Significance
During HIV maturation, protease cleaves the Gag polyprotein in immature particles, which culminates in dramatic morphological changes and assembly of mature, infectious particles. The final cleavage step occurs between the C-terminal domain (CTD) of the CA capsid protein and the SP1 spacer peptide. A potential therapeutic strategy is blockage of the CTD-SP1 protease binding, and the first-in-class maturation inhibitor was bevirimat. Unfortunately, therapeutic development was abrogated by resistance mutants. To date, the detailed mechanism of action of bevirimat has not been determined. Therefore, we performed electron cryomicroscopy and electron diffraction of thin 3D microcrystals of CTD-SP1 with and without bound bevirimat. The atomic resolution structures revealed the detailed mechanism for binding of bevirimat, which will inform further drug discovery efforts.
Keywords: HIV-1, Gag, maturation inhibitors, drug discovery, cryoEM
Abstract
HIV-1 protease (PR) cleavage of the Gag polyprotein triggers the assembly of mature, infectious particles. Final cleavage of Gag occurs at the junction helix between the capsid protein CA and the SP1 spacer peptide. Here we used MicroED to delineate the binding interactions of the maturation inhibitor bevirimat (BVM) using very thin frozen-hydrated, 3D microcrystals of a CTD-SP1 Gag construct with and without bound BVM. The 2.9-Å MicroED structure revealed that a single BVM molecule stabilizes the six-helix bundle via both electrostatic interactions with the dimethylsuccinyl moiety and hydrophobic interactions with the pentacyclic triterpenoid ring. These results provide insight into the mechanism of action of BVM and related maturation inhibitors that will inform further drug discovery efforts. This study also demonstrates the capabilities of MicroED for structure-based drug design.
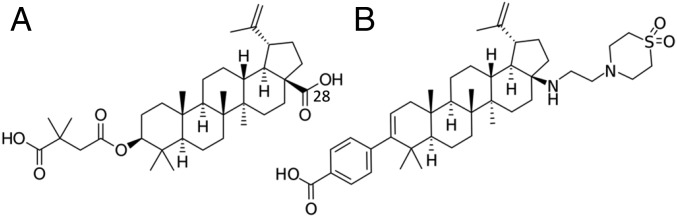
HIV-1 Gag and Gag-Pol assemble and bind the plasma membrane of infected cells (1). The packing interactions between Gag hexamers induce positive membrane curvature, and engagement of cellular endosomal sorting complexes required for transport results in budding and release of immature HIV-1 virus particles composed of the membrane-encased shell formed by the Gag lattice and the viral RNA. In the immature virion, Gag hexamers form a fissured, spherical shell with quasi-hexagonal symmetry on the inner leaflet of the viral envelope (2–4). HIV-1 viral protease (PR) is liberated from Gag-Pol by autoproteolysis at a late stage of viral assembly, which ensures that Gag molecules are not processed before assembly of the lattice. PR cleaves five sites in Gag at different rates in the following order: SP1-NC > MA-CA ∼ SP2-P6 > NC-SP2 > CA-SP1 (5–10). The final CA-SP1 cleavage triggers the dramatic morphological changes between immature and mature infectious HIV-1 particles that contain the characteristic conical capsid (11, 12). Inhibition of Gag cleavage effectively prevents HIV-1 replication, and two therapeutic strategies have been pursued: protease inhibitors that bind directly to PR and maturation inhibitors, which interfere with PR cleavage of Gag. The first characterized maturation inhibitor was bevirimat (BVM) (Fig. 1A), which acts by preventing PR cleavage between residues L363 and A364 in the junction helix formed by the C-terminal domain of CA and the SP1 spacer peptide, thereby stabilizing the immature lattice (13, 14). However, the mechanism by which BVM stabilizes the immature lattice is unclear. Although BVM entered clinical trials, efficacy was thwarted by the presence of preexisting resistant viruses containing SP1 polymorphisms (15, 16). Second-generation maturation inhibitors (e.g., Fig. 1B) have improved resistance profiles (17, 18), but rational drug development has been impeded by a lack of high-resolution structural data on maturation inhibitor binding to Gag.
Fig. 1.
Schematic chemical representations of (A) BVM and (B) BMS-955176, a second-generation maturation inhibitor. BMS-955176 preserves the terminal carboxyl of BVM and includes a C-28 extension that may provide additional stability to the six-helix bundle in the case of resistance mutations. Adapted with permission from ref. 47. Copyright 2016 American Chemical Society.
In 2016, compelling evidence for the predicted six-helix bundle structure of the junction helices was provided by electron cryotomography and subtomogram averaging of ΔMA-Gag, nonenveloped viruslike particles (VLPs) (19), X-ray crystallography of a Gag construct composed of the C-terminal domain (CTD) of CA and SP1 (20), and solid-state NMR spectroscopy of ΔMA-Gag VLPs (21). The cryoEM and X-ray structures revealed that the protease cleavage sites are sequestered on the interior of the six-helix bundle. The cryoEM map also showed that a single molecule of the maturation inhibitor bevirimat binds at the center of each six-helix bundle. To achieve a resolution of 3.9 Å, sixfold averaging was applied to the cryoEM map, which prevented characterization of the interactions between BVM and the CTD-SP1 junction helices.
To examine the molecular basis for BVM binding, we used electron diffraction from frozen-hydrated, 3D microcrystals, so-called MicroED (microcrystal electron diffraction) (22–26), to solve the structures of drug-free and BVM-bound CTD-SP1 to resolutions of 3.0 and 2.9 Å, respectively, without applying sixfold symmetry (Table 1).
Table 1.
MicroED data collection and refinement statistics
| Drug-free CTD-SP1* | CTD-SP1-BVM† | |
| Data collection | ||
| Space group | C121 | C121 |
| Cell dimensions | ||
| a, b, c, Å | 70.6, 122.5, 78.7 | 71.4, 123.0, 81.8 |
| α, β, γ, ° | 90.0, 90.0, 96.5 | 90.0, 90.0, 95.0 |
| Resolution, Å | 27.5–3.0 (3.2–3.0) | 27.4–2.9 (3.1–2.9) |
| Rpim | 0.255 (1.324) | 0.180 (0.860) |
| <I/σI> | 3.0 (0.7) | 3.7 (1.1) |
| CC1/2 | 0.90 (0.35) | 0.96 (0.10) |
| Completeness, % | 79.8 (78.7) | 83.4 (82.3) |
| Redundancy | 5.7 (5.7) | 7.5 (7.5) |
| Refinement | ||
| Resolution, Å | 19.9–3.0 (3.2–3.0) | 19.9–2.9 (3.0–2.9) |
| No. reflections | 10,480 (1,006) | 11,635 (1,137) |
| Rwork/Rfree | 0.254/0.292 | 0.236/0.280 |
| No. protein atoms | 7,410 | 7,814 |
| ADP, Å2 | ||
| Mean | 41.1 | 52.6 |
| Minimum/maximum | 6.8/177.9 | 19.2/160.5 |
| rms deviations | ||
| Bond lengths, Å | 0.003 | 0.002 |
| Bond angles, ° | 0.561 | 0.474 |
| MolProbity clashscore | 1.48 | 0.38 |
Values in parentheses are for the highest-resolution shell.
Data from six crystals.
Data from five crystals.
Results
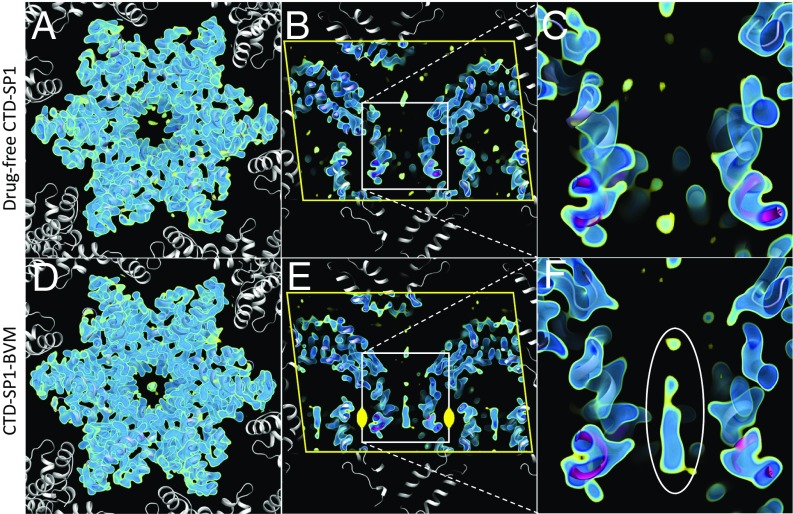
The CTD-SP1 crystals were of the same form that was used to solve the drug-free X-ray crystallographic structure to 3.27-Å resolution (20). The CTD-SP1 Gag construct was expressed and purified as described previously (20), and the crystallization conditions were adapted to grow microcrystals <1 μm thick (as described in Methods and SI Appendix, Fig. S1). The asymmetric unit contains one CTD-SP1 hexamer, and layers of the C2 crystals approximate the HIV-1 Gag immature lattice (Fig. 2). Molecular replacement phasing of the CTD-SP1-BVM MicroED data with the CTD X-ray structure (PDB 5I4T) resulted in a map that contains BVM density at the center of the six-helix bundle formed by the CTD-SP1 junction helices (Fig. 2 D–F). The density attributed to BVM is absent in our MicroED map of drug-free CTD-SP1 (Fig. 2 A–C). The location of the single BVM molecule within the six-helix bundle is consistent with a 3.9-Å map of HIV-1 Gag VLPs determined by electron cryotomography and subtomogram averaging (19). In the case of the cryoEM map of VLPs, the BVM density was a featureless ellipsoid due to sixfold averaging required to achieve a resolution of 3.9 Å for the protein density. However, sixfold averaging was not applied in the determination of the CTD-SP1-BVM MicroED map, and the BVM density is asymmetric with a dominant feature consistent with en face (Fig. 3C) and profile (Fig. 3D) views of the pentacyclic triterpenoid moiety of BVM. The presence of a unique mode of binding is presumably a consequence of the twofold symmetric crystallographic environment of the binding site (Fig. 2E).
Fig. 2.
MicroED maps of drug-free (3.0 Å) and BVM-bound (2.9 Å) CTD-SP1. Maps include multiple contour levels from 1.2σ (yellow) to 2.0σ (dark blue). Indicated in B and E are crystallographic symmetry mates (white), unit cell edges (yellow line), and the twofold symmetry operator (yellow). (A and D) Views along the hexamer axis showing the CTD domain and packing that mimics the immature HIV-1 lattice. (B and E) Cutaway of the CTD-SP1 hexamers along the hexamer axis (yellow line, unit cell boundary). (C and F) Close-ups showing the center of the six-helix bundles (slate blue, CTD; magenta, SP1), with BVM density present in F (white ellipse).
