direction of a Light Source?
Spherical Synechocystis cells physically sense light direction by acting as a microlens that focuses the incoming directional light on the portion of the membrane at the opposite side of the cell. Cells then actively move away from the bright spot, resulting in positive phototaxis (27). Although not spherical in shape, S. elongatus also lenses light. However, it is not clear yet whether this lensing effect is used by the cell to determine the direction of incoming light. Interestingly, PixJSe localizes to the cell poles in a pattern that is similar to that of chemoreceptors in E. coli and some other prokaryotic species (57). Delocalization of PixJSe and its redistribution throughout the cell membrane was concurrent with the abolition of phototaxis (Fig. 5). Similar polar localization of the PixJ homolog was reported in T. elongatus (58). We hypothesize that lensing of incoming light on localized photoreceptor complexes enhances directional light detection and orientation to enable phototaxis.
Methods
Bacterial Strains, Growth Conditions, and DNA Manipulation.
The plasmids and strains used in this study are described in SI Appendix, Tables S2 and S3. All S. elongatus strains were cultured in BG-11 medium as previously described (43), illuminated with 70–150 µmol photons⋅m−2⋅s−1 fluorescent light. Plasmids were constructed using the GeneArt Seamless Cloning and Assembly Kit (Life Technologies) and propagated in E. coli DH5α with antibiotics. Cyanobacterial mutants were generated by transforming with knockout vectors or corresponding PCC 7942 UGS library plasmids (41, 42). Cys → Ala substitutions were introduced into the GAF domains by site-directed mutagenesis (Pfu Turbo DNA polymerase; Agilent), and all constructs were verified by sequencing. Homogenous segregation of alleles was confirmed by PCR for all knockout mutants.
UTEX 3055 Isolation.
S. elongatus UTEX 3055 was isolated from Waller Creek at The University of Texas at Austin at the following GPS coordinates: latitude 30°17′5.636ʺN, longitude 97°44′4.501ʺW. Samples were given three designations based on collection location: P1, collected from a small pool of water in rocks; M1, collected from soil particles and water from embankment mud; and R1, collected by scraping blue-green growth off rocks at the waterline. A 4-mL aliquot from each sample location was used to inoculate each of nine 50-mL glass culture tubes containing 40 mL of sterile Kratz–Myers C medium (KMC) (36). The tubes were placed on a temperature gradient (38.5–51 °C) under a 12-h light:12-h dark diel cycle and bubbled with 1.5% CO2 in air. Cultures were screened by microscopy for the presence of putative strains of Synechococcus spp. Based on the results of this screening, serial dilutions (1:10–1:1,000) from the P1 isolate grown at 42 °C were plated on KMC agar supplemented with 25 µg/mL cycloheximide. Unialgal colonies were picked from plates after 17 d and were used to inoculate 10-mL volumes of KMC. All P1 isolates were maintained at 42 °C under a 12-h:12-h diel cycle. The internal transcribed spacer (ITS) regions of unialgal isolates were amplified by PCR and sequenced (59). BLAST searches against the National Center for Biotechnological Information database identified one unialgal isolate with 99% similarity to the ITS region of PCC 7942.
The unialgal strain, archived as AMC2389, was made axenic through streaking for single colonies; the lack of other bacteria was verified using BG-11 plates containing 0.04% (wt/vol) glucose and 5% (vol/vol) LB broth. The axenic strain was archived as AMC2388. To avoid domestication, UTEX 3055 culturing was alternated in liquid and on solid medium, and the strain was revived from the same frozen stock every 6 mo. We observed that continuous liquid culture promotes loss of biofilm formation, and passaging exclusively on standard hard-agar plates promotes loss of phototaxis.
Sequencing, Assembly, and Annotation of UTEX 3055.
