Significance
Constitutive HIF-2α activity has been well demonstrated to promote cancer progression. A newly designed small molecule (PT2399) has been proven to inhibit HIF-2α through binding its PAS-B domain in recent studies. In the present study, we generated a mouse model featuring a cavity-filling point mutation (S305M), which fails to bind PT2399 in vivo. In addition to response to hypoxia stress, we show that HIF-2α PAS-B domain also mediates response to metabolic stress such as high fat diet, and PT2399 could ameliorate metabolic condition in obese animals, suggesting potential therapeutic applications for HIF-2α antagonists in metabolic disorders.
Keywords: HIF-2α antagonist, hypoxia, adipogenesis, obesity, nonalcoholic fatty liver disease
Abstract
Hypoxia-inducible factors (HIFs) are transcription factors in the basic helix–loop–helix PER-ARNT-SIM (bHLH-PAS) protein family that contain internal hydrophobic cavities within their PAS-A and PAS-B domains. Among HIFs, the HIF-2α PAS-B domain contains a relatively large cavity exploited for the development of specific artificial ligands such as PT2399. Administration of PT2399 could suppress HIF-2α target gene expression without affecting HIF-1 activity in mice under hypoxia conditions. A single mutation (S305M) within the HIF-2α PAS-B domain suppressed HIF-2α activity while conferring resistance to PT2399 in vivo, indicating the vital role of PAS-B domain in HIF-2α hypoxia response. In contrast, the mutant mice did not phenocopy PT2399 intervention in wild-type mice under metabolic stress. Under a high-fat diet (HFD), the mutant mice exert enhanced adipogenesis and obtain larger adipose mass and body weight gain compared to wild type. However, administration of PT2399 along with HFD feeding sufficiently suppressed HFD-induced body weight and adipose mass increase through suppression of adipogenesis and lipogenesis. The accompanying decreased lipid accumulation in the liver and improved glucose tolerance in wild-type mice were not observed in the mutant mice indicating negative regulation of HIF-2α on obesity and a complex role for the PAS-B domain in metabolic regulation. Notably, short-term administration of PT2399 to obese mice decreased adipose mass and improved metabolic condition. These results indicate a regulatory role for HIF-2α in obesity progression and suggest a therapeutic opportunity for PT2399 in obesity and associated metabolic disorders.
Hypoxia-inducible factors (HIFs) are a group of mammalian basic helix–loop–helix-PER-ARNT-SIM (bHLH-PAS) proteins that work as transcription factors responding to oxygen and other stress (1–4). The HIFs function as heterodimers composed of a stable expressed beta subunit HIF-1β (5) and an oxygen regulated alpha subunit including HIF-1α (6) and HIF-2α (7). Each subunit contains two PAS domains, designated PAS-A and PAS-B, which all contribute to the heterodimerization of the α and β subunit (8, 9). PAS domains frequently act as sensors where ligands bind to internal cavities induce allosteric changes affecting protein interactions at the surface (10, 11). As HIF dimerization is essential for DNA binding to initiate transcription, destabilization of protein–protein interactions in the system represents an optimal therapeutic approach for disease treatment, especially cancer (12).
Comparing to other HIFs, NMR and X-ray crystallography showed that HIF-2α PAS-B domain contains a fairly large hydrated cavity (290 Å3) that is amenable to ligand binding (8, 13). We previously identified an artificial ligand that binds within the pocket leading to structural and functional changes that suppress HIF-2α DNA-binding activity in cultured cells (14, 15), promoting the development of HIF-2α–specific antagonist PT2385 and its newest derivative PT2399 (16–18). Recent studies confirmed that PT2399 could bind the HIF-2α PAS-B domain with a Kd of 42.5 nM and presented high specificity and efficiency for treatment of clear cell renal cell carcinoma (ccRCC) (16, 17), and a clinical study with PT2385 is showing a favorable safety profile and is active in patients with heavily pretreated ccRCC (19). HIF-2α antagonist was designed to bind with PAS-B domain, and we have shown that a single amino acid substitution (S304M) within the human HIF-2α PAS-B domain could significantly attenuate binding of exogenous ligands including PT2399 through in vitro studies (17, 20), while in vivo validation remains poorly investigated.
In addition to cancer development, HIFs have been indicated as major regulators in multiple disease progression such as peripheral arterial disease, pulmonary arterial hypertension, and sleep apnea. (21, 22). Notably, regulatory roles of HIFs in obesity and insulin signaling have been revealed in recent studies (23, 24). Both HIF-1α and HIF-2α proteins were found to stably increase in adipose tissue of high-fat diet (HFD)-induced obese mice (24, 25). Both in vivo and in vitro studies have well presented the deleterious role of HIF-1α in regulation of adipose function (24, 26, 27). However, the involvement of HIF-2α remains obscure and controversial (25, 28, 29). Thus, in the current study, we generated the analogous S305M mutation within HIF-2α PAS-B domain in mice to explore the adaptive response of HIF-2α under hypoxia and metabolic stress; with the HIF-2α antagonist PT2399, we validated its on-target effect in vivo and explored its potential therapeutic approach for obesity and associated metabolic disorders.
Results
Mice Homozygous for the Epas1S305M Knock-In Allele Respond to Hypoxia but Not to PT2399.