Fig. 3.
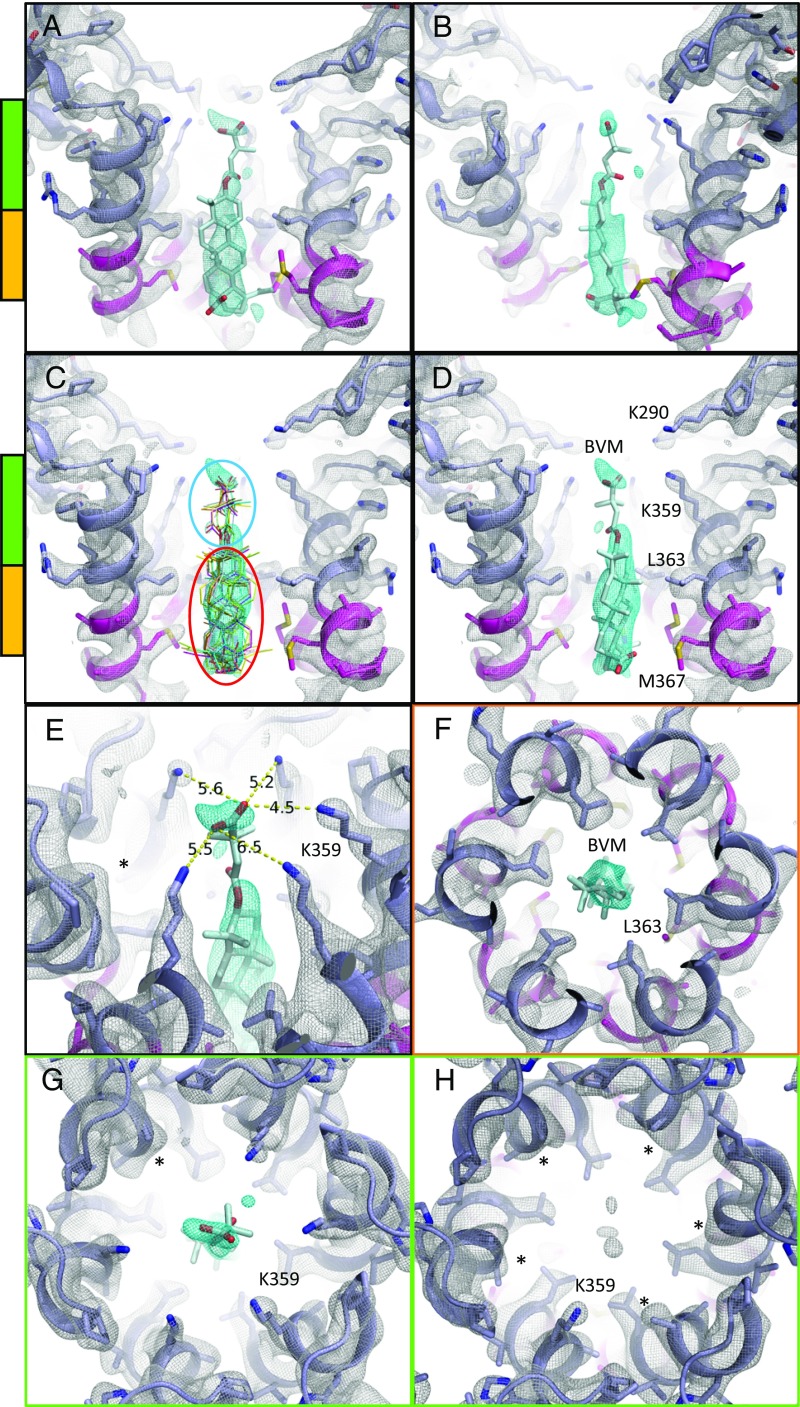
MicroED map of CTD-SP1-BVM suggests a mode of drug binding. CTD-SP1-BVM map contoured at 1.2σ above the mean (a single map is shown with protein density colored white and BVM density in cyan). Green and orange bars indicate the axial positions shown in F–H. BVM was never included in the model used for map calculation. (A) En face and (B) profile views of BVM density within the six-helix bundle formed at the junction of CTD (slate blue) and SP1 (magenta). (C) Eleven BVM conformers (48) docked into the map show that the rigid pentacyclic triterpenoid (red ellipse) fits well into the primary drug density, while the terminal carboxyl of each conformer fits within the satellite peak. Flexibility of the dimethylsuccinyl group (blue ellipse) is likely responsible for the break between the primary and satellite drug densities. (D) A single BVM conformer shows that drug binding spans the length of the junction helices and positions the dimethylsuccinyl carboxyl at the center of the K359 ring of lysines. (E) The K359 side chain densities and B factors vary among the protomers, supporting a preferred BVM binding pose with associated, specific K359 interactions. The side chain of one K359 (*) was not modeled due to a complete lack of density. (F) L363 of the CTD-SP1 protease cleavage site contributes the preponderance of hydrophobic interactions with BVM. The opposite axial orientation (BVM-down) leaves the satellite peak unoccupied and positions the C-28 carboxyl of the pentacyclic triterpenoid distant from K359 (Fig. 4B). (G) Axial view of the BVM binding site showing density for five of six K359 side chains near the terminal BVM carboxylic acid. (H) Axial view of drug-free CTD-SP1 showing the absence of density for five of six K359 side chains.
We define the axial orientation of the bound drug according to the location of the dimethylsuccinyl group, either oriented toward (BVM-up, Fig. 4A) or away from (BVM-down, Fig. 4B) the CA domain. There is a distinct satellite density directed toward the CA domain, which we ascribe to the dimethylsuccinyl moiety (Fig. 3 A–E). This BVM-up orientation places the carboxylic acid of the dimethylsuccinyl moiety in proximity to a ring of lysine residues (K359). Interestingly, the extent of the density of the K359 side chains varies between protomers in the BVM-bound map. In particular, the side chain density is better defined for the K359 residue that is closest to the density peak that we ascribe to the dimethylsuccinyl (Fig. 3 E and G). In contrast, density for the K359 side chain is absent in five of six protomers in the drug-free CTD-SP1 map (Fig. 3H), likely due to conformational flexibility.
Fig. 4.
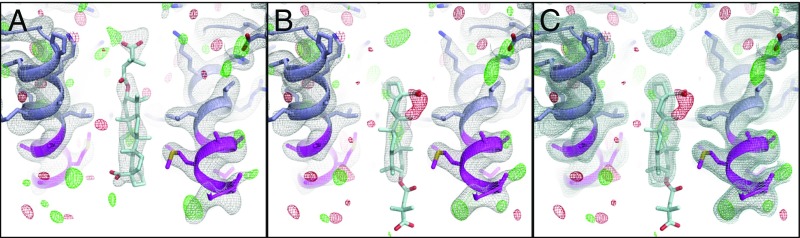
MicroED map suggests the orientation of the bound bevirimat. (A and B) CTD-SP1-BVM 2Fo–Fc (gray, contoured at 1.0σ) and Fo–Fc (green and red, contoured at ±3.0σ) maps with BVM modeled in different axial orientations. (A) In the up orientation, the dimethylsuccinyl group points toward the CTD. (B) In the down orientation it points away from the CTD. (A) In the up orientation, the pentacyclic triterpenoid moiety occupies the large map feature near the CTD-SP1 protease cleavage site, and the carboxyl of BVM is proximal to the ring of K359 lysines. (B) In the down orientation, when the pentacyclic triterpenoid is positioned in the primary drug density, there is no density for the dimethylsuccinyl group, and the C-28 carboxyl is positioned far from K359. During refinement, without BVM, some maps include weak extensions of the primary drug density peak (>0.6σ) out of the helical bundle, that is, away from the CTD. However, in most maps, the drug density terminates abruptly. Following refinement with BVM in the down orientation, no density is present for the dimethylsuccinyl, which could be attributable to high mobility. (C) Maps are the same as in B with an additional 2Fo–Fc contour level at 0.6σ above the mean.
Refinement of the structure with BVM in the up orientation also supports our interpretation (Fig. 4A), whereas refinement with BVM in the down orientation positions the C-28 carboxyl more than 7 Å away from the nearest K359 Nζ, and the dimethylsuccinyl extends out of the junction helix bundle (Fig. 4 B and C).
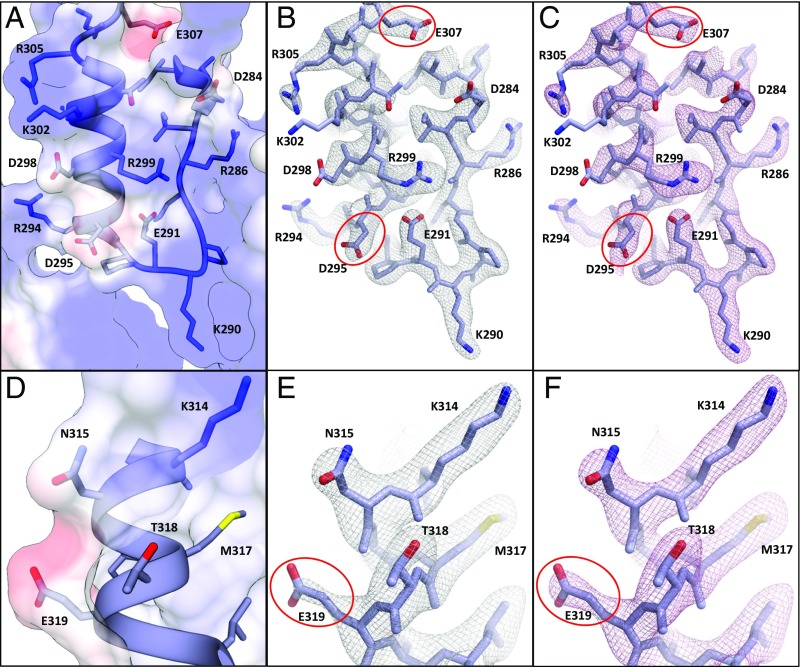
The presence of carboxylic acid groups at each end of BVM (Fig. 1A) makes consideration of the physics of electron scattering relevant to our analysis of BVM binding to Gag because carboxylates frequently have diminished density in maps derived from cryoEM (27–30). Electron diffraction is a result of elastic scattering due to Coulomb forces between the incident electron beam and the crystal and therefore is dependent on the charge distribution in the crystal. Unlike atomic scattering factors for X-rays, electron scattering factors for some atoms are strongly dependent on scattering angle. In the case of negatively charged oxygen (O−), the atomic scattering factor is negative at low resolution (less than about 8 Å) and positive at higher resolution. To examine these effects on our maps, we examined the resolution dependence of our Coulomb potential maps (Fig. 5 A and D). Including low-resolution intensities (20–2.9 Å) resulted in MicroED maps with a negative contribution to acidic side chains (Fig. 5 B and E) (30). In contrast, maps calculated only with data in the higher-resolution range (8–2.9 Å), in which scattering factors for O− are positive, were unaffected (Fig. 5 C and F). The electron scattering factor for uncharged oxygen is fairly constant; therefore, neutralization of acidic side chains by direct interaction with, or proximity to, basic residues eliminates or minimizes the resolution dependence of the density of these side chains (e.g., E291 in a salt bridge with R299) (Fig. 5 B and C). Similarly, such Coulomb potential neutralization of the BVM dimethylsuccinyl carboxylic acid by the nearby ring of K359 lysine residues (Fig. 3E) likely accounts for the presence of density for this moiety.
Fig. 5.
Coulomb potential map of CTD-SP1-BVM at 2.9-Å resolution. (A and D) The CTD-SP1-BVM Coulomb potential surface [−8 kcal/mol e− (red) to 8 kcal/mol e− (blue)] calculated from the model (49) shows significant negative charge for solvent-exposed acidic residues (D295, E307, and E319) and neutralization in the case of an acidic residue involved in a salt bridge (E291). (B, C, E, and F) CTD-SP1-BVM MicroED maps showing acidic and basic residues in a variety of environments. Electron atomic scattering factors are strongly dependent on charge at low resolution, and excluding these reflections results in additional side chain density for residues containing O−; e.g., note the reduced density of D295, E307, and E319 (red ellipses) in B and E (map calculated from 20–2.9 Å, gray mesh) compared with C and F (map calculated from 8–2.9 Å, pink mesh). Scattering factors for neutral oxygen are much less dependent on resolution and are positive at all scattering angles; therefore, densities are nearly identical for acidic side chains involved in salt bridges (e.g., E291 in B and C) or neutral oxygen-containing side chains (e.g., N315 and T318 in E and F) whether the full or high-resolution range is used in map calculations.
The absence of acidic side chain density can also be due to mobility and/or radiation damage. O− is more susceptible to radiation damage by electrons than neutral and positively charged atoms, as shown in the analysis of radiation sensitivity of acidic side chains in cryoEM experiments (31, 32). These studies also demonstrated that radiation damage to acidic side chains is dose-dependent, and its manifestation in the density varies with resolution. At the average electron doses (∼3 e−/Å2) and resolutions observed in this study, we expect radiation damage to have only minor effects on the density.
Discussion
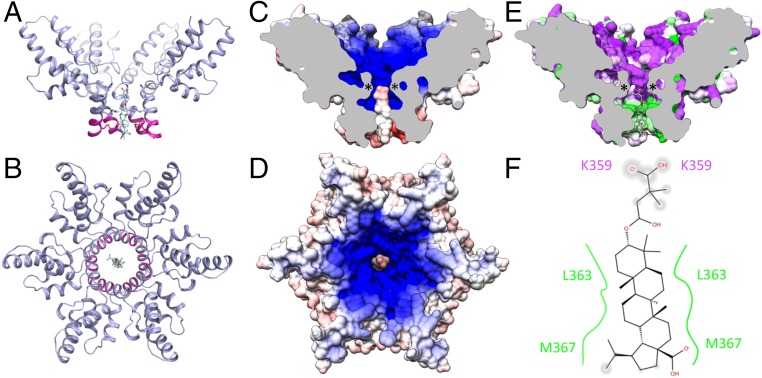
BVM is known to act by stabilizing the immature Gag lattice (14). The location of the pentacyclic triterpenoid adjacent to the junction helices clearly indicates that hydrophobic interactions are important for the mechanism of BVM binding (Figs. 3 A–D and 6 E and F). Specifically, L363 and M367 from each of the six protomers contribute the majority of the BVM binding interactions (Fig. 3 C, D, and F). The BVM dimethylsuccinyl ester is also thought to contribute to the activity (18), and we propose that BVM stabilizes Gag hexamers by electrostatically anchoring K359 of the CACTD domain via the dimethylsuccinyl carboxylate (Fig. 6 C and D) and binding together the junction helices via hydrophobic interactions between L363 and M367 from each protomer and the BVM pentacyclic triterpenoid moiety (Fig. 6 E and F). This BVM binding mechanism could accommodate lattice and hexamer distortions due to the nonspecific nature of the hydrophobic interactions and the electrostatic attraction with the six K359 residues, which is amplified by a second coaxial ring of lysine residues (K290) (Fig. 3 C and D). This mechanism of BVM binding and Gag stabilization is also consistent with the structures of second-generation HIV-1 maturation inhibitors in which the dimethylsuccinyl ester is replaced with benzoic acid (Fig. 1B). C-3 benzoic acid substitution places a carboxyl in proximity to K359 while preserving the hydrophobic interactions between the junction helices and the pentacyclic triterpenoid. The fact that the C-3 benzoic acid substituted compound exhibited antiviral activity comparable to BVM and had no effect on the potency against the BVM Gag escape mutant V370A provides additional support for BVM binding with the succinic acid positioned in the vicinity of K359 and away from V370. In addition, the BVM-up orientation positions C-28 of BVM near M367, and C-28 derivatives that extend BVM analogs toward the C-terminal end of SP-1 have the highest potency against HIV-1 with the Gag V370A polymorphism (Fig. 7) (16–18). BVM analogs with C-28 extensions may provide additional, compensatory interactions at the C-terminal end of the six-helix bundle that are sufficient to prevent protease access to the junction helix cleavage site.
Fig. 6.
BVM binding to Gag is mediated by electrostatic and hydrophobic interactions. Ribbon model of the CTD (slate blue) and SP1 (magenta) six-helix bundle with bound BVM, viewed from the (A) side and (B) top. (C and D) CTD-SP1 and BVM electrostatic surface potentials show that the negative charge of the dimethylsuccinyl carboxyl resides at the center of the positive charge generated by the dual-ring of lysines (K290 and K359). (E) BVM binding also includes hydrophobic interactions between the junction helices of each Gag protomer and the pentacyclic triterpenoid of BVM [hydrophobic (green) to polar (purple)]. Asterisks in C and E show the locations of K359. (F) Schematic 2D representation of BVM binding interactions showing hydrophobic (green) and polar (purple) residues. PR cleavage occurs at the L363 residues, which contribute the majority of the hydrophobic interactions with BVM.
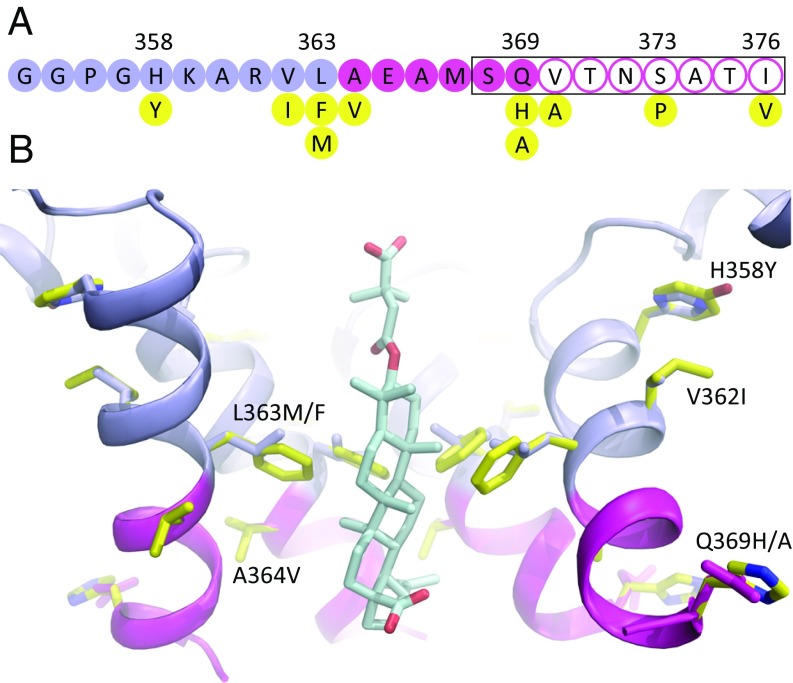
Fig. 7.
BVM resistance mutations and polymorphisms. (A) Single amino acid residues of the CTD (slate blue) and SP1 (magenta) construct of Gag, showing resistance mutations and polymorphisms (yellow) (34–36). The black rectangle shows the region of the junction with high sequence variability in the Los Alamos sequence database (20). Filled circles indicate residues resolved in the MicroED map, and residues with open circles were not resolved. (B) MicroED structure showing the CTD (blue) and SP1 (magenta) regions including the structure of BVM. Four subunits in the hexamer are displayed for clarity. Sites of Gag BVM resistance mutations and polymorphisms (yellow) (H358Y, L363M/F, V362I, A364V, and Q369H/A) are shown in the context of the wild-type structure, and their side chains are displayed with the rotamer configurations of the wild-type residues.
We realize that our interpretation of the binding mechanism of BVM to the six-helix bundle is based on a comparison of MicroED structures with and without the drug. For instance, photoaffinity labeling of detergent-stripped VLPs suggested that BVM may bind in the down orientation (33). This would be consistent with more distal mutants such as S373P and I376V conferring low-level BVM resistance (34–36). Likewise, the orientation may be influenced by the chemical properties of R-group substitutions. In the context of assembly of the immature hexagonal Gag lattice at the plasma membrane, it may be possible that other factors such as binding of viral RNA may influence the up–down distribution of BVM orientations as well as susceptibility to resistance. For instance, V20I confers strong resistance to bevirimat, even though valine and isoleucine are both hydrophobic, β-branched amino acids. The main difference is that the extra methylene increases the volume of isoleucine by ∼20% (165 versus 139 Å3) (37). Notably, the MicroED structure shows that this residue resides on the exterior of the helical bundle, where it would appear to have minimal effect on the stability of α-helical packing within the bundle. Regardless of uncertainties in the mechanisms of resistance, our results provide atomic resolution results that provide insight into the molecular basis for BVM binding.
Binding a single BVM molecule to the Gag hexamer presents challenges for structure-based drug design, but this mode of binding is likely responsible for the efficacy of BVM and related maturation inhibitors that bind to Gag, despite structural heterogeneity inherent in the quasi-hexagonal immature lattice. The CTD-SP1-BVM structure provides additional insights into BVM binding and activity that will inform further development of HIV-1 maturation inhibitors.
The CTD-SP1-BVM structure also demonstrates the capability of MicroED for structure-based drug design and discovery even in challenging cases (i.e., low crystallographic symmetry and diffraction to moderate resolution). The increasing availability of electron microscopes and the ability to obtain microcrystals, either by screening under low dose in an electron microscope or generated by mechanical disruption of larger crystals (38), make MicroED an attractive option for high-resolution macromolecular structure determination, even for proteins that are far too small for analysis by single-particle cryoEM methods.
Methods
Purification and Crystallization of CTD-SP1 and CTD-SP1-BVM.
The protein was expressed and purified as described previously (20). The conditions for growing microcrystals were adapted from the conditions to grow larger crystals for X-ray crystallography (20). The crystallization solution contained 0.1 M bis-Tris propane (pH: 7.0), 1.1–1.2 M LiSO4, and 0.8–1.0 mg/mL Gag CTD-SP1. The drug molar excess was 48×, assuming 1 BVM molecule/Gag hexamer; 600 µL of the crystallization solution were added to the well of a hanging drop plate. The protein solution was mixed with BVM (e.g., for a 1:8 protein:drug ratio, 10 μL of 1 mg/mL protein were mixed with 10 µL of a solution of 0.64 mM BVM in DMSO and water). An aliquot (1 μL) of the protein–BVM solution was mixed with 2 µL of the well solution. The crystallization plates were incubated at 17 °C, and platelike microcrystals appeared after 1–2 d. The same conditions were used to grow drug-free crystals. Initial screening of the crystals was performed by applying a 3-mm continuous carbon-coated EM grid to the crystallization drop for 90 s, blotting the grid with filter paper, and allowing the grid to dry at room temperature. Successful transfer of the crystals to the grid was first confirmed by light microscopy at 40× magnification (SI Appendix, Fig. S1). The grid was then examined in an FEI TF20 electron microscope operating at 200 kV. The unstained crystals were sufficiently thick to be readily observed from 10,000–40,000× magnification, and successful crystallization was confirmed by electron diffraction.
MicroED Sample Preparation, Data Collection, and Processing.
Quantifoil EM grids were glow-discharged for 30 s on both sides, and the grid was placed onto the crystallization droplet with the carbon film side facing the droplet. After a 5- to 10-min incubation, tweezers were used to separate the grid from the droplet, and an additional 1.5 μL of solution from the same droplet was pipetted onto the back side of the grid. The grid was then mounted in an FEI Vitrobot, which was used to plunge-freeze the grid into an ethane slush, with the following settings: blotting time of 7–15 s, force of 24, and 40% humidity. The grid was transferred to a Gatan 626 cryostage, and continuous-rotation MicroED data were recorded using an FEI TF20 microscope equipped with a field emission electron source and a bottom-mounted TVIPS TemCam-F416 complementary metal oxide semiconductor camera operating in rolling shutter mode (23). Diffraction patterns were recorded using 200-keV electrons with a camera length of 2 m. The electron dose rate was ∼0.01 e−∙Å−2∙s−1, and the crystals were continuously rotated unidirectionally at a rate of 0.062°/s. Eight-second frames were recorded during the movie, and data typically spanned a tilt range of 45°–60°. Diffraction images were corrected to account for negative pixel values (39); then they were indexed and integrated in MOSFLM (40). Integrated intensities from six and five crystals for the drug-free and BVM-bound samples, respectively, were scaled and merged in AIMLESS (41).
MicroED Structure Determination and Map Calculation.
Phases for the BVM-bound and drug-free MicroED reflections were determined by molecular replacement with Phaser (42) using the X-ray structure of hexameric CTD-SP1 (PDB 5I4T) as the search model. Cycles of real-space and reciprocal space refinements were performed with Coot (43) and phenix.refine (44), using electron scattering factors (45). Torsion angle noncrystallographic symmetry (NCS) restraints were used in the early rounds of refinement. In solving the structure of CTD-SP1-BVM, the drug was never included in the model during refinements that were used for map calculations. Maximum-entropy maps were calculated in Phenix and showed plausible improvements in map detail and thus were used to aid the interpretation of the maps.
We also solved the structure of CTD-SP1-BVM independent of the CTD-SP1 X-ray structure. We generated an all-atom hexameric model of the CTD domain by combination of the X-ray crystal structure of the Gag CTD dimer [PDB 4COP (46)] and the backbone model of the CTD hexamer from the Gag CA structure determined by a combination of tomography and helical reconstruction. We aligned a copy of the 4COP monomer to each protomer of the 4D1K (46) CTD hexamer and performed molecular replacement with Phaser using the all-atom hexameric CTD as the search model. The top Phaser solution (TFZ = 9.4) showed well-defined density for the CTD and clear α-helical density for SP1. We generated an ideal helix in Coot within the best SP1 density and then generated the initial six-helix bundle by application of sixfold NCS. We then performed rounds of building and refinement in Coot and Phenix. The resultant refined BVM-bound CTD-SP1 model and associated maps recapitulated those resulting from molecular replacement with the CTD-SP1 X-ray structure.
Supplementary Material
Acknowledgments
We gratefully acknowledge Eric Freed for a critical reading of the manuscript. Some molecular graphics and analyses were performed with the University of California, San Francisco Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institute of General Medical Sciences Grant P41 GM103311). The T.G. laboratory is funded by the Howard Hughes Medical Institute. This study was previously funded by US National Institutes of Health Grants R01 GM066087 and P50 GM082545 and is now funded by Grant R01 GM128507 (to M.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. Z.A. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates were deposited in the Protein Data Bank (drug-free CTD-SP1: PDB ID 6N3J; CTD-SP1 with bound bevirimat: PDB ID 6N3U). Because a unique bevirimat binding pose could not be determined, the drug is not included in the PDB coordinate file. MicroED maps were deposited in the EMDataBank (drug-free CTD-SP1: EMDB-0335: CTD-SP1 with bound bevirimat: EMDB-0337).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806806115/-/DCSupplemental.