The UTEX 3055 genome was sequenced on the Illumina MiSeq platform using a paired-end library construction with a 300-bp insert size as well as with PacBio RS Single Molecule Real-Time sequencing. A 2.76-Mbp chromosome and an 89-kb plasmid were assembled from PacBio sequence data using Canu (60). Illumina MiSeq reads were mapped to the assembly, and unmapped reads were extracted and assembled into a 24-kb plasmid using SPAdes (61). The assembly was corrected for small errors obtained with Illumina data using Pilon (62). Contigs were circularized using Circlator (63). The finished assembly was annotated by JGI (64), and annotation was further refined manually.
Phototaxis Assay.
BG-11 medium solidified with 0.3% agarose (wt/vol) and 10 mM sodium thiosulfate was used for phototaxis assays. Wild-type S. elongatus and mutants grown in liquid BG-11 were adjusted to an OD750 of 0.6–1.0, and 2-µL samples of culture were spotted at specific positions on the surface of agarose plates. After inoculation, the plates were placed in a dark box with one side opening toward a fluorescent light. Photographs were taken after 3–5 d of incubation. Phototaxis to LED light was performed as shown in SI Appendix, Fig. S11A.
Expression and Purification of GAF-Domain Protein.
E. coli strain LMG194, which contains plasmid pPL-PCB for synthesis of PVB, was transformed with pAM5498 for expression of PixJSeGAF2-His or with pBAD_Cph1 (56) as a positive control. Overnight cultures were used to inoculate 100 mL of LB medium containing 50 µg/mL ampicillin and 12 µg/mL kanamycin to an OD600 of 0.5. After about 8 h of growth at 37 °C, this 100-mL culture was added to 900 mL of LB medium supplemented with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) to induce PCB expression. After a 1-h incubation, L-arabinose was added to a final concentration of 0.04% (wt/vol) to induce PixJSeGAF2-His expression (0.02% for Cph1), and incubation was continued at 37 °C for 5 h. Cells were harvested by centrifugation at 3,795 × g for 10 min at 4 °C, and pellets were frozen at −20 °C or were immediately resuspended in lysis buffer (50 mM Tris, 500 mM NaCl, and 20 mM imidazole, pH 8.0). Cells were lysed with a homogenizer (Emulsiflex C3; Avestin) at 15,000–20,000 psi for 10 min and then were subjected to centrifugation at 32,500 × g for 40 min to remove cell debris. The soluble fraction of lysates was incubated with an Ni-NTA gravity column (Qiagen). The unbound proteins were removed by washing with 50 mL lysis buffer, and bound protein was recovered with 10 mL of elution buffer (50 mM Tris, 500 mM NaCl, and 250 mM imidazole, pH 8.0).
Zinc Blots.
Chromophore incorporation was assayed as previously reported (46). SDS/PAGE gels were soaked in 100 mM zinc acetate with gentle shaking for 30 min in the dark. The zinc-impregnated gel was then irradiated with 305-nm UV light, and the fluorescent bands were recorded with a FluorChem HD2 system (Alpha Innotech).
Spectroscopy.
Cuvettes containing a 500-µL protein solution were exposed to blue or green LED light for 2 min, and then absorption was measured immediately with a UV-Vis spectrophotometer (NanoDrop 2000c; Thermo Scientific). All measurements were performed in the dark at room temperature.
Microscopy.
An Olympus IX71 inverted microscope with an attached WeatherStation environmental chamber was used for imaging and time-lapse movie production. Images were captured using a CoolSnap HQ2 CCD camera (Photometrics).
For images of PixJSe-YFP localization, a 2-µL sample of culture grown to an OD750 of 0.6 was fixed on an 1.2% agarose (wt/vol) pad in BG-11 for imaging. TRITC filters (excitation 555/28 nm and emission 617/73 nm) were used to image cyanobacterial autofluorescence. YFP filters (excitation 500/20 nm and emission 535/30 nm) were used to image PixJSe-YFP protein localization. Series of Z-stack images were taken and deconvolved using the softWoRx imaging program (Applied Precision). For time-lapse movies 2-µL cell samples were placed on a pad of 0.3% (wt/vol) agarose in BG-11, left to absorb into the agarose for 5 min, and then covered with a coverslip (SI Appendix, Fig. S4A). A 10× or 20× objective was used for monitoring projections of colony fingers. For tracking single-cell motility, a 40× or 100× objective was used (SI Appendix, Fig. S4B). Lateral illumination was provided by a white LED. Images were acquired at 0.5-s or 2-s intervals. To image cells in the dark, the LED light was turned off, and images were captured using the microscope’s transillumination lighting system (a 100-W halogen lamp), with a limited exposure time of 0.01–0.05 s.