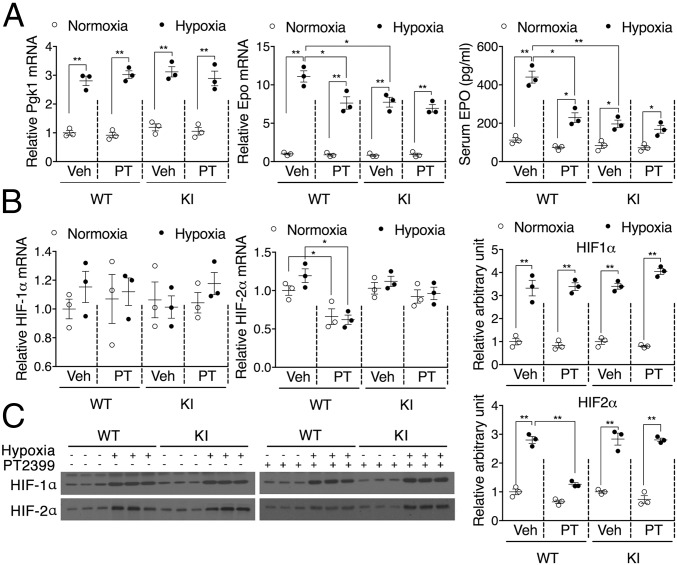
We previously reported that a single amino acid substitution (S304M) within the human HIF-2α PAS-B domain could attenuate binding of exogenous ligands such as PT2399 (17, 20). Here, we introduced the analogous S305M mutation (SI Appendix, Fig. S1) into the murine Epas1 gene locus. The resultant homozygous Epas1S305M/S305M knock-in (KI) mice are viable and healthy. To characterize their hypoxic response, WT and KI mice were housed at 9% O2 for 6 h. Hypoxic induction of both the HIF-1α selective target gene Pgk1 and the HIF-2α selective target gene Epo was observed in the kidneys of WT and KI mice. While Pgk1 expression was unaffected by the HIF-2α mutation, the magnitude of Epo induction was selectively suppressed in the KI mice (Fig. 1A). The effects of the KI mutation were mimicked in WT mice by administration of PT2399, which selectively attenuated the induction of Epo mRNA and protein accumulation. However, PT2399 treatment had no additional effect on Epo expression in the KI mice (Fig. 1A), demonstrating that the Epas1S305M allele confers resistance to HIF-2α antagonists in vivo. As expected, PT2399 had no effect on Pgk1 expression in either strain (Fig. 1A). Also of note, PT2399-induced decreases in HIF-2α expression were observed in WT mice at both the mRNA (Fig. 1B) and protein (Fig. 1C) levels. These on-target effects were not observed in the KI mice, suggesting that direct feedback regulation of the Epas1 gene may complement heterodimer antagonism by exogenous ligands in vivo.
Fig. 1.
Mice homozygous for the Epas1S305M KI allele respond to hypoxia but not to PT2399. WT and KI mice were pretreated with PT2399 at 30 mg/kg for 1 wk followed by hypoxia induction at 9% O2 for 6 h. (A) Hypoxic induction of a HIF-1 target gene (Pgk1) and expression of the HIF-2 target Epo at both the mRNA (kidney) and protein (serum) levels in WT and KI mice. (B) Expression of HIF-1α and HIF-2α mRNA in WT and KI mice. (C) HIF-2α protein accumulation in WT and KI mice. Data are presented as mean ± SEM. Significance between every two groups was calculated using unpaired two-tailed Student’s t test unless otherwise indicated. *P < 0.05, **P < 0.01; n = 3 per group.
Alteration of PAS-B Domain Promotes Adipogenesis Activity in Mice.
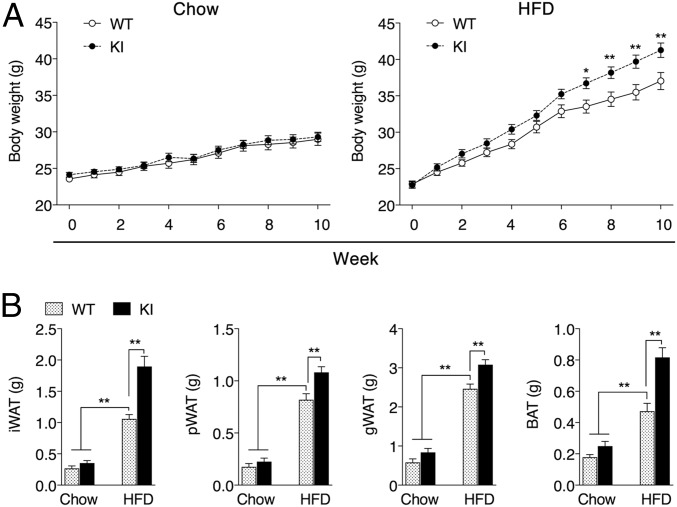
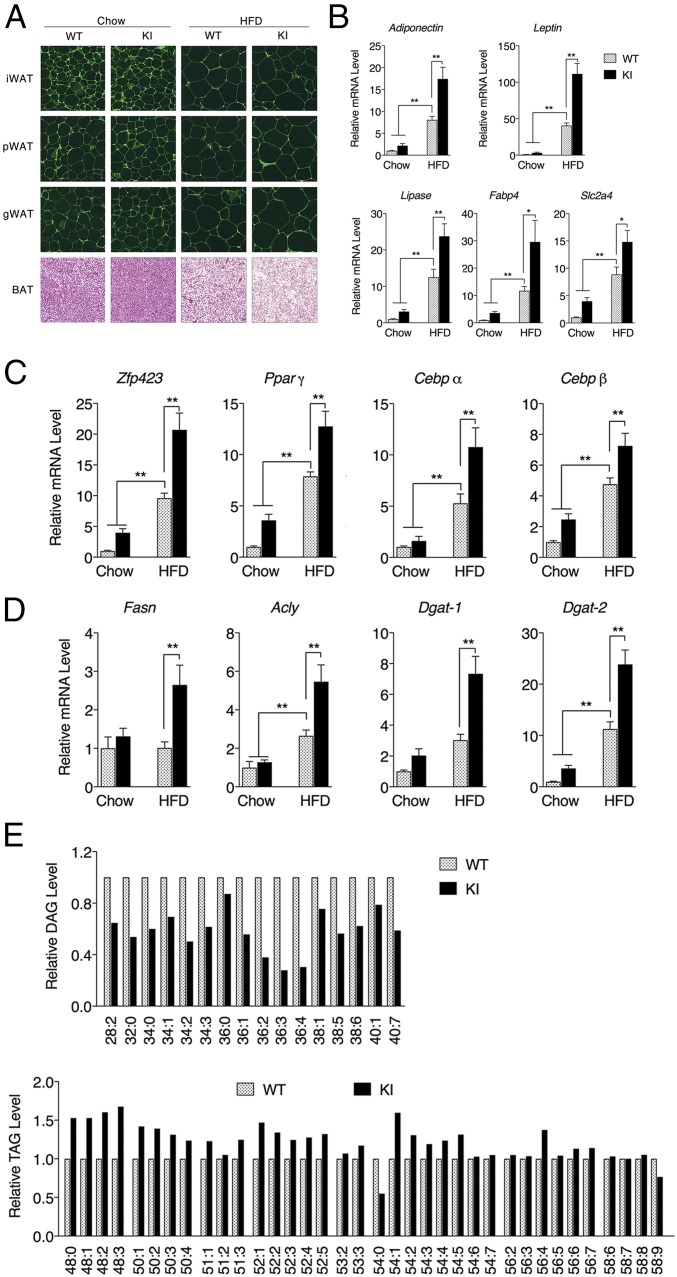
