Significance
The nervous system consumes a large quotient of oxygen and as such is at risk for high levels of oxidative DNA damage. A key DNA-repair factor, apurinic/apyrimidinic endonuclease 1 (APE1), is essential for repair of oxidative DNA lesions, although the specific role(s) for this enzyme in the nervous system is unknown. Surprisingly, mice lacking APE1 throughout neurogenesis were viable and showed little discernible phenotype at birth. However, after birth, when tissue oxygenation shifts from the placenta to respiration, loss of APE1 led to rapid and pronounced genome instability, resulting in widespread apoptosis, demyelination, thermoregulation defects, and brain tumors. Our findings reveal unrestrained oxidative DNA damage in the nervous system can result in specific pathology implicated in many human diseases.
Keywords: APE1, DNA damage, nervous system, development
Abstract
Frequent oxidative modification of the neural genome is a by-product of the high oxygen consumption of the nervous system. Rapid correction of oxidative DNA lesions is essential, as genome stability is a paramount determinant of neural homeostasis. Apurinic/apyrimidinic endonuclease 1 (APE1; also known as “APEX1” or “REF1”) is a key enzyme for the repair of oxidative DNA damage, although the specific role(s) for this enzyme in the development and maintenance of the nervous system is largely unknown. Here, using conditional inactivation of murine Ape1, we identify critical roles for this protein in the brain selectively after birth, coinciding with tissue oxygenation shifting from a placental supply to respiration. