Significance
The adult mammalian heart does not functionally repair itself after injury. Therefore, identification of cardiac stem cell (CSC) populations is of great interest for regenerative intervention. However, the significance of such CSC populations remains heavily debated. Using single-cell mRNA sequencing and genetic lineage tracing, we interrogate the existence of CSCs with unbiased mouse models of proliferation. Cycling cardiomyocytes were only robustly observed in the early postnatal growth phase, while cycling cells in homoeostatic and damaged adult myocardium consisted mainly of various noncardiomyocyte cell types. Injury-activated cardiac fibroblasts that acquire a gene expression profile similar to that of neonatal cardiac fibroblasts signal—in an autocrine fashion—to prevent cardiac rupture. We find no evidence for the existence of a quiescent CSC population.
Keywords: stem cells, lineage tracing, cardiac regeneration, fibroblasts, single-cell transcriptomics
Abstract
The significance of cardiac stem cell (CSC) populations for cardiac regeneration remains disputed. Here, we apply the most direct definition of stem cell function (the ability to replace lost tissue through cell division) to interrogate the existence of CSCs. By single-cell mRNA sequencing and genetic lineage tracing using two Ki67 knockin mouse models, we map all proliferating cells and their progeny in homoeostatic and regenerating murine hearts. Cycling cardiomyocytes were only robustly observed in the early postnatal growth phase, while cycling cells in homoeostatic and damaged adult myocardium represented various noncardiomyocyte cell types. Proliferative postdamage fibroblasts expressing follistatin-like protein 1 (FSTL1) closely resemble neonatal cardiac fibroblasts and form the fibrotic scar. Genetic deletion of Fstl1 in cardiac fibroblasts results in postdamage cardiac rupture. We find no evidence for the existence of a quiescent CSC population, for transdifferentiation of other cell types toward cardiomyocytes, or for proliferation of significant numbers of cardiomyocytes in response to cardiac injury.
Possible cell cycle reentry of adult cardiomyocytes or derivation of new cardiomyocytes from putative resident progenitor cells upon myocardial infarction (MI) has been intensively investigated. Although with inconsistent results, these studies contributed to the general consensus that throughout adult mammalian life a small fraction of cardiomyocytes is replaced (1–3). Both the differentiation from a stem cell compartment and the division of preexisting myocytes have been suggested as the source of new cardiomyocytes (4, 5). Nonetheless, the capacity of the adult mammalian heart to functionally regenerate upon injury remains controversial (6–10).
Studies on cardiac stem cells (CSCs) have relied heavily on specific stem cell markers that have been defined in unrelated stem cell systems such as c-KIT (4, 11), SCA-1 (12), or ABCG2, a marker of so-called cardiac side population cells (13, 14). The validity of each of these markers has been disputed (8, 9, 15–17). Also, anticipated changes in cellular metabolism or DNA label retention have been used to identify actively cycling cardiomyocytes, yet these methods do not allow visualization of the cellular offspring (18, 19). Furthermore, in these studies, the proliferative capacity of resident noncardiomyocyte cell lineages has been largely neglected. To address these issues, we sought to generate an unbiased map of proliferating cells and their progeny in neonatal, adult, and postdamage murine hearts. Since the single defining characteristic of a stem cell is its ability to produce functional daughter cells by cell division (20), we posit that the most unbiased way of interrogating the involvement of stem cells in any biological growth or repair process is to genetically lineage trace all cells that proliferate during that pertinent biological process.
While absent from cells resting in the G0 phase (Fig. 1A), Ki67 is specifically expressed by all actively cycling cells in the cell cycle stages of G1, S, G2, and M (21). It is therefore widely used in basic cell and developmental biology, in clinical oncology, and in pathology as a general proliferation marker (22). We have generated Mki67 knockin mice (23, 24) to perform genetic lineage tracing (25) and comprehensively determine the offspring of any cell that becomes proliferative in neonatal, adult homoeostatic, and adult damaged heart. With these genetic models, we set out to ask whether homoeostatic or damaged hearts harbor stem cells, under the premise that stem cells would have to enter the cell cycle to produce progeny replacing lost cells (20).
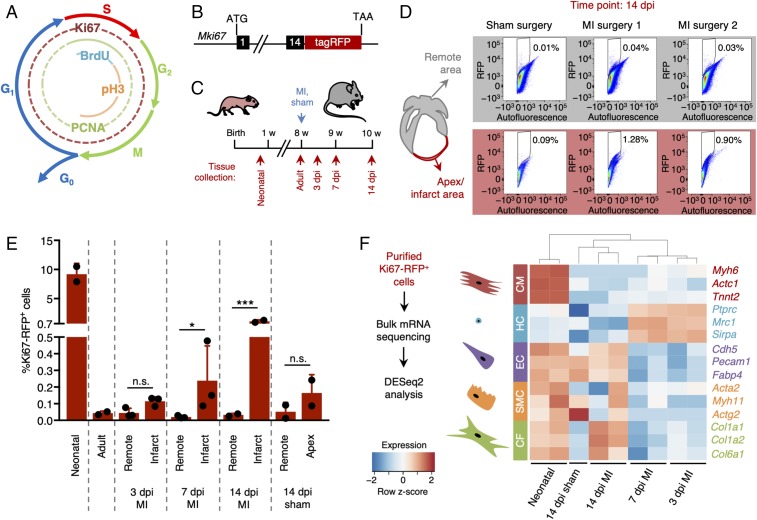
Fig. 1.
Quantification and characterization of cardiac cell proliferation following injury. (A) Diagram showing the markers of the different phases of the cell cycle. (B) Schematic representation of the reporter mouse model expressing RFP-tagged Ki67 (Mki67TagRFP). (C) Experimental timeline of myocardial infarction (MI) surgery and tissue collection 3, 7, and 14 d post-MI injury (dpi). (D and E) Quantification of Ki67-RFP+ cells after MI or sham surgery in both remote (gray) and infarcted zone (red). The gating and sorting strategies are described in SI Appendix, Fig. S1. (D) Representative flow cytometry scatter plots 14 dpi. (E) Quantification of Ki67-RFP+ cells in neonatal, adult, infarct, remote and apex areas 3, 7, and 14 dpi (n = 2–3 mice per condition). All error bars represent ±SD. Asterisks indicate significance (Student’s t test: n.s., not significant, P ≥ 0.05; *P < 0.05; ***P < 0.001). (F) Bulk mRNA sequencing strategy and gene expression levels of different cardiac cell types identified in the purified Ki67-RFP+ populations. Cardiomyocytes were enriched in neonatal hearts. Hematopoietic cells were enriched 3 and 7 dpi, while resident lineages such as endothelial cells and fibroblasts were more abundant 14 dpi. CF, cardiac fibroblasts; CM, cardiomyocytes; EC, endothelial cells; HC, hematopoietic cells; SMC, smooth muscle cells.
Results
Cardiac Cell Proliferation Is Triggered in Response to Myocardial Damage.
We first aimed to determine the levels of cardiac cell proliferation in murine hearts at different postnatal time points and under different physiologic conditions. We therefore utilized a mouse model that expresses a red fluorescent protein (TagRFP) linked to the Ki67 protein (23) allowing for quantification and isolation of actively cycling live cells using flow cytometry (Fig. 1B and SI Appendix, Fig. S1). To analyze proliferating cardiac cells under homoeostatic conditions, we collected hearts of 1- and 8-wk-old Mki67TagRFP mice (Fig. 1C). To evaluate cardiac cell proliferation upon injury, we subjected 8-wk-old Mki67TagRFP mice to an MI by permanent ligation of the proximal left anterior descending (LAD) coronary artery and collected hearts 3, 7, and 14 d later (Fig. 1C). The percentage of Ki67-RFP+ cells in the hearts of 1-wk-old neonatal mice was about 10%, while less than 0.05% of cells purified from the hearts of adult mice were positive for Ki67-RFP (Fig. 1 D and E). In mice subjected to MI surgery, the ratio of Ki67-RFP+ cells in the infarcted area was comparable to the corresponding region of hearts (apex) collected from sham-operated mice 3 d after the injury (Fig. 1 D and E). The percentage of Ki67-RFP+ cells doubled at 7 d post-MI and reached 1% enrichment at 14 d following the injury (Fig. 1E). The percentage of Ki67-RFP+ cells in the remote tissue of both damaged and control hearts remained below 0.05% over the course of the experiment (Fig. 1 D and E).