References
- 1.Sundquist WI, Kräusslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson L-A, et al. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson L-A, et al. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright ER, et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettit SC, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiegers K, et al. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 8.Coren LV, et al. Mutational analysis of the C-terminal Gag cleavage sites in human immunodeficiency virus type 1. J Virol. 2007;81:10047–10054. doi: 10.1128/JVI.02496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Marco A, et al. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010;6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S-K, Potempa M, Swanstrom R. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem. 2012;287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamson CS. Protease-mediated maturation of HIV: Inhibitors of protease and the maturation process. Mol Biol Int. 2012;2012:604261. doi: 10.1155/2012/604261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, et al. PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci USA. 2003;100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol. 2011;85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamson CS, Waki K, Ablan SD, Salzwedel K, Freed EO. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457) J Virol. 2009;83:4884–4894. doi: 10.1128/JVI.02659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swidorski JJ, et al. Inhibitors of HIV-1 maturation: Development of structure-activity relationship for C-28 amides based on C-3 benzoic acid-modified triterpenoids. Bioorg Med Chem Lett. 2016;26:1925–1930. doi: 10.1016/j.bmcl.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Urano E, et al. Alkyl amine bevirimat derivatives are potent and broadly active HIV-1 maturation inhibitors. Antimicrob Agents Chemother. 2015;60:190–197. doi: 10.1128/AAC.02121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Gu Q, Morris-Natschke SL, Chen C-H, Lee K-H. Incorporation of privileged structures into bevirimat can improve activity against wild-type and bevirimat-resistant HIV-1. J Med Chem. 2016;59:9262–9268. doi: 10.1021/acs.jmedchem.6b00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schur FKM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 20.Wagner JM, et al. Crystal structure of an HIV assembly and maturation switch. eLife. 2016;5:e17063. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayro MJ, Ganser-Pornillos BK, Zadrozny KK, Yeager M, Tycko R. Helical conformation in the CA-SP1 junction of the immature HIV-1 lattice determined from solid-state NMR of virus-like particles. J Am Chem Soc. 2016;138:12029–12032. doi: 10.1021/jacs.6b07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi D, Nannenga BL, Iadanza MG, Gonen T. Three-dimensional electron crystallography of protein microcrystals. eLife. 2013;2:e01345. doi: 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nannenga BL, Shi D, Leslie AGW, Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods. 2014;11:927–930. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nannenga BL, Shi D, Hattne J, Reyes FE, Gonen T. Structure of catalase determined by MicroED. eLife. 2014;3:e03600. doi: 10.7554/eLife.03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez JA, Eisenberg DS, Gonen T. Taking the measure of MicroED. Curr Opin Struct Biol. 2017;46:79–86. doi: 10.1016/j.sbi.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 28.Kühlbrandt W, Wang DN, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuoka K, et al. The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: Implication of the charge distribution. J Mol Biol. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 30.Yonekura K, Kato K, Ogasawara M, Tomita M, Toyoshima C. Electron crystallography of ultrathin 3D protein crystals: Atomic model with charges. Proc Natl Acad Sci USA. 2015;112:3368–3373. doi: 10.1073/pnas.1500724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci USA. 2014;111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hattne J, et al. Analysis of global and site-specific radiation damage in cryo-EM. Structure. 2018;26:759–756. doi: 10.1016/j.str.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen AT, et al. The prototype HIV-1 maturation inhibitor, bevirimat, binds to the CA-SP1 cleavage site in immature Gag particles. Retrovirology. 2011;8:101. doi: 10.1186/1742-4690-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Baelen K, et al. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob Agents Chemother. 2009;53:2185–2188. doi: 10.1128/AAC.01650-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margot NA, Gibbs CS, Miller MD. Phenotypic susceptibility to bevirimat in isolates from HIV-1-infected patients without prior exposure to bevirimat. Antimicrob Agents Chemother. 2010;54:2345–2353. doi: 10.1128/AAC.01784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knapp DJHFK, et al. In vitro selection of clinically relevant bevirimat resistance mutations revealed by “deep” sequencing of serially passaged, quasispecies-containing recombinant HIV-1. J Clin Microbiol. 2011;49:201–208. doi: 10.1128/JCM.01868-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harpaz Y, Gerstein M, Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 38.de la Cruz MJ, et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat Methods. 2017;14:399–402. doi: 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hattne J, Shi D, de la Cruz MJ, Reyes FE, Gonen T. Modeling truncated pixel values of faint reflections in MicroED images. J Appl Cryst. 2016;49:1029–1034. doi: 10.1107/S1600576716007196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell HR, Battye TGG, Kontogiannis L, Johnson O, Leslie AGW. Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm. Nat Protoc. 2017;12:1310–1325. doi: 10.1038/nprot.2017.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowton K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colliex C, et al. Electron diffraction. In: Prince E, editor. International Tables for Crystallography, Vol C: Mathematical, Physical and Chemical Tables. International Union of Crystallography; Chester, UK: 2006. pp. 259–429. [Google Scholar]
- 46.Bharat TAM, et al. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc Natl Acad Sci USA. 2014;111:8233–8238. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regueiro-Ren A, et al. Discovery of BMS-955176, a second generation HIV-1 maturation inhibitor with broad spectrum antiviral activity. ACS Med Chem Lett. 2016;7:568–572. doi: 10.1021/acsmedchemlett.6b00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miteva MA, Guyon F, Tufféry P. Frog2: Efficient 3D conformation ensemble generator for small compounds. Nucleic Acids Res. 2010;38:W622–W627. doi: 10.1093/nar/gkq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.