Lensing Simulation.
The optical field distribution of blue light (460 nm) propagating through a single cell was calculated using FDTD simulations as previously described (27). A space grid of Δx = Δy = 5 nm was used for the simulation of a 5 × 5 µm square array. The refractive index of the cell was approximated to be 1.4, thus leading to an effective refractive index of 1.05 of the cell relative to that of water. The cell length of 3.25 µm with an area of 3.69 µm2 was determined by averaging microscopy images of cells (62 and 77 cells, respectively).
Supplementary Material
Acknowledgments
We thank Dr. J. S. Parkinson for providing the E. coli strain used for receptor localization, members of the James W. Golden and S.S.G. laboratories for helpful discussions, and the University of California, San Diego qBio Hacker Laboratory for instruments and equipment used in phototaxis assays. The research reported in this publication was supported by NIH Grant R35 GM118290 (to S.S.G.); National Science Foundation (NSF) and US‐Israel Binational Science Foundation Grant NSF‐BSF 2012823 (to S.S.G); NSF Grant 1201881 (to D.R.N.); an Excellence Initiative of the German Research Foundation Spemann Graduate School Award (to A.J.); and Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy Grant DOE DE-FG02-09ER16117 (to J.C.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CP033061, CP033062, and CP033063).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812871115/-/DCSupplemental.
References
- 1.Jékely G. Evolution of phototaxis. Philos Trans R Soc Lond B Biol Sci. 2009;364:2795–2808. doi: 10.1098/rstb.2009.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage JP, Hellingwerf KJ. Light-induced behavioral responses (;phototaxis’) in prokaryotes. Photosynth Res. 2003;76:145–155. doi: 10.1023/A:1024974111818. [DOI] [PubMed] [Google Scholar]
- 3.Wilde A, Mullineaux CW. Light-controlled motility in prokaryotes and the problem of directional light perception. FEMS Microbiol Rev. 2017;41:900–922. doi: 10.1093/femsre/fux045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaya D. Light matters: Phototaxis and signal transduction in unicellular cyanobacteria. Mol Microbiol. 2004;53:745–754. doi: 10.1111/j.1365-2958.2004.04160.x. [DOI] [PubMed] [Google Scholar]
- 5.Schuergers N, Mullineaux CW, Wilde A. Cyanobacteria in motion. Curr Opin Plant Biol. 2017;37:109–115. doi: 10.1016/j.pbi.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 7.Sourjik V, Wingreen NS. Responding to chemical gradients: Bacterial chemotaxis. Curr Opin Cell Biol. 2012;24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi S, Sourjik V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol. 2018;45:22–29. doi: 10.1016/j.mib.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Yoshihara S, Katayama M, Geng X, Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 11.Bhaya D, Takahashi A, Grossman AR. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci USA. 2001;98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA. 2011;108:11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose Y, Narikawa R, Katayama M, Ikeuchi M. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc Natl Acad Sci USA. 2010;107:8854–8859. doi: 10.1073/pnas.1000177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012;51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 15.Rockwell NC, Martin SS, Lagarias JC. Red/green cyanobacteriochromes: Sensors of color and power. Biochemistry. 2012;51:9667–9677. doi: 10.1021/bi3013565. [DOI] [PubMed] [Google Scholar]
- 16.Ikeuchi M, Ishizuka T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 17.Yoshihara S, Suzuki F, Fujita H, Geng XX, Ikeuchi M. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2000;41:1299–1304. doi: 10.1093/pcp/pce010. [DOI] [PubMed] [Google Scholar]
- 18.Ishizuka T, et al. Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol. 2006;47:1251–1261. doi: 10.1093/pcp/pcj095. [DOI] [PubMed] [Google Scholar]
- 19.Ishizuka T, Narikawa R, Kohchi T, Katayama M, Ikeuchi M. Cyanobacteriochrome TePixJ of Thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 2007;48:1385–1390. doi: 10.1093/pcp/pcm106. [DOI] [PubMed] [Google Scholar]
- 20.Okajima K, et al. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J Biochem. 2005;137:741–750. doi: 10.1093/jb/mvi089. [DOI] [PubMed] [Google Scholar]
- 21.Sugimoto Y, Nakamura H, Ren S, Hori K, Masuda S. Genetics of the blue light-dependent signal cascade that controls phototaxis in the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 2017;58:458–465. doi: 10.1093/pcp/pcw218. [DOI] [PubMed] [Google Scholar]
- 22.Narikawa R, et al. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011;52:2214–2224. doi: 10.1093/pcp/pcr155. [DOI] [PubMed] [Google Scholar]
- 23.Song J-Y, et al. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA. 2011;108:10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiedler B, Börner T, Wilde A. Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: Role of different photoreceptors. Photochem Photobiol. 2005;81:1481–1488. doi: 10.1562/2005-06-28-RA-592. [DOI] [PubMed] [Google Scholar]
- 25.Wilde A, Fiedler B, Börner T. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol Microbiol. 2002;44:981–988. doi: 10.1046/j.1365-2958.2002.02923.x. [DOI] [PubMed] [Google Scholar]
- 26.Savakis P, et al. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol. 2012;85:239–251. doi: 10.1111/j.1365-2958.2012.08106.x. [DOI] [PubMed] [Google Scholar]
- 27.Schuergers N, et al. Cyanobacteria use micro-optics to sense light direction. eLife. 2016;5:e12620. doi: 10.7554/eLife.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen SE, Golden SS. Circadian rhythms in cyanobacteria. Microbiol Mol Biol Rev. 2015;79:373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golden SS. Light-responsive gene expression in cyanobacteria. J Bacteriol. 1995;177:1651–1654. doi: 10.1128/jb.177.7.1651-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustos SA, Golden SS. Light-regulated expression of the psbD gene family in Synechococcus sp. strain PCC 7942: Evidence for the role of duplicated psbD genes in cyanobacteria. Mol Gen Genet. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]
- 31.Angermayr SA, Gorchs Rovira A, Hellingwerf KJ. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol. 2015;33:352–361. doi: 10.1016/j.tibtech.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Schatz D, et al. Self-suppression of biofilm formation in the cyanobacterium Synechococcus elongatus. Environ Microbiol. 2013;15:1786–1794. doi: 10.1111/1462-2920.12070. [DOI] [PubMed] [Google Scholar]
- 33.Parnasa R, et al. Small secreted proteins enable biofilm development in the cyanobacterium Synechococcus elongatus. Sci Rep. 2016;6:32209. doi: 10.1038/srep32209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondou Y, Nakazawa M, Higashi S, Watanabe M, Manabe K. Equal-quantum action spectra indicate fluence-rate-selective action of multiple photoreceptors for photomovement of the thermophilic cyanobacterium Synechococcus elongatus. Photochem Photobiol. 2001;73:90–95. doi: 10.1562/0031-8655(2001)073<0090:eqasif>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 35.Welkie DG, et al. A hard day’s night: Cyanobacteria in diel cycles. Trends Microbiol. December 5, 2018 doi: 10.1016/j.tim.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kratz WA, Jack M. Nutrition and growth of several blue‐green algae. Am J Bot. 1955;42:282–287. [Google Scholar]
- 37.Herdman M, Castenholz RW, Waterbury JB, Rosmarie R. Form-genus XIII. In: Boone DR, Castenholz RW, Garrity GM, editors. Synechococcus. Bergey’s Manual of Systematic Bacteriology. 2nd Ed. Springer; New York: 2001. pp. 544–546. [Google Scholar]
- 38.Sugita C, et al. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: Gene content and organization. Photosynth Res. 2007;93:55–67. doi: 10.1007/s11120-006-9122-4. [DOI] [PubMed] [Google Scholar]
- 39.Burriesci M, Bhaya D. Tracking phototactic responses and modeling motility of Synechocystis sp. strain PCC6803. J Photochem Photobiol B. 2008;91:77–86. doi: 10.1016/j.jphotobiol.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Varuni P, Menon SN, Menon GI. Phototaxis as a collective phenomenon in cyanobacterial colonies. Sci Rep. 2017;7:17799. doi: 10.1038/s41598-017-18160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holtman CK, et al. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 2005;12:103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Holtman CK, Taton A, Golden SS. Functional Genomics and Evolution of Photosynthetic Systems, Advances in Photosynthesis and Respiration. Springer; Dordrecht: 2012. Functional analysis of the Synechococcus elongatus PCC 7942 Genome; pp. 119–137. [Google Scholar]
- 43.Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- 44.Ishizuka T, et al. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry. 2011;50:953–961. doi: 10.1021/bi101626t. [DOI] [PubMed] [Google Scholar]
- 45.Rockwell NC, Martin SS, Lagarias JC. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry. 2012;51:3576–3585. doi: 10.1021/bi300171s. [DOI] [PubMed] [Google Scholar]
- 46.Berkelman TR, Lagarias JC. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 47.Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA. 2001;98:10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fushimi K, et al. Cyanobacteriochrome photoreceptors lacking the canonical Cys residue. Biochemistry. 2016;55:6981–6995. doi: 10.1021/acs.biochem.6b00940. [DOI] [PubMed] [Google Scholar]
- 49.Rockwell NC, et al. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry. 2008;47:7304–7316. doi: 10.1021/bi800088t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taton A, Ma AT, Ota M, Golden SS, Golden JW. NOT gate genetic circuits to control gene expression in cyanobacteria. ACS Synth Biol. 2017;6:2175–2182. doi: 10.1021/acssynbio.7b00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu PY, et al. An optofluidic imaging system to measure the biophysical signature of single waterborne bacteria. Lab Chip. 2014;14:4237–4243. doi: 10.1039/c4lc00783b. [DOI] [PubMed] [Google Scholar]
- 52.Whitman WB. Insights into the life of an oxygenic phototroph. Proc Natl Acad Sci USA. 2015;112:14747–14748. doi: 10.1073/pnas.1520098112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Sourjik V. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol Microbiol. 2012;86:1482–1489. doi: 10.1111/mmi.12070. [DOI] [PubMed] [Google Scholar]
- 54.Paulick A, et al. Mechanism of bidirectional thermotaxis in Escherichia coli. eLife. 2017;6:e26607. doi: 10.7554/eLife.26607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng W-O, Grossman AR, Bhaya D. Multiple light inputs control phototaxis in Synechocystis sp. strain PCC6803. J Bacteriol. 2003;185:1599–1607. doi: 10.1128/JB.185.5.1599-1607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell EL, et al. Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. J Bacteriol. 2015;197:782–791. doi: 10.1128/JB.02374-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gestwicki JE, et al. Evolutionary conservation of methyl-accepting chemotaxis protein location in bacteria and archaea. J Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondou Y, et al. Bipolar localization of putative photoreceptor protein for phototaxis in thermophilic cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2002;43:1585–1588. doi: 10.1093/pcp/pcf176. [DOI] [PubMed] [Google Scholar]
- 59.Boyer SL, Flechtner VR, Johansen JR. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol. 2001;18:1057–1069. doi: 10.1093/oxfordjournals.molbev.a003877. [DOI] [PubMed] [Google Scholar]
- 60.Koren S, et al. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker BJ, et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunt M, et al. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huntemann M, et al. The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4) Stand Genomic Sci. 2015;10:86. doi: 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.