We processed the purified Ki67-RFP+ cells for mRNA sequencing and analyzed their gene expression profiles using the DESeq2 algorithm (26). Principal-component analysis showed very similar expression profiles of Ki67-RFP+ cells collected 3 and 7 d after MI (SI Appendix, Fig. S2). Proliferative cells isolated from the hearts of neonatal or adult mice 14 d following injury, however, were distinctively different (SI Appendix, Fig. S2). Neonatal Ki67-RFP+ cells were enriched for cardiomyocyte markers (Myh6, Actc1, and Tnnt2), while Ki67-RFP+ cells from all other conditions showed enrichment for transcripts of noncardiomyocyte markers (Fig. 1F). Ki67-RFP+ cells collected within 1 wk after injury were enriched for transcripts of hematopoietic genes such as Ptprc (encoding CD45), Mrc1, and Sirpa, suggesting the presence of a proliferative inflammatory infiltrate (Fig. 1F). Genes specific for resident cell types such as endothelial cells (Cdh5, Pecam1, and Fabp4) and cardiac fibroblasts (Col1a1, Col1a2, and Col6a1) were more abundant in Ki67-RFP+ cells at 14 d post-MI injury (dpi) (Fig. 1F). Expression of the smooth muscle cell marker genes Acta2, Actg2, and Myh11 did not change upon damage in our dataset (Fig. 1F).
Single-Cell Transcriptome Analysis Uncovers Distinct Proliferative Populations Within the Murine Heart.
Fewer than 35% of cardiac ventricular cells are cardiomyocytes; the remainder belong to the endothelial, hematopoietic, and fibroblast lineages (27). A recent study has shown that single-cell transcriptome data can readily be generated from all cardiac cell lineages in the adult murine heart (28). To generate a comprehensive transcriptome atlas of proliferative cells during postnatal growth, adult homoeostasis, and cardiac regeneration, we therefore aimed to profile individual Ki67-RFP+ and Ki67-RFP− cardiac cells as well as unlabeled Mki67wt/wt cardiac cells in a variety of settings: in 1-wk old mice, when murine cardiomyocytes still have significant proliferative capacity (1), in 8- to 10-wk old mice when cardiomyocytes should be homoeostatic, and in adult mice after cardiac injury (Fig. 2A). To collect cardiac cells from infarcted hearts, we chose the time point with the highest number of Ki67-RFP+ cells and at which most previous CSC studies have focused (11, 15, 29): 14 d after MI surgery (Fig. 2A). We performed single-cell mRNA sequencing using the sorting and robot-assisted transcriptome sequencing (SORT-seq) method (30) and analyzed the sequencing data using the RaceID2 algorithm (31) (Fig. 2B). To exclude cells with low-quality RNA, we removed all cells expressing fewer than 1,000 transcripts. We then clustered all 1,939 cells that passed this threshold based on transcriptome similarities assessed by 1 – Pearson’s correlation and grouped by k-medoids clustering (31). This k-medoids clustering compartmentalized the cells into nine different major cell clusters, as visualized by t-distributed stochastic neighbor embedding (t-SNE) (Fig. 2C and SI Appendix, Fig. S3). Based on differential gene expression of some well-characterized markers, we found that all major cardiac cell types were present in the single-cell dataset (Fig. 2 C–E and SI Appendix, Fig. S3): clusters 8 and 9 comprised 437 cardiomyocytes enriched for transcripts of Myh6, Actc1, and Tnnt2; clusters 6 and 7 contained a total of 282 cardiac fibroblasts expressing Pdgfra, Col1a1, and Tcf21; 458 hematopoietic cells enriched for Ptrpc, Mrc1, and Sirpa were assigned to clusters 1 and 4; 705 endothelial cells expressing Pecam1, Cdh5, and Fabp4 were present in clusters 2 and 3; and 57 cells in cluster 5 were enriched for smooth muscle genes Acta2, Myh11, and Actg2. Of note, while SIRPA has been reported to be expressed by human embryonic stem cell-derived cardiac progenitors (32), we only found significant Sirpa expression in hematopoietic cells in our dataset.
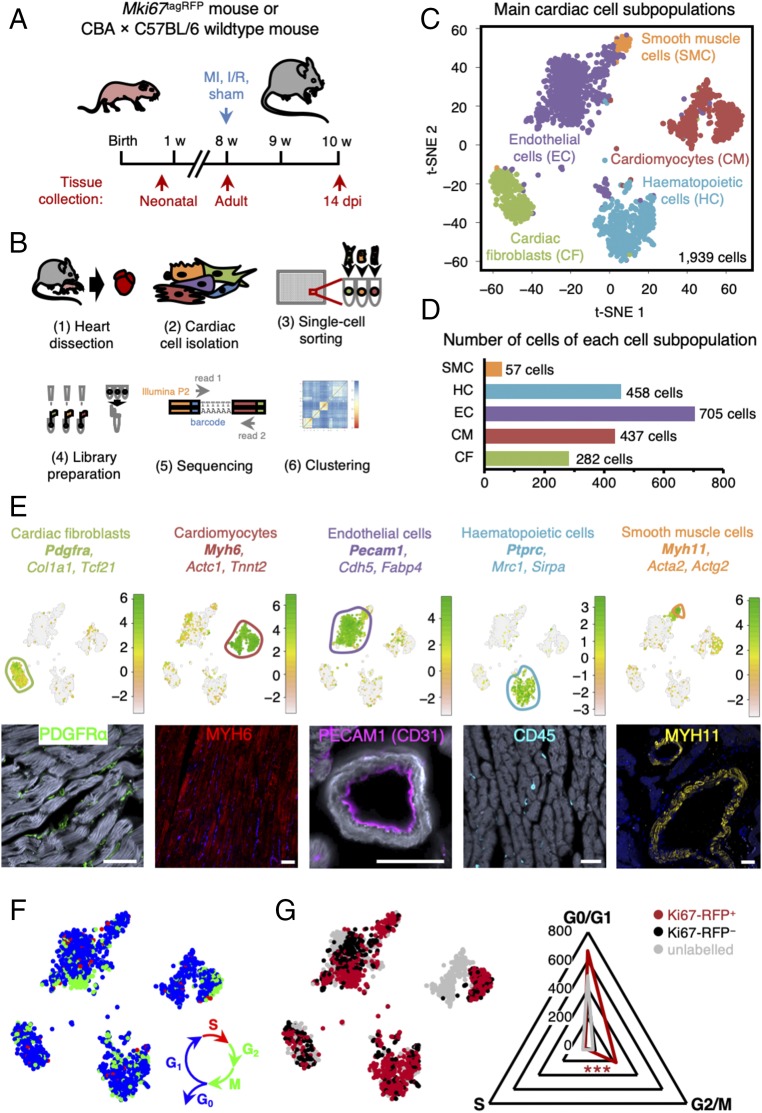
Fig. 2.
Single-cell transcriptome analysis uncovers distinct proliferative populations within the murine heart. (A) Experimental timeline for tissue collection of hearts from wild-type and Mki67RFP mice, either neonatal or adults, 14 d after sham, ischemia/reperfusion (I/R), or MI surgery (n = 2–4 mice per condition). (B) Schematic representation of SORT-seq workflow. Hearts were isolated (1) and digested into single-cell suspension (2), and Ki67-RFP+ and Ki67-RFP− cells were sorted into 384-well plates containing primers, dNTPs, and spike-ins (3). Retrotranscription mix was distributed using Nanodrop II, and material was pooled and amplified (4) before pair-end sequencing (5). Cells were clustered using RaceID2 (6). (C) Clustering of cardiac cells and cell-to-cell distances visualized by t-distributed stochastic neighbor-embedding (t-SNE) map, highlighting identified major cardiac cell types. (D) Numbers of cells assigned to each cardiac cell lineage. (E) t-SNE map highlighting identified cell types based on previously described cellular markers (logarithmic scale of transcript expression). Markers expression is shown in Lower panel by immunofluorescent staining. (Scale bars: 50 μm.) (F) t-SNE map displaying cell cycle stage of each cell [S (red), G2/M (green), G0/G1 (blue)] assigned by the cyclone algorithm. (G) t-SNE map showing the Ki67-RFP status from the flow cytometry data; Ki67-RFP+ (red), Ki67-RFP− (black), or Mki67wt/wt cells without TagRFP construct (gray) and radar plot showing Ki67-RFP+ cells enriched for the cycling G2/M stage according to the cyclone algorithm. Asterisks indicate significance (χ2 test: ***P < 0.001).
To validate that sequenced Ki67-RFP+ cells were proliferative, we used the cyclone algorithm (33) to assign cell cycle stages to each individual cell in our filtered dataset (Fig. 2F). While present throughout the cell cycle (and absent from quiescent cells arrested in G0) (22), Ki67 is most highly expressed in the G2 stage (34, 35). Consistent with these previous studies, cells in S and G2/M phase were enriched in the fraction of Ki67-RFP+ cells (Fig. 2G; χ2 test, P < 0.001), confirming that they were actively cycling.