KI mice are generally indistinguishable from WT mice when maintained under typical laboratory settings. For example, when fed a standard chow diet (10% fat), total body weight (Fig. 2A) and fat pad masses (Fig. 2B) of the KI mice are equivalent to their WT counterparts. However, when challenged with an HFD (60%), KI mice gain more weight than WT mice (Fig. 2A) despite consuming the same amount of food (SI Appendix, Fig. S2). These differences are largely due to the increase in adipose tissue mass (Fig. 2B). Increased white adipose tissue (WAT) expansion during obesity progression was not due to the further enlargement of preexisting adipocytes (Fig. 3A). Instead, KI mice fed HFD have increased expression of genes encoding adipocyte markers and adipogenesis promoting factors (Fig. 3 B and C), consistent with enhanced adipogenesis (30, 31). The differentiation of primary stromal vascular fraction (SVF) isolated from inguinal white adipose tissue (iWAT) of KI mice was likewise accompanied by increased expression of adipogenic factors and enhanced lipid accumulation relative to WT SVF (SI Appendix, Fig. S3). This increased adipogenic potential in KI mice iWAT could even be observed in mice fed a standard chow diet following exposure to 6 °C to induce browning (SI Appendix, Fig. S4). Along with an enhanced adipogenesic program, WAT from KI mice also displayed increased expression of lipogenic enzymes (Fig. 3D), including Dgat-1 and -2, which catalyze the formation of triacylglycerides (TAGs) from diacylglycerols (DAGs) and Acyl-CoA (32). Hepatic lipidomic analysis of KI mice fed HFD also reveals reduced levels of DAGs and increased levels of TAGs relative to WT mice (Fig. 3E and SI Appendix, Table S1). In analyzing the localization of HIF-2α in adipose tissue under a normal diet, we found that KI mice obtained more HIF-2α protein in the nucleus (SI Appendix, Fig. S5), we suspect that the mutation may promote HIF-2α nuclear translocation in adipocytes, which was exaggerated under HFD feeding accompanied by more stabilized HIF-2α protein due to the potential hypoxia condition (SI Appendix, Fig. S5). Compared with lean mice fed a standard chow diet, both WT and KI mice subjected to HFD-induced obesity manifested impaired glucose tolerance as early as 5 wk. However, no significant difference was observed between WT and KI mice with respect to their glucose tolerance, fasting glucose levels, or insulin sensitivity (SI Appendix, Fig. S6). Together, these results indicate that the cavity within the HIF-2α PAS-B domain contributes to the adipogenic program initiated in response to the HFD challenge. Altering the ligand-binding potential of this domain in the KI mice augments adigogenesis and lipogenesis without further exacerbation of glucose responsiveness, suggesting a relatively healthy increase in additional adipose expansion.
Fig. 2.
A metabolic phenotype is observed in KI mice fed a HFD. (A) Body weight of WT and KI mice fed standard chow (10% fat) or HFD (60% fat) for 10 wk beginning at 8 wk of age. (B) Adipose tissue mass of iWAT, pWAT, gonadal white adipose tissue (gWAT), and BAT after 10 wk of feeding. Data are presented as mean ± SEM. Significance between every two groups was calculated using unpaired two-tailed Student’s t test. Two-way ANOVA was performed for comparison of two groups over time followed by Sidak’s multiple test. *P < 0.05, **P < 0.01; n = 10 per group.
Fig. 3.
Expression of the Epas1S305M allele promotes increased adipogenesis in KI mice. (A) HE staining of iWAT, pWAT, gWAT, and BAT for WT or KI mice fed standard chow or HFD for 10 wk beginning at 8 wk of age. (False color was used to highlight cell boundaries.) (Magnification: A, 200×.) mRNA accumulation levels of genes in iWAT encoding adipocyte markers (B), adipogenesis promoting factors (C), or lipogenic enzymes (D) following 10 wk of feeding were measured by qPCR. n = 8 per group. (E) Relative hepatic DAG and TAG levels between WT and KI mice fed on HFD for 12 wk (n = 4 per group). Data are presented as mean ± SEM. Significance between every two groups was calculated using unpaired two-tailed Student’s t test unless otherwise indicated. *P < 0.05, **P < 0.01.
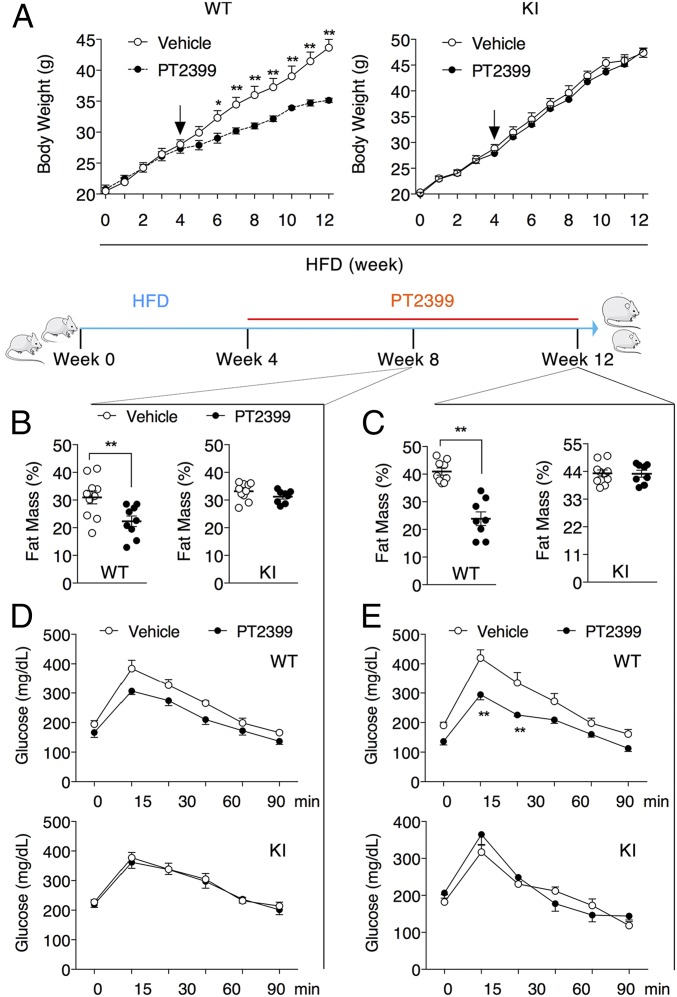
PT2399 Ameliorates Metabolic Dysfunction in WT Mice, but Not KI Mice, Fed HFD.
Incorporation of the S305M cavity-filling mutation antagonized renal HIF-2α activity in the setting of hypoxic stress (Fig. 1). To investigate whether incorporation of the S305M cavity-filling mutation similarly antagonizes HIF-2α activity in the setting of HFD, we attempted to recapitulate these effects in WT mice by administering the HIF-2α antagonist PT2399. Previous studies reported that PT2399 treatment had no effect on body weight when mice were fed a standard chow diet (16). In contrast, PT2399 treatment had a dramatic impact on body weight increase in WT mice fed on HFD. These changes were not due to a difference in food consumption or other “off-target” effects as PT2399 did not alter body weight in KI mice fed HFD (Fig. 4A and SI Appendix, Fig. S7 A–C). Decreases in fat mass were observed in as little as 4 wk of PT2399 treatment in WT, but not KI mice (Fig. 4B). Following 8 wk of PT2399 administration, there were substantial changes in body composition (Fig. 4C) and fat pad mass (SI Appendix, Fig. S7 D and E) only in the WT mice. Gene expression analysis in iWAT isolated from these mice revealed that in addition to decreases in the mRNA levels of HIF-2α target genes (SI Appendix, Fig. S8A), PT2399 treatment decreased the levels of adipocyte markers and adipogenic promoting factors, as well as genes encoding lipogenic and lipolysis enzymes (SI Appendix, Fig. S8 B–E). As the size of adipocytes in iWAT and perirenal white adipose tissue (pWAT) were largely unaffected by PT2399 (SI Appendix, Fig. S8F), these data suggest that HIF-2α inhibition suppresses adipogenesis in vivo, perhaps through the ZFP423/PPARγ axis (33). Likewise, the brown adipose tissue (BAT) mass also decreased in PT2399-treated WT mice fed on HFD (SI Appendix, Fig. S8G), accumulating less lipid (SI Appendix, Fig. S8H) while retaining higher expression levels of characteristic BAT genes (SI Appendix, Fig. S8I). PT2399-induced decrease in body weight was accompanied by decrease in glucose tolerance (Fig. 4 D and E), fasting glucose, fasting insulin, and serum leptin only in the WT mice (SI Appendix, Fig. S7 F–H).
Fig. 4.
The HIF-2 antagonist PT2399 ameliorates metabolic dysfunction in WT mice, but not KI mice. (A) Body weight of WT and KI mice maintained on HFD for 12 wk beginning at 8 wk of age. Vehicle or 30 mg/kg PT2399 was administered twice a day by oral gavage for the final 8 wk (arrows indicate the initiation of PT2399 treatment). Body composition of WT and KI mice following treatment with vehicle or PT2399 for 4 wk (B) or 8 wk (C). OGTT of WT and KI mice following treatment for with vehicle or PT2399 for 4 wk (D) or 8 wk (E). Data are presented as mean ± SEM. Significance between every two groups was calculated using unpaired two-tailed Student’s t test. Two-way ANOVA was performed for comparison of two groups over time followed by Sidak’s multiple test. *P < 0.05, **P < 0.01; n = 8 per group.
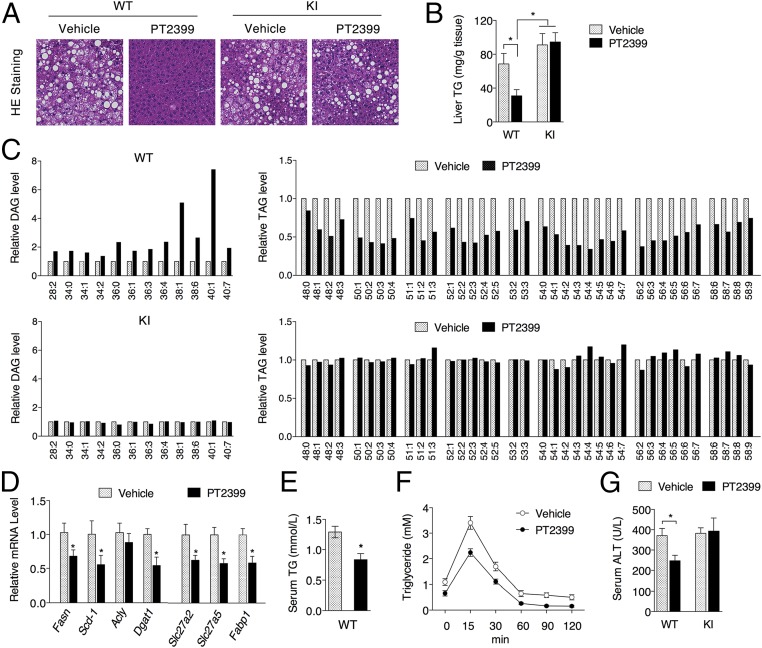
PT2399 Ameliorates Fatty Liver in Mice Fed on HFD.
Nonalcoholic fatty liver disease (NAFLD) is recognized as the most common cause of chronic liver disease worldwide, and obesity and associated metabolic disorders have been considered major risk factors for NAFLD (34, 35). To determine if this overall improvement in metabolic function extends to other organs, we examined the effects of PT2399 treatment on the livers of WT and KI mice fed HFD. PT2399 treatment for 8 wk resulted in a decrease in triglycerides in the livers of WT, but not KI, mice (Fig. 5 A and B). Lipidomic profiling revealed increases in DAG levels and decreases in TAG levels only in the PT2399-treated WT mice (Fig. 5C and SI Appendix, Table S2), consistent with reduced expression of lipid synthesis genes (Fasn, Scd-1, Dgat1, and Dgat2) in the livers of PT2399-treated WT mice (Fig. 5D). PT2399 treatment also lowered serum TG levels in WT mice on HFD (Fig. 5E) and enhanced TG clearance (Fig. 5F) despite down-regulation of lipid absorption genes (Slc27a2, Slc27a5, and Fabp1; Fig. 5D). Serum alanine aminotransferase (ALT) activity was also reduced (Fig. 5G), reflecting improved overall liver function.
Fig. 5.
PT2399 treatment reduces fatty liver symptoms only in WT mice fed HFD. WT and KI mice fed HFD for 12 wk were administered vehicle or 30 mg/kg PT2399 twice a day for the final 8 wk. HE staining of liver tissues (A) and liver TG levels (B) from WT and KI mice. (Magnification: A, 200×.) (C) Relative liver DAG and TAG levels in WT and KI mice (n = 4 per group). (D) qPCR analysis of lipid metabolism genes in the liver. (E) Serum TG levels of WT mice. (F) TG clearance of WT mice following 7 wk of PT2399 treatment. (G) Serum ALT levels from WT or KI mice. n = 8 per group. Data are presented as mean ± SEM. Significance between every two groups was calculated using unpaired two-tailed Student’s t test. Two-way ANOVA was performed for comparison of two groups over time followed by Sidak’s multiple test. *P < 0.05, **P < 0.01.
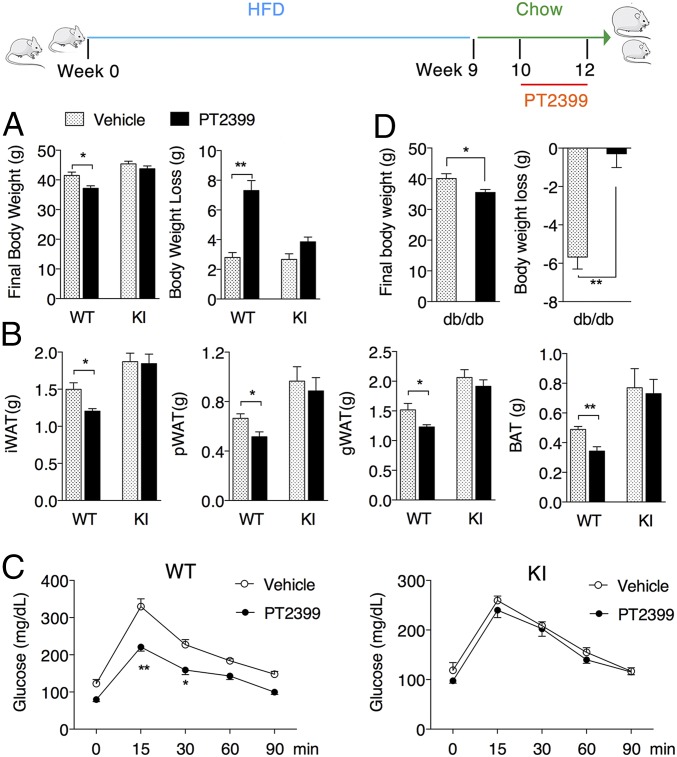
PT2399 Treatment Enhances Weight Loss in Obese Mice.
To determine if HIF-2α inhibition could provide benefits in a treatment, rather than prophylactic setting, we first induced obesity by feeding mice with HFD for 9 wk followed by a switch to normal chow and PT2399 treatment. Switching the diet slightly induced weight loss in both WT and KI mice with vehicle administration, yet PT2399 promoted more significant weight loss (Fig. 6A), as well as reductions in fat pad mass (Fig. 6B) and lipid accumulation in the livers of obese WT mice but not obese KI mice (SI Appendix, Fig. S9A). Again, PT2399 treatment improved glucose tolerance (Fig. 6C), liver TG values, and serum ALT levels (SI Appendix, Fig. S9 B and C) only in the obese WT mice, reflecting overall improvement of their metabolic condition. In another commonly used obese model, PT2399 treatment consistently promoted weight loss in db/db mice under normal diet condition (Fig. 6D). These data demonstrated potential therapeutic effects of PT2399 on obesity and associated disorders in mice through inhibition of HIF-2α.
Fig. 6.
PT2399 treatment enhances weight loss in obese mice. Obesity was induced in WT and KI mice fed HFD for 9 wk beginning at 8 wk of age. Mice were transitioned to a standard chow diet for 1 wk and then administered vehicle or 30 mg/kg PT2399 twice a day for 2 wk. (A) Final body weight and body weight loss of WT and KI obese mice following treatment. (B) Tissue mass of iWAT, pWAT, gWAT, and BAT following treatment. (C) OGTT of WT and KI mice following treatment. (D) db/db mice under normal diet were treated with 30 mg/kg PT2399 twice a day for 4 wk, starting at the age of 8 wk, and final body weight and body weight loss were recorded. Data are presented as mean ± SEM. Significance between every two groups was calculated using unpaired two-tailed Student’s t test. Two-way ANOVA was performed for comparison of two groups over time followed by Sidak’s multiple test. *P < 0.05, **P < 0.01; n = 8 per group.
Discussion
As well-known bHLH-PAS proteins, HIFs are central to the genomic response to hypoxia, which drives a pivotal role of HIFs in promoting tumor progression (22). Although HIFs have attracted increasing attention for being therapeutic targets, limited progress has been made, and HIF2α was even considered undruggable (36). Although it is clear that both PAS domains of HIFs contribute to heterodimerization, the overall structural, biochemical, and protein interaction characteristics suggest that PAS-B commonly functions as signaling domain in addition to contributing dimerization, whereas PAS-A functions primarily as an essential dimerization domain (37). Following characterizing the PAS-B domain of HIF-2α, we were able to develop drug-like chemicals such as PT2399 that can specifically bind within PAS-B domain (14, 20). Previous studies have validated the on-target effect of PT2399 against HIF-2α through cultured cancer cells; here, we further demonstrate the on-target effect of PT2399 in mice using PAS-B domain S305M mutation mice as negative control.
PAS domains have been reported to bind cofactors such as heme or flavin mononucleotide. The ability of the HIF transcription factors to respond to environmental and metabolic stresses requires mechanisms to sense changes in the cellular milieu. For example, regulatory dioxygenases can “sense” the cellular microenvironment by virtue of their substrate and cofactor requirements and modulate HIF stability and activity through hydroxylation of the HIF-α subunit (38–40), and PAS domains have also been reported to directly bind cofactors such as heme or flavin mononucleotide (11). Structure analysis of HIF-2α PAS-B domain revealed the presence of an unusually large buried cavity, which is rare and only observed in 0.3% of protein structures (13, 14), indicating a unique role of PAS-B domain for regulating HIF-2α stability and functional activity.
Although we have previously shown that the HIF-2α PAS-B domain can be regulated by exogenous ligands, here we demonstrate that this ligand binding cavity also contributes to metabolic stress response in addition to hypoxia. Although the introduction of the S305M mutation compromises the artificial ligand binding capacity of this domain, there were no overt phenotypes observed in the KI mice maintained under standard laboratory conditions. However, HIF-2α−dependent responses were altered following exposure to both hypoxia and metabolic stress. Interestingly, the consequences of this mutation were dependent on the nature of the challenge. Introduction of the cavity-filling S305M mutation suppressed hypoxic activation of HIF-2α in the kidneys while HIF-2 activity was enhanced, rather than inhibited, in the context of the HFD response, partly attributed by higher HIF-2α nucleus localization in KI mice under HFD feeding. The disparate outcomes observed here may indicate that tissue-selective contributions to HIF-2α regulation are dependent upon the local presence of one or more endogenous ligand(s). This paradigm establishes support for further investigation of cavities residing in each of the HIF PAS domains (41–43) as they too might contribute to the ability of HIFs to sense and respond to environmental and metabolic signals.
Since obesity-associated extensive fat expansion is accompanied by dramatically lower oxygen diffusion, stabilized HIFs protein were thereby observed in the adipose tissue (44, 45). Mice with adipocyte-targeted HIF-1β deletion were lean with reduced fat formation and were protected from HFD-induced obesity and glucose intolerance (28, 46); genetic or pharmacologic inhibition of HIF-1α showed benefits against HFD induced obesity and inflammatory response (24), suggesting the involvement of HIFs protein in regulating obesity progression. Here, we showed that globally blocking HIF-2α PAS-B domain with PT2399 in mice would prevent HFD-induced obesity and promote body weight loss in obese mice, suggesting potential preventive and therapeutic benefits against obesity by targeting HIF-2α. Although the mutation of PAS-B domain lost the binding capacity of PT2399, the KI mice exerted quite opposite phenotype as WT mice with PT2399 under HFD challenge, highlighting the vital role of PAS-B domain in response to metabolic stress. Further investigations are warranted to identify the natural binding molecules of PAS-B domain in regulating diverse activities.
In addition to body weight gain, obesity is consistently associated with NAFLD and glucose intolerance. Weight loss by blocking PAS-B domain was accompanied with reduced liver fat accumulation, improved glucose tolerance, as well as triglyceride clearance. The overall benefits may be attributed to the general weight loss in mice, or direct modulation of gene profiling on lipogenesis and fatty acid uptake, since direct activation of HIF-2α promoting fatty liver progression has been well presented (47), and inhibition of HIF-2α in the intestine was found to confer systemic improvements of metabolic function in mice on HFD (48). However, obesity-associated inflammation response, and elevated serum free fatty acid level have all been indicated as contributors to fatty liver progression (49). Therefore, the amelioration of fatty liver may be a consequence of global benefits of HIF-2α inhibition, and the benefits in both adipocytes and liver tissue may eventually contribute to improved glucose tolerance and insulin sensitivity in obese mice.
Taken together, the current study reveals an important role of PAS-B domain in regulating HIF-2α response to hypoxia and metabolic stresses, highlighting the importance of HIF-2α in regulating obesity progression and provides a potential therapeutic approach for treatment of obesity and associated metabolic syndrome.
Materials and Methods
PT2399 was provided by Peloton Therapeutics. Mice expressing the Epas1S305M KI allele were generated by the University of Texas Southwestern Transgenic Core Facility using CompoZr Custom Zinc Finger Nucleases designed by Sigma-Aldrich. All animal experiments were performed with the approval of the University of Texas Southwestern (UTSW) Institutional Animal Care and Use Committee. All experimental conditions are further available in SI Appendix, including reagents, primary cell culture, immunoblotting, systemic metabolic assay, liver chemistry, and lipidomics analysis.
Supplementary Material
Acknowledgments
We thank A. Pudasaini (UTSW) and M. Shao (UTSW) for assistance with the mouse studies, J. McDonald (UTSW) and M. Mitsche (UTSW) for help with the lipodomics analysis, and Peloton Therapeutics for providing PT2399. This work was supported by Robert A. Welch Foundation Grant I-1568 (to R.K.B.) and Cancer Prevention and Research Institute of Texas Grant RP130513 (to R.K.B.). This study was also supported by National Basic Research Program Grant 2015CB553602 and National Natural Science Foundation of China Grants 81571050 and 31570777. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR 15437-01 from the National Center for Research Resources. R.K.B. is the Michael L. Rosenberg Scholar in Medical Research.
Footnotes
Conflict of interest statement: R.K.B. owns equity in Peloton Therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810897115/-/DCSupplemental.
References
- 1.Semenza GL. Hypoxia-inducible factors: Coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 2017;36:252–259. doi: 10.15252/embj.201695204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schito L, Semenza GL. Hypoxia-inducible factors: Master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Lin N, Simon MC. Hypoxia-inducible factors: Key regulators of myeloid cells during inflammation. J Clin Invest. 2016;126:3661–3671. doi: 10.1172/JCI84426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 8.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, et al. Functions of the Per/ARNT/Sim domains of the hypoxia-inducible factor. J Biol Chem. 2005;280:36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 10.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH-PAS proteins in cancer. Nat Rev Cancer. 2013;13:827–841. doi: 10.1038/nrc3621. [DOI] [PubMed] [Google Scholar]
- 13.Scheuermann TH, et al. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheuermann TH, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9:271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheuermann TH, et al. Isoform-selective and stereoselective inhibition of hypoxia inducible factor-2. J Med Chem. 2015;58:5930–5941. doi: 10.1021/acs.jmedchem.5b00529. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho H, et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature. 2016;539:107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace EM, et al. A small-molecule antagonist of HIF2α is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 2016;76:5491–5500. doi: 10.1158/0008-5472.CAN-16-0473. [DOI] [PubMed] [Google Scholar]
- 19.Courtney KD, et al. Phase I dose-escalation trial of PT2385, a first-in-class hypoxia-inducible factor-2α antagonist in patients with previously treated advanced clear cell renal cell carcinoma. J Clin Oncol. 2017;36:867–874. doi: 10.1200/JCO.2017.74.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers JL, et al. Development of inhibitors of the PAS-B domain of the HIF-2α transcription factor. J Med Chem. 2013;56:1739–1747. doi: 10.1021/jm301847z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 23.Wei K, et al. A liver Hif-2α-Irs2 pathway sensitizes hepatic insulin signaling and is modulated by Vegf inhibition. Nat Med. 2013;19:1331–1337. doi: 10.1038/nm.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YS, et al. Increased adipocyte O2 consumption triggers HIF-1α, causing inflammation and insulin resistance in obesity. Cell. 2014;157:1339–1352. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choe SS, et al. Macrophage HIF-2α ameliorates adipose tissue inflammation and insulin resistance in obesity. Diabetes. 2014;63:3359–3371. doi: 10.2337/db13-1965. [DOI] [PubMed] [Google Scholar]
- 26.Shin MK, et al. Metabolic consequences of high-fat diet are attenuated by suppression of HIF-1α. PLoS One. 2012;7:e46562. doi: 10.1371/journal.pone.0046562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun K, Halberg N, Khan M, Magalang UJ, Scherer PE. Selective inhibition of hypoxia-inducible factor 1α ameliorates adipose tissue dysfunction. Mol Cell Biol. 2013;33:904–917. doi: 10.1128/MCB.00951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang C, et al. Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes. 2011;60:2484–2495. doi: 10.2337/db11-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Q, et al. Activation of hypoxia-inducible factor-2 in adipocytes results in pathological cardiac hypertrophy. J Am Heart Assoc. 2013;2:e000548. doi: 10.1161/JAHA.113.000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimba S, Wada T, Hara S, Tezuka M. EPAS1 promotes adipose differentiation in 3T3-L1 cells. J Biol Chem. 2004;279:40946–40953. doi: 10.1074/jbc.M400840200. [DOI] [PubMed] [Google Scholar]
- 31.Rutkowski JM, Stern JH, Scherer PE. The cell biology of fat expansion. J Cell Biol. 2015;208:501–512. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villanueva CJ, et al. Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology. 2009;50:434–442. doi: 10.1002/hep.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RK, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharabi Y, Eldad A. Nonalcoholic fatty liver disease is associated with hyperlipidemia and obesity. Am J Med. 2000;109:171. doi: 10.1016/s0002-9343(00)00434-4. [DOI] [PubMed] [Google Scholar]
- 35.Marchesini G, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 36.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao N, Whitelaw ML, Shearwin KE, Dodd IB, Chapman-Smith A. Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Res. 2011;39:3695–3709. doi: 10.1093/nar/gkq1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 39.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 40.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardoso R, et al. Identification of Cys255 in HIF-1α as a novel site for development of covalent inhibitors of HIF-1α/ARNT PasB domain protein-protein interaction. Protein Sci. 2012;21:1885–1896. doi: 10.1002/pro.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Y, Scheuermann TH, Partch CL, Tomchick DR, Gardner KH. Coiled-coil coactivators play a structural role mediating interactions in hypoxia-inducible factor heterodimerization. J Biol Chem. 2015;290:7707–7721. doi: 10.1074/jbc.M114.632786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 44.Ye J, Gao Z, Yin J, He Q. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am J Physiol Endocrinol Metab. 2007;293:E1118–E1128. doi: 10.1152/ajpendo.00435.2007. [DOI] [PubMed] [Google Scholar]
- 45.Rausch ME, Weisberg S, Vardhana P, Tortoriello DV. Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. Int J Obes. 2008;32:451–463. doi: 10.1038/sj.ijo.0803744. [DOI] [PubMed] [Google Scholar]
- 46.Lee KY, Gesta S, Boucher J, Wang XL, Kahn CR. The differential role of Hif1β/Arnt and the hypoxic response in adipose function, fibrosis, and inflammation. Cell Metab. 2011;14:491–503. doi: 10.1016/j.cmet.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rankin EB, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29:4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie C, et al. Activation of intestinal hypoxia-inducible factor 2α during obesity contributes to hepatic steatosis. Nat Med. 2017;23:1298–1308. doi: 10.1038/nm.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: Biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.