Most Ki67-RFP+ neonatal cells were identified as cardiomyocytes, while no Ki67-RFP+ cardiomyocytes were found in the datasets compiled from either homoeostatic or injured adult hearts (SI Appendix, Fig. S4A). In homoeostatic adult heart, Ki67-RFP+ cells mapped to noncardiomyocyte lineages, in particular to the hematopoietic and endothelial cell lineages (SI Appendix, Fig. S4A). Ki67-RFP+ cells present in the infarcted region 14 dpi were mainly fibroblasts and hematopoietic cells (SI Appendix, Fig. S4A), while Ki67-RFP+ cells in the remote region additionally represented mostly the endothelial and hematopoietic lineages (SI Appendix, Fig. S4A). Collectively, these data show that there is a significant number of proliferative cardiomyocytes in neonatal hearts, but not in homoeostatic or injured adult hearts.
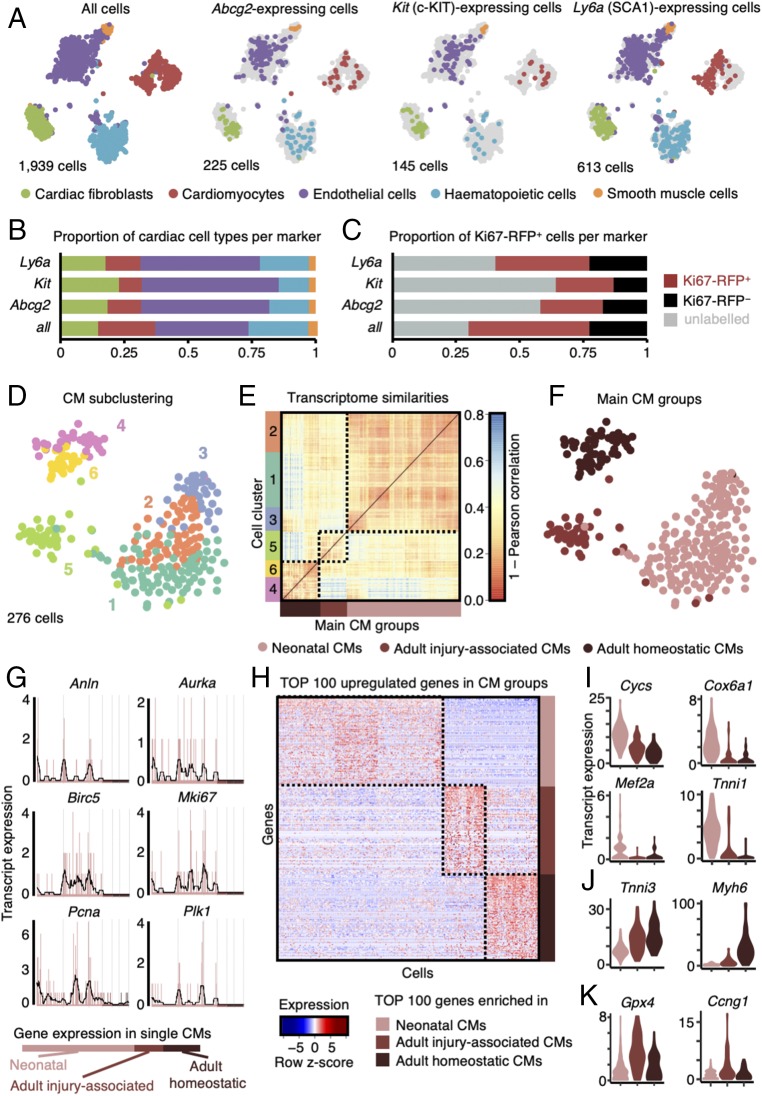
Since we did not find evidence for a distinct CSC population expressing proposed markers such as Abcg2 (side population marker), Kit (encoding c-KIT), and Ly6a (encoding SCA-1), we investigated the presence of cells expressing at least one transcript of these markers within all cardiac cell types (Fig. 3 A–C). We identified 225 cells expressing Abcg2, which were scattered within all main cardiac cell types found in our dataset (Fig. 3 A and B) (13, 14). Most of the 145 Kit-expressing cells were endothelial cells (Fig. 3 A and B) (15), in contrast to earlier reports (4, 11). Some fibroblasts, which are usually not excluded when purifying putative c-KIT+ CSCs (4, 11, 36), also expressed Kit (Fig. 3 A and B). About 32% of all cells (613 cells of a total of 1,939 cells) in our dataset expressed Ly6a (Fig. 3A). Ly6a-expressing cells were assigned to all cardiac cell types, while absent in neonatal cardiomyocytes (12) (Fig. 3 A and B). Cells positive for a putative CSC marker (i.e., Abcg2, Kit, or Ly6a) were identified in both Ki67-RFP+ and Ki67-RFP− cardiac cell populations (Fig. 3C). Gata4, a marker of regenerating cardiomyocytes in zebrafish (37), was expressed in most cardiac lineages in a total of 214 cells (SI Appendix, Fig. S5). The cardiomyocyte lineage marker Nkx2-5 was only enriched in neonatal and adult cardiomyocytes (138 cells; SI Appendix, Fig. S5), as shown before (38, 39). We further found expression of the fetal cardiac progenitor marker Isl1 (40) in 87 cells belonging to the noncardiomyocyte lineages (cardiac fibroblasts, endothelial cells, and smooth muscle cells), but not in cardiomyocytes (SI Appendix, Fig. S5). These data imply that cell culture, transplantation, and lineage-tracing experiments relying on these putative CSC markers should be interpreted with great caution (17).
Fig. 3.
Expression patterns of putative CSC markers and cardiomyocytes. (A) t-SNE map displaying stem cell marker (Abcg2, Kit, and Ly6a) expressing cells in major cell type clusters; CF, cardiac fibroblasts; CM, cardiomyocytes; EC, endothelial cells; HC, hematopoietic cells; SMC, smooth muscle cells. (B) Quantification of marker-based cell fraction per cell type reveals most stem cell markers cover endothelial cells but can be found in all resident populations. (C) Quantification of marker-based cell fraction in sorted populations (unlabeled, Ki67-RFP+, and Ki67-RFP−). (D) t-SNE map of cardiomyocyte subclusters (n = 6) identified using the RaceID2 algorithm. (E) Heatmap representation of the transcriptome similarities between cardiomyocytes from the six identified subclusters combined into three main groups of cardiomyocytes. (F) t-SNE map highlighting the cells assigned to each of the three main groups of cardiomyocyte subclusters. (G) Expression profile of representative proliferation markers in neonatal and adult (homoeostatic or injury-associated) cardiomyocytes. Gene expression is shown on the y axis as transcript counts per cell on x axis with the running mean in black. (H) Heatmap representation of top 100 up-regulated genes in neonatal and adult (homoeostatic or injury-associated) cardiomyocytes. (I–K) Violin plots showing the expression of representative genes up-regulated in neonatal cardiomyocytes (I), adult homoeostatic cardiomyocytes (J), and adult injury-associated cardiomyocytes (K).
To determine whether unique cell clusters appeared following MI in our single-cell mRNA-sequencing dataset, we quantified the number of cells from each experimental condition present within a given cluster (SI Appendix, Fig. S4 B and C and Dataset S1). Cardiomyocytes clustered into two main clusters (clusters 8 and 9) irrespective of the cell cycle stage (actively cycling vs. quiescent) (SI Appendix, Fig. S4 B and C and Dataset S1). Endothelial cells clustered into clusters 2 and 3 irrespective of the experimental condition of origin (Fig. 2F, SI Appendix, Fig. S4 B and C, and Dataset S1). The majority of hematopoietic cells in the dataset were purified from injured adult hearts (clusters 1 and 4); however, cluster 1 mainly contained hematopoietic cells present in homoeostatic (adult or neonatal) hearts (SI Appendix, Fig. S4 B and C and Dataset S1). Fibroblast cluster 7 was predominantly composed of cells purified from homoeostatic adult hearts (65%). Surprisingly, while the majority of cells assigned to fibroblast cluster 6 were purified from injured adult heart (48%), 17% of the cells originated from neonatal hearts (SI Appendix, pie chart Insets in SI Appendix, Fig. S4C). This suggests that in response to ischemic injury, some activated fibroblasts adopt a neonatal-like state. The fibroblasts in cluster 6 were predominantly Ki67-RFP+ (Fig. 2F, SI Appendix, Fig. S4 B and C, and Dataset S1) and had up-regulated levels of genes involved in extracellular matrix remodeling such as Cola1a2, Col3a1, Col5a1, and Sparc, as well as genes encoding secreted proteins such as Sfrp2 and Dkk3 (Dataset S1).
To explore the cardiomyocyte fraction in more detail, we extracted all 437 cardiomyocytes out of the complete dataset and performed cell type-specific subclustering. We reran the extracted cells through the RaceID2 algorithm with a higher cutoff of 2,000 transcripts per cell to allow for refined cell clustering. The 276 cardiomyocytes retained after quality filtering grouped into six k-medoids clusters (Fig. 3D). Neonatal cardiomyocytes were enriched in clusters 1, 2, and 3, while adult injury-associated cardiomyocytes were mainly found in cluster 5 and adult homoeostatic cardiomyocytes in clusters 4 and 6 (Fig. 3 D–F and Dataset S2). We found robust expression of Mki67 and other markers of proliferation (Anln, Aurka, Birc5, Pcna, and Plk1) used by Soonpaa et al. (2) in neonatal cardiomyocytes (Fig. 3G), in agreement with our bulk mRNA-sequencing data (Fig. 1F). However, expression of these proliferation markers was largely negligible in adult cardiomyocytes (Fig. 1F). We performed differential gene expression analysis by comparing neonatal and adult (homoeostatic and injury-associated) cardiomyocyte clusters to each other (Fig. 3H). Among the 100 most up-regulated genes in neonatal cardiomyocytes were genes related to the mitochondrial electron transport chain (Cycs, Cox6a1) and cardiac development (Mef2a, Tnni1) (Fig. 3I and Dataset S3). Genes up-regulated in adult homoeostatic cardiomyocytes included sarcomere assembly (Tnni3) and classical myosin marker (Myh6) (Fig. 3J and Dataset S3). Adult injury-associated cardiomyocytes up-regulated genes involved in oxidative damage response (Gpx4) and damage-induced cell cycle arrest (Ccng1) (Fig. 3K and Dataset S3). Taken together, these data provide no evidence for the existence of a distinct CSC population in neonatal or adult (homoeostatic or postdamage) murine hearts and demonstrate that immature neonatal cardiomyocytes are readily proliferative. However, the presence of proliferative cardiomyocytes in adult murine hearts remains low even after ischemic injury, as indicated by the rare Ki67-RFP+ cardiomyocyte found in our single-cell dataset collected from adult hearts (SI Appendix, Fig. S4A).
Ki67 Lineage Tracing Demonstrates de Novo Generation of Cardiomyocytes in the Neonatal Murine Heart.
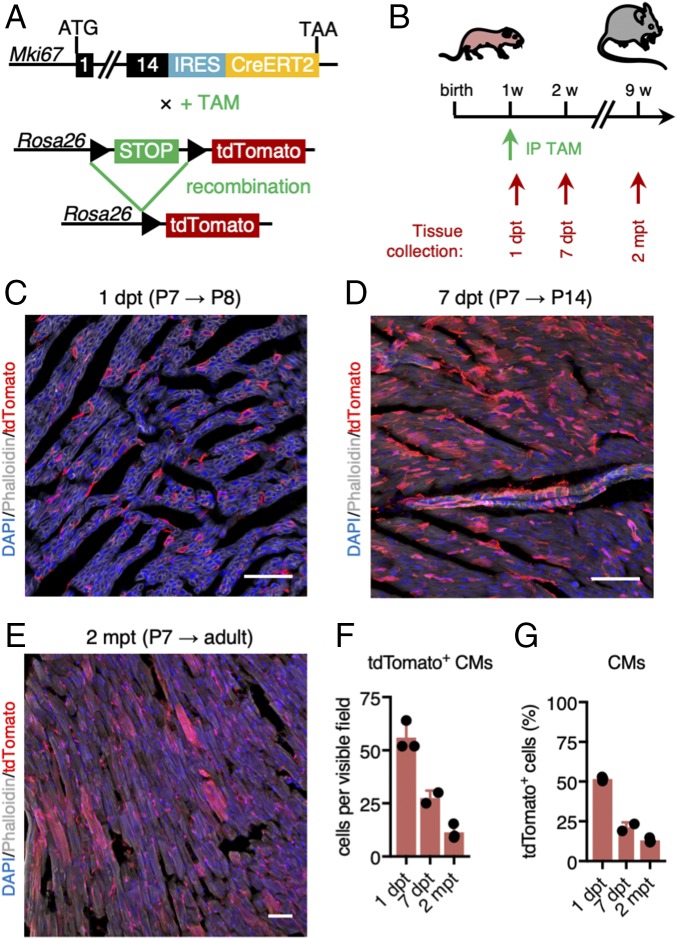
To identify the progeny of all proliferative cardiac cell types, we performed genetic lineage tracing of Mki67-expressing cells (25). We used mice in which an internal ribosome entry site (IRES)–Cre recombinase is fused to a tamoxifen-inducible version of the human estrogen receptor (IRES-CreERT2) expression cassette, inserted downstream of the Mki67 protein coding region (Mki67IRES-CreERT2) (24). We crossed these mice with Rosa26-CAG-loxP-stop-loxP-(LSL)-tdTomato reporter mice (hereafter LSL-tdTomato mice; Fig. 4A) (41). To assess initial labeling of proliferating Mki67-expressing cardiac cells, we first treated 1-wk-old [postnatal day 7 (P7)] Mki67IRES-CreERT2 × LSL-tdTomato mice with tamoxifen and collected tissue after 1 d [1 d post tamoxifen (dpt)] (Fig. 4B). We found many tdTomato+ cardiomyocytes 1 dpt (51.75 ± 0.77% of all tdTomato+ cells, i.e., 568 of an estimated 4,000 cardiomyocytes analyzed were tdTomato+; Fig. 4 C and F), confirming our single-cell transcriptome data. To trace the progeny of proliferating cells, we also collected hearts 1 wk (7 dpt) and 2 mo (2 mpt) after tamoxifen treatment at P7 (Fig. 4B). At 7 dpt, 21.23 ± 2.32% of all tdTomato+ cells were cardiomyocytes (Fig. 4 D, F, and G), while tdTomato+ cardiomyocytes represented 13.01 ± 0.87% of all reporter-labeled cells at 2 mpt (Fig. 4 E–G). This demonstrated that our approach also readily labels proliferative cardiomyocytes, but that the bulk of the cells produced from P7 proliferative (noncardiomyocyte) cells long-term represented noncardiomyocyte lineages.
Fig. 4.
Ki67 lineage tracing demonstrates de novo generation of cardiomyocytes in the neonatal murine heart. (A) Genetic cross between Mki67IRES-CreERT2 and LSL-tdTomato reporter mice to lineage trace Mki67-expressing cells. TAM, tamoxifen. (B) Neonatal hearts were collected 1 and 7 d post tamoxifen (dpt) as well as 8 wk post tamoxifen injection. Tamoxifen was injected at the age of 1 wk. IP, i.p. injection. (C–E) tdTomato labeling 1 d (C), 7 d (D), and 2 mo (E) after tamoxifen injection at postnatal day 7 (P7). Nuclei were stained with DAPI (blue). Phalloidin (green or gray) was used to stain polymerized F-actin. (Scale bars: 500 μm.) (F and G) Number of tdTomato+ cardiomyocytes per visible field (F) and percentage of tdTomato+ cardiomyocytes of all tdTomato+ cells (G) at 1 dpt (P8; n = 3 mice), 7 dpt (P14; n = 2 mice), and 2 mo post tamoxifen (mpt) (n = 3 mice).
Cardiac Cells of Noncardiomyocyte Lineages Are Continuously Replenished During Adult Homoeostasis in Murine Hearts.
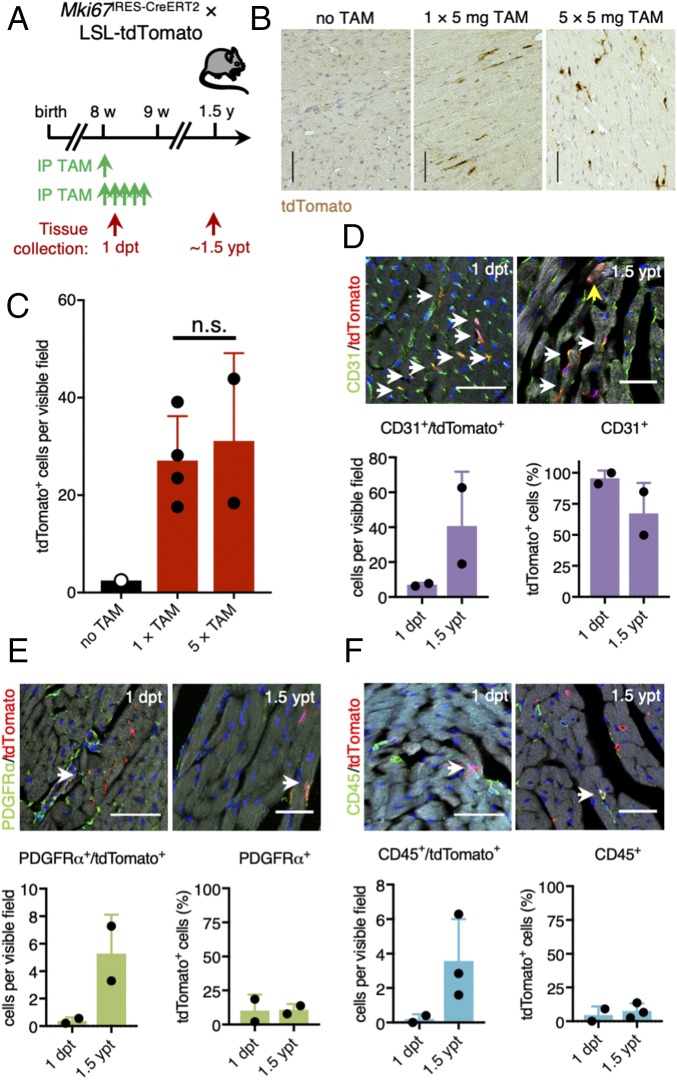
To trace proliferating cardiac cells during adult homoeostasis, we next treated 8-wk-old Mki67IRES-CreERT2 × LSL-tdTomato mice with tamoxifen and collected hearts 1 dpt and 1.5 y post tamoxifen exposure (1.5 ypt) (Fig. 5A). Reporter labeling in heart tissue was similar in mice injected one or five times with tamoxifen, with ∼30 tdTomato+ cells per 20× field (Fig. 5 B and C). In agreement with previous studies and a recent consensus statement describing modest cardiomyocyte turnover in adult mouse hearts (1, 5, 10), we found no tdTomato+ cardiomyocytes 1 dpt and only very few tdTomato+ cardiomyocytes 1.5 ypt (0.16% of all tdTomato+ cells or 11 of an estimated 8 million cardiomyocytes analyzed were tdTomato+; Fig. 5 B and C). The vast majority of proliferative cardiac cells labeled with tdTomato 1 d after tamoxifen injection were CD31+ endothelial cells (Fig. 5D), while the remaining tdTomato+ cells were either PDGFRα+ fibroblasts (Fig. 5E) or CD45+ hematopoietic cells (Fig. 5F). We found a similar abundance of these cell types among all tdTomato+ cells 1.5 ypt (Fig. 5 D–F), suggesting a continuous cellular turnover of these noncardiomyocyte lineages in the adult homoeostatic murine heart.
Fig. 5.
Continuous cellular turnover of noncardiomyocyte lineages during adult homoeostasis of the murine heart. (A) Timeline of tamoxifen injection and tissue collection in adult Mki67IRES-CreERT2 × LSL-tdTomato mice. (B and C) Representative images of stained paraffin sections (B) and corresponding quantification (C) of tdTomato labeling 1.5 y after tamoxifen exposure (1.5 ypt). Mice were either injected once with 5 mg of tamoxifen (n = 4 mice) or five times with 5 mg of tamoxifen (n = 2 mice). Noninjected mice were used as control (n = 1 mouse). (Scale bars: 100 μm.) (D–F) Representative images and quantification of costainings of tdTomato-traced cells (red) at 1 dpt and 1.5 ypt and CD31+ (D), PDGFRα+ (E), or CD45+ cells (F) (green) (n = 2–3 mice). ypt, years post tamoxifen. White arrows point at double-positive cells. The yellow arrow shows one of the tdTomato-labeled cardiomyocytes we found across sections. Nuclei were counterstained with DAPI (blue). Phalloidin (gray) was used to stain polymerized F-actin. [Scale bars: 50 μm (Upper) and 30 μm (Lower).]
Ki67 Lineage Tracing Reveals Proliferative Response by Noncardiomyocyte Cell Lineages Following Ischemic Injury.
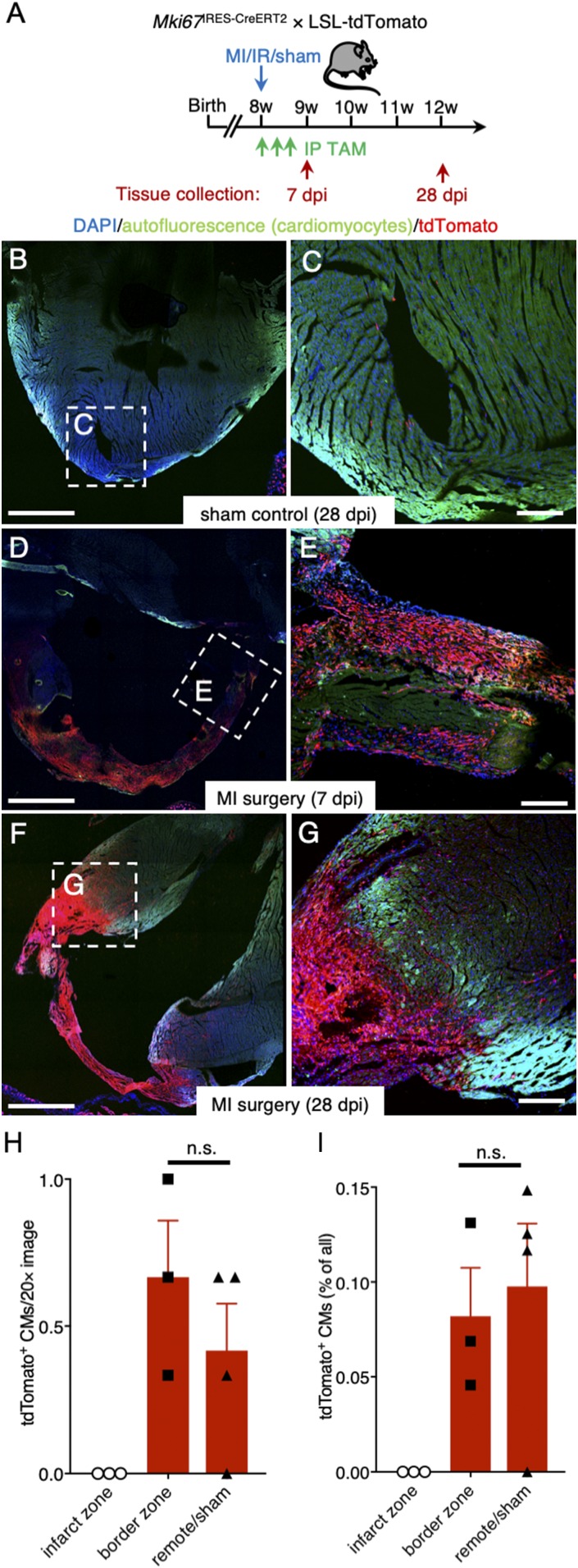
It has been proposed that endogenous quiescent CSCs reenter the cell cycle in response to cardiac injury and subsequently regenerate the myocardium (11). To trace such proliferating cells following ischemic injury, we performed MI surgery on 8-wk-old adult Mki67IRES-CreERT2 × LSL-tdTomato mice that received tamoxifen every other day for a total of three doses following the MI surgery (Fig. 6A). We chose these injection time points to label proliferative cardiac cells in the acute phase of the injury response of the myocardium, when proliferation of CSCs and de novo generation of cardiomyocytes had been described in previous studies (11, 42). Sham control hearts displayed tdTomato labeling similar to tracings found in homoeostatic adult hearts, while hearts collected 1 wk following MI showed abundant tdTomato+ cells in the infarcted area (Fig. 6 B and C). One month following MI, scar tissue was almost entirely composed of tdTomato+ cells, and robust tdTomato labeling was present in the border zone (Fig. 6 D and E). No tdTomato+ cardiomyocytes were found in the infarct zone, and we found no significant difference in the percentage of tdTomato+ cardiomyocytes in the MI border zone compared with the remote area, or to the hearts of sham mice (Fig. 6 F–I; Student’s t test, P ≥ 0.05). Thus, ischemic injury triggers a dramatic proliferative response in the infarcted myocardium including in the border zone but does not stimulate significant generation of new cardiomyocytes.
Fig. 6.
Ki67 lineage tracing reveals proliferative response following ischemic injury. (A) Experimental timeline. (B and C) Visualization of rare tdTomato+ cells in cryosections of sham control mice 28 dpi (n = 1 mouse). (D–G) Large accumulation of tdTomato+ cells in the infarct zone 7 dpi (D and E; n = 2 mice) and 28 dpi (F and G; n = 3 mice). The myocardium was visualized using cardiomyocyte autofluorescence; CM (green); nuclei were counterstained with DAPI (blue). (H) Quantification of tdTomato+ cardiomyocytes (CMs) per section scored (n = 3–4 mice). Student’s t test: n.s., not significant. (I) Percentage of tdTomato+ cardiomyocytes (n = 3–4 mice). Student’s t test: n.s. [Scale bars: 1 mm (B, D, and F) and 200 μm (C, E, and G).]
FSTL1 Expression Is Characteristic to a Population of Injury-Activated Cardiac Fibroblasts.
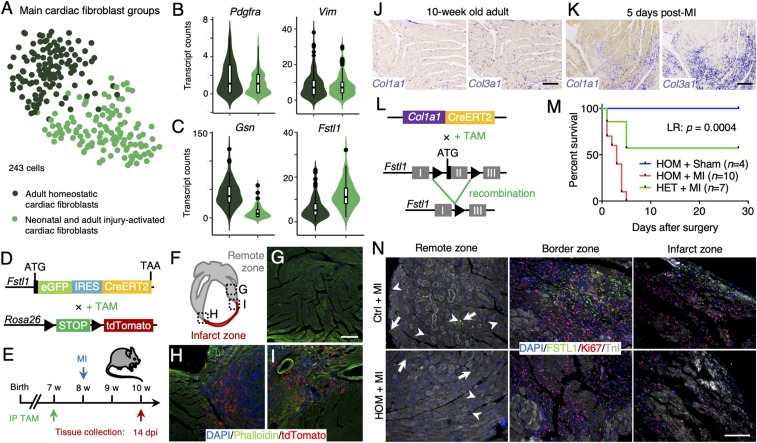
To more carefully document the role of cardiac fibroblasts in postdamage repair, we went back to our single-cell transcriptome dataset and performed refined cell type-specific subclustering. We first extracted the cardiac fibroblast fraction from our initial analysis (257 cells) and reran these through the RaceID2 algorithm with a higher cutoff of 2,000 transcripts per cell to allow for refined cell clustering (as done for the cardiomyocyte subclustering). After filtering, the 243 remaining cardiac fibroblasts clustered into 11 subclusters (SI Appendix, Fig. S6A). Cardiac fibroblasts purified from homoeostatic adult heart were mainly found in clusters 1, 3, 5, and 7, while neonatal fibroblasts and in the neonatal-like injury-associated fibroblasts were enriched in clusters 2, 4, and 8–11 (SI Appendix, Fig. S6 A and B). We combined these clusters and performed differential gene expression analysis (Fig. 7A). Lineage markers Pdgfra and Vim were robustly expressed in all fibroblasts (Fig. 7B) (27, 43, 44), while Gsn, a gene previously associated with cardiac remodeling after MI (45), was up-regulated in clusters enriched for adult homoeostatic fibroblasts (Fig. 7C and Dataset S4). Fibroblasts also robustly expressed Fstl1, a marker that was recently described as a cardiomyogenic factor of epicardial origin expressed in the adult murine heart (29). Expression of Fstl1 was significantly up-regulated in the clusters containing neonatal fibroblasts and neonatal-like injury-associated fibroblasts, but Fstl1 was also expressed by the fibroblasts in the healthy adult heart (Fig. 7C and Dataset S4).
Fig. 7.
FSTL1 expression is characteristic to a population of injury-activated fibroblasts and is crucial in preventing cardiac rupture upon damage. (A) t-SNE map of cardiac fibroblast subclusters identified using the RaceID2 algorithm projecting the different experimental conditions. (B and C) Violin plots showing the expression of Pdgfra, Vim (B), and Gsn and Fstl1 (C) in neonatal and injury-activated fibroblasts (light green) and homoeostatic adult fibroblasts (dark green). (D) Schematic representation of the generation of mice expressing eGFP and CreERT2 by cassette insertion within the Fstl1 protein coding region, enabling lineage tracing by crossing it with LSL-tdTomato mice. (E) Experimental timeline. (F–I) Progeny (red) of Fstl1-expressing fibroblasts is enriched in the scar tissue 14 dpi (H and I), while tdTomato labeling in the remote area remains low (G). Nuclei were counterstained with DAPI (blue) and F-actin visualized by phalloidin (green). (J and K) In situ hybridization of Col1a1 and Col3a1 in adult homoeostatic heart (J) and 5 d after MI (K). (L) Schematic representation of the generation of conditional knockout (cKO) mice by crossing Col1a1-CreERT2 mice with Fstl1flox/flox mice, resulting in the depletion of Fstl1 exon 2 upon tamoxifen induction. (M) Kaplan–Meier survival curve of homozygous cKO mice after MI (red), homozygous cKO mice after sham surgery (blue), and heterozygous cKO mice after MI (green) over the course of 30 d. (N) Immunofluorescence stainings of FSTL1 (green), Ki67 (red), and TnI (white) in damaged control and homozygous cKO hearts in remote, border, and infarct zone. Nuclei were counterstained with DAPI (blue). (Scale bars: 100 μm.)
To further explore this observation, we utilized a mouse model carrying an eGFP-IRES-CreERT2 expression cassette inserted into the Fstl1 locus (46) (Fig. 7D). As suggested by our single-cell RNA-sequencing dataset, eGFP+ cells in the neonatal hearts of these mice were cardiac fibroblasts residing in the myocardium (SI Appendix, Fig. S6 C–I). In contrast to a previous report (29), we did not find any eGFP expression in the epicardium (SI Appendix, Fig. S6H); eGFP+ cells in the adult heart were PDGFRα+ cardiac fibroblasts (SI Appendix, Fig. S6J).
Next, we asked whether Fstl1-eGFP+ fibroblasts residing in the myocardium were a source for the fibroblast populations that appear in the infarcted area following cardiac ischemic injury. We therefore generated Fstl1eGFP-IRES-CreERT2 × LSL-tdTomato mice to enable genetic lineage tracing of FSTL1+ cells (Fig. 7D). To exclusively label cardiac fibroblasts residing in the homoeostatic myocardium and thereby avoid possible Cre-induced recombination in cells expressing Fstl1 in response to ischemic injury, we injected tamoxifen 1 wk before induction of the MI and collected the tissue 2 wk following surgery (Fig. 7E). In mice undergoing sham surgery, no significant numbers of tdTomato+ cells were found. We made similar observations in the remote myocardium of injured hearts (Fig. 7 F and G). However, robust numbers of tdTomato+ fibroblasts were found in the infarcted region of injured hearts (Fig. 7 F, H, and I), confirming that these cells are indeed derived from preexisting cardiac fibroblasts infiltrating the infarct following cardiomyocyte apoptosis. Finally, we stained tissue sections from infarcted hearts with an antibody specific to FSTL1. As expected, confocal microscopy revealed that most FSTL1 immunoreactivity was present in the infarcted region of the myocardium and the border zone (SI Appendix, Fig. S6 K–N).
Collagens such as Col1a1 and Col3a1 were robustly expressed by cardiac fibroblasts (Fig. 7J) and were strongly up-regulated in response to cardiac injury (Dataset S4). Fibroblasts expressing these collagens made up a large fraction of the fibrotic scar (Fig. 7K). To elucidate the functional role of FSTL1 in the formation of the fibrotic scar, we generated Col1a1-CreERT2 × Fstl1flox/flox mice to enable conditional knockout (cKO) of Fstl1 in cardiac fibroblasts (Fig. 7L) (46). We injected cKO and control mice with tamoxifen 2 wk before LAD occlusion to specifically remove Fstl1 from the Col1a1-expressing fibroblast population. While loss of Fstl1 did not seem to have an effect under homoeostatic conditions (Fig. 7M), it resulted in a high mortality rate within the first 5 d following LAD occlusion with all cKO mice dying due to cardiac rupture, a rupture of the left ventricular-free wall (Fig. 7M). Heterozygous cKO or wild-type control animals, with one or two remaining functional Fstl1 alleles, had a significantly lower mortality rate (Fig. 7M; log rank test, P = 0.0004). Characterization of the fibrotic scar in homozygous cKO mice demonstrated fewer Ki67+ proliferating cells in both the border and the infarct zones compared with control (Fig. 7N). Collectively, these data imply that FSTL1 is a marker of injury-activated fibroblasts that—in an autocrine fashion—signals to prevent cardiac rupture upon damage.
Discussion
We have generated an exhaustive map of proliferative cardiac cells and their progeny during three critical conditions of postnatal life. Here, we assessed cardiac cell proliferation in an unbiased fashion. Most studies on cardiac regeneration focus on cell cycle reentry of cardiomyocytes or the existence of resident stem cells, thereby restricting the analysis by the use of generic stem cell markers or of DNA label retention as a surrogate stem cell marker (4, 5, 11, 12, 18). Hence, we think that our dataset allows resolution of current controversies on cardiac regeneration (6–9, 17).
While our data demonstrate that neonatal murine cardiomyocytes are actively cycling (Figs. 1 and 2), we did not find evidence for significant de novo generation of cardiomyocytes in the homoeostatic or regenerating adult murine heart through cell division (Figs. 4 and 5). In accordance with this, recent studies by the laboratories of Jonas Frisén and Olaf Bergmann used radiocarbon dating to demonstrate that the number of cardiomyocytes in human and mice is mainly established in the perinatal period and turnover rates decrease throughout life (1, 47). These data were confirmed by other means (2, 18). Of note, significant induction of proliferation of adult postmitotic cardiomyocytes has only been achieved in vivo by overexpressing cell cycle regulators that are usually solely expressed by fetal cardiomyocytes (48).
It has been proposed that endogenous quiescent CSCs reenter the cell cycle in response to cardiac injury (9, 11, 36). We find no evidence for such CSCs that would contribute to the myocardial lineage upon damage. Furthermore, we find that proposed CSC markers are widely expressed by noncardiomyocyte cardiac lineages (Fig. 3). Putative c-KIT+ CSCs, for example, are often purified by removing CD45+ hematopoietic cells and CD31+ endothelial cells (4, 11, 36). However, cardiac fibroblasts in our single-cell transcriptome dataset also express Kit transcripts. This demonstrates that Kit is expressed by a wide range of cardiac cell populations, which may explain some of the opposing results published on the role of c-KIT+ cells for cardiac regeneration (4, 8, 9, 11, 15, 36).
In contrast to previous studies utilizing label incorporation (i.e., BrdU) into the DNA of S-phase cells to assess proliferation of putative CSCs or cardiomyocytes (4, 11, 42), we utilize genetic lineage tracing of Ki67+ cardiac cells to investigate evidence for de novo generation of cardiomyocytes in response to myocardial damage. Our data do not reveal significant numbers of newly formed cardiomyocytes (Fig. 5), in line with a recent study assessing cardiomyocyte expansion during murine heart injury (49). While we did not find evidence for cardiac regeneration through actively cycling cardiomyocytes or CSCs, we charted the robust injury response in the adult mammalian heart that essentially consists of proliferation of noncardiomyocyte cardiac cell lineages (Figs. 1, 2, and 5). The majority of proliferative cells in the adult homoeostatic heart are CD31+ cells of the vasculature (Fig. 5). However, hematopoietic cells as well as cardiac fibroblasts robustly enter the cell cycle in the infarcted areas of the myocardium of injured adult mice (Figs. 6 and 7). Cardiac fibroblasts proliferated in response to ischemic injury and adopt a gene expression profile similar to that of neonatal cardiac fibroblasts. This phenomenon has previously been described for adult murine skin epidermis, where dermal fibroblasts are reprogrammed to a neonatal-like state in response to sustained activation of epidermal Wnt/β-catenin signaling (50). A subsequent study showed that adult fibroblast remodeling significantly improved epidermal regeneration upon wounding of adult murine skin (51).
Our single-cell transcriptome dataset further showed that the proposed epicardial-specific cardiomyogenic factor FSTL1 (29) is prominently expressed by the cardiac fibroblast lineage. We do not find evidence for significant epicardial expression of FSTL1 under any of the conditions analyzed. We confirmed our single-cell data using a Fstl1-eGFP reporter mouse as well as by histological staining (Fig. 7 and SI Appendix, Fig. S6) (46). Using genetic lineage tracing, we subsequently demonstrate that Fstl1-expressing fibroblasts contribute principally to the myocardial remodeling occurring in response to cardiac injury. Finally, we show that the genetic deletion of Fstl1 in myocardial fibroblasts results in a severe phenotype with high mortality due to cardiac rupture upon injury (Fig. 7M), a phenomenon similar to that described in another study with a less pronounced phenotype (52). This demonstrates the importance of autocrine FSTL1 signaling in scar formation.
Overall, our results imply that all markers used in previous studies to identify CSCs lack appropriate specificity in the heart. In addition, we find that postinfarct generation of new cardiomyocytes through proliferation of any type of cardiac cells is negligible, compared with the proliferative response of noncardiomyocytes. We find proliferating cells within all other cardiac cell lineages, suggesting that these lineages are not maintained by distinct stem cells, but rather through cell division within the cell lineage. The proliferative response of cardiac fibroblasts in particular is important for infarct healing, yet it does not generate new cardiomyocytes, but rather a postinfarction fibrotic scar.
Materials and Methods
Mice.
Generation and genotyping of Mki67IRES-CreERT2 (24), Mki67TagRFP (23), Fstl1eGFP-IRES-CreERT2, and Fstl1flox/flox (46). LSL-tdTomato [B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J] (41) and Col1a1-CreERT2 [B6.Cg-Tg(Col1a1cre/ERT2)1Crm/J] (53) were described elsewhere. All alleles were maintained on a mixed C57BL/6 background. All mouse experiments were conducted under a project license granted by the Central Committee Animal Experimentation of the Dutch government and approved by the Hubrecht Institute Animal Welfare Body and by the Animal Experimental Committee of the Academic Medical Center, Amsterdam University Medical Centers. Littermates were used as controls, as indicated for each experiment. Both male and female mice were used in all experiments, except for mRNA-sequencing experiments, which were preferentially performed with male mice.
Surgical Procedures.
MI was induced by permanent occlusion of the LAD coronary artery. Ischemia/reperfusion (I/R) was performed by LAD occlusion for 1 h (ischemic period), followed by LAD reperfusion. Detailed surgical procedures are described in SI Appendix, SI Materials and Methods.
Genetic Lineage-Tracing Experiments.
Genetic lineage tracing of Mki67-expressing cells or Fstl1-expressing cells was induced by treating, respectively, Mki67IRES-CreERT2 × LSL-tdTomato or Fstl1eGFP-IRES-CreERT2 × LSL-tdTomato with tamoxifen (Sigma-Aldrich) dissolved in sunflower oil via the i.p. route. Details on the experimental procedures, treatment regime, and tamoxifen dosage injected are provided in SI Appendix, SI Materials and Methods.
cKO Experiment.
For cKO of Fstl1 in cardiac fibroblasts, 8-wk-old Col1a1-CreERT2 × Fstl1flox/flox mice were injected with 10 mg/kg tamoxifen for 4 consecutive days. Two weeks after the start of tamoxifen injection, mice underwent permanent ligation of the LAD artery. Survival rate was monitored or mice were sedated with CO2 and killed up to 30 d after surgery.
Cardiac Cell Isolation.
Mouse hearts were dissected, the atria were removed, and, when suitable, hearts were divided up into remote and apex (i.e., for MI or I/R surgery, border zone and infarcted zone). Whole ventricles were prepped from dissected hearts of neonatal mice. Tissue dissection was either performed as recently described (28) or as detailed in SI Appendix, SI Materials and Methods.
Flow Cytometric Purification.
4′,6-Diamidino-2-phenylindole (DAPI) was added immediately before flow sorting. DAPI-negative and MitoTracker-positive living cells were either sorted into TRIzol reagent (Thermo Scientific) for bulk mRNA sequencing or into 384-well plates containing 96 or 384 unique molecular identifier barcode primer sets, ERCC92 spike-ins (Agilent) and dNTPs (Promega) for single-cell mRNA-sequencing (SORT-seq) (30) using a flow sorter (FACSAriaII, FACSFusion, or FACSJazz; all BD). Sorted samples were stored at −80 °C until further processing.
mRNA Sequencing.
Samples were lysed and RNA from each bulk sort sample or single cell was barcoded and processed using the CEL-Seq2 technique (30, 31, 54). Details on the sequencings performed are described in SI Appendix, SI Materials and Methods. Bioinformatics analysis of sequenced libraries was performed as described below.
Histology.
Histology on cryosections and paraffin sections was performed using standard methods as detailed in SI Appendix, SI Materials and Methods.
The following antibodies or labeling agents were used for immunostainings: rat anti-CD45 (30F11; eBiosciences), goat anti-mFSTL1 (AF1738; R&D Systems), goat anti-mouse PDGFRα (AF1062; R&D Systems), phalloidin Alexa Fluor 488 (A12379; Life Technologies), phalloidin Atto 647N (Sigma), rabbit anti-RFP/tdTomato (600–401-379; Rockland), mouse anti-Ki67 (556003; BD Biosciences), Alexa Fluor 568 donkey anti-rabbit IgG (A10042; Life Technologies), and Alexa Fluor 568 donkey anti-goat IgG (A11057; Life Technologies).
In Situ Hybridization.
In situ hybridization (ISH) was performed on 8-μm paraffin sections of mouse hearts, as described (46). Detailed ISH procedures are described in SI Appendix, SI Materials and Methods.
Imaging.
Images of cryosections were acquired on an inverted confocal laser scanning microscope (Leica SP8X and SP8). Paraffin sections were imaged using a Leica DM4000 or DM6000 optical microscope. ISH sections were imaged using a Leica DM5000 microscope.
Bioinformatics Analysis.
DNA library sequencing, mapping to the mouse reference genome, and quantification of transcript abundance were performed as described elsewhere (30). Bulk sequencing libraries were analyzed using the DESeq2 package (26). Cell clusters and gene expression levels across clusters were generated using the RaceID2 algorithm (31) from single-cell libraries. All bioinformatics analysis was performed using R, version 3.4.0 (R Foundation; https://www.r-project.org), and RStudio, version 1.0.143 (https://www.rstudio.com). Detailed bioinformatics analysis is described in SI Appendix, SI Materials and Methods.
Statistics.
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to the sample allocation during experiments and outcome assessment. No animals were excluded from analysis. All data are presented as mean ± SD, unless stated otherwise. Statistically significant differences between the mean of two conditions or experimental groups were determined using an unpaired two-tailed Student’s t test. A χ2 test (χ2) was performed to determine statistical significance for the data presented in Fig. 2F. A log-rank (Mantel–Cox) test was performed to determine statistical significance for the data presented in Fig. 7M. Data were obtained from at least two biological replicates per condition and from at least two independent experiments to ensure reproducibility. Kaplan–Meier survival curves were generated, and all statistical analyses were performed using GraphPad Prism, version 7.0, software.
Data Resources.
Differentially expressed genes of the initial cardiac cell clustering, of the cardiomyocyte subclustering, of the main cardiomyocyte clusters, and of the cardiac fibroblast subclustering are provided in Datasets S1–S4, respectively. RNA-sequencing data were deposited in the Gene Expression Omnibus (GEO) database (accession number GSE102048), except for some single-cell transcriptome data of cardiac cells isolated from wild-type (Mki67wt/wt) mice, which were published elsewhere (28).
Supplementary Material
Acknowledgments
We thank Benedetta Artegiani and Talya Dayton for discussions and critical reading of the manuscript; Reinier van den Linden for help with flow cytometry; Judith Vivié for help with SORT-seq; the Utrecht Sequencing Facility for sequencing; Mauro Muraro and Anna Alemany Arias for help with bioinformatics analysis; and Anko de Graaff and the Hubrecht Imaging Centre for assistance with microscopy. This work was supported by a Cardiovasculair Onderzoek Nederland (CVON)–Human Stem Cells for Cardiac Repair (HUSTCARE) project grant (to H.C.) and a CVON-HUSTCARE Young Talent Grant (to K.K.). K.K. is a long-term fellow of the Human Frontier Science Program Organisation (LT771/2015) and was the recipient of a VENI grant from the Netherlands Organisation for Scientific Research (NWO-ZonMW) (016.166.140).
Footnotes
Conflict of interest statement: H.C. is co-principal investigator (co-PI) on a Dutch grant on organs-on-a-chip with C.L.M. as co-PI, however, on an unrelated topic.
Data deposition: The RNA-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE102048).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805829115/-/DCSupplemental.
References
- 1.Alkass K, et al. No evidence for cardiomyocyte number expansion in preadolescent mice. Cell. 2015;163:1026–1036. doi: 10.1016/j.cell.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 2.Soonpaa MH, et al. Cardiomyocyte cell-cycle activity during preadolescence. Cell. 2015;163:781–782. doi: 10.1016/j.cell.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 3.Naqvi N, et al. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell. 2014;157:795–807. doi: 10.1016/j.cell.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 5.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Berlo JH, Molkentin JD. An emerging consensus on cardiac regeneration. Nat Med. 2014;20:1386–1393. doi: 10.1038/nm.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uygur A, Lee RT. Mechanisms of cardiac regeneration. Dev Cell. 2016;36:362–374. doi: 10.1016/j.devcel.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Berlo JH, et al. van Berlo et al. reply. Nature. 2018;555:E18. doi: 10.1038/nature25772. [DOI] [PubMed] [Google Scholar]
- 9.Vicinanza C, et al. Kitcre knock-in mice fail to fate-map cardiac stem cells. Nature. 2018;555:E1–E5. doi: 10.1038/nature25771. [DOI] [PubMed] [Google Scholar]
- 10.Eschenhagen T, et al. Cardiomyocyte regeneration: A consensus statement. Circulation. 2017;136:680–686. doi: 10.1161/CIRCULATIONAHA.117.029343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison GM, et al. Adult c-kitpos cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 12.Uchida S, et al. Sca1-derived cells are a source of myocardial renewal in the murine adult heart. Stem Cell Rep. 2013;1:397–410. doi: 10.1016/j.stemcr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfister O, et al. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circ Res. 2008;103:825–835. doi: 10.1161/CIRCRESAHA.108.174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yellamilli A, van Berlo JH. The role of cardiac side population cells in cardiac regeneration. Front Cell Dev Biol. 2016;4:102. doi: 10.3389/fcell.2016.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jesty SA, et al. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci USA. 2012;109:13380–13385. doi: 10.1073/pnas.1208114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai CL, Molkentin JD. The elusive progenitor cell in cardiac regeneration: Slip slidin’ away. Circ Res. 2017;120:400–406. doi: 10.1161/CIRCRESAHA.116.309710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura W, et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature. 2015;523:226–230. doi: 10.1038/nature14582. [DOI] [PubMed] [Google Scholar]
- 19.Nakada Y, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–227. doi: 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- 20.Clevers H. STEM CELLS. What is an adult stem cell? Science. 2015;350:1319–1320. doi: 10.1126/science.aad7016. [DOI] [PubMed] [Google Scholar]
- 21.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 23.Basak O, et al. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014;33:2057–2068. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basak O, et al. Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc Natl Acad Sci USA. 2018;115:E610–E619. doi: 10.1073/pnas.1715911114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto AR, et al. Revisiting cardiac cellular composition. Circ Res. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gladka MM, et al. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation. 2018;138:166–180. doi: 10.1161/CIRCULATIONAHA.117.030742. [DOI] [PubMed] [Google Scholar]
- 29.Wei K, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muraro MJ, et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grün D, et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skelton RJ, et al. SIRPA, VCAM1 and CD34 identify discrete lineages during early human cardiovascular development. Stem Cell Res (Amst) 2014;13:172–179. doi: 10.1016/j.scr.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 33.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 34.Bruno S, Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 1992;25:31–40. doi: 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]
- 35.Bruno S, Crissman HA, Bauer KD, Darzynkiewicz Z. Changes in cell nuclei during S phase: Progressive chromatin condensation and altered expression of the proliferation-associated nuclear proteins Ki-67, cyclin (PCNA), p105, and p34. Exp Cell Res. 1991;196:99–106. doi: 10.1016/0014-4827(91)90460-c. [DOI] [PubMed] [Google Scholar]
- 36.Vicinanza C, et al. Adult cardiac stem cells are multipotent and robustly myogenic: c-kit expression is necessary but not sufficient for their identification. Cell Death Differ. 2017;24:2101–2116. doi: 10.1038/cdd.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- 39.Ranjbarvaziri S, et al. Generation of Nkx2-5/CreER transgenic mice for inducible Cre expression in developing hearts. Genesis. 2017;55:e23041. doi: 10.1002/dvg.23041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart N, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tallquist MD, Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol. 2017;14:484–491. doi: 10.1038/nrcardio.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeisberg EM, Kalluri R. Origins of cardiac fibroblasts. Circ Res. 2010;107:1304–1312. doi: 10.1161/CIRCRESAHA.110.231910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li GH, et al. Gelsolin regulates cardiac remodeling after myocardial infarction through DNase I-mediated apoptosis. Circ Res. 2009;104:896–904. doi: 10.1161/CIRCRESAHA.108.172882. [DOI] [PubMed] [Google Scholar]
- 46.Sylva M, et al. The BMP antagonist follistatin-like 1 is required for skeletal and lung organogenesis. PLoS One. 2011;6:e22616. doi: 10.1371/journal.pone.0022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergmann O, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 48.Mohamed TMA, et al. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173:104–116.e12. doi: 10.1016/j.cell.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sereti KI, et al. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat Commun. 2018;9:754. doi: 10.1038/s41467-018-02891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins CA, Kretzschmar K, Watt FM. Reprogramming adult dermis to a neonatal state through epidermal activation of β-catenin. Development. 2011;138:5189–5199. doi: 10.1242/dev.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Driskell RR, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruyama S, et al. Follistatin-like 1 promotes cardiac fibroblast activation and protects the heart from rupture. EMBO Mol Med. 2016;8:949–966. doi: 10.15252/emmm.201506151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JE, Nakashima K, de Crombrugghe B. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: A new tool to examine physiology and disease of postnatal bone and tooth. Am J Pathol. 2004;165:1875–1882. doi: 10.1016/S0002-9440(10)63240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hashimshony T, et al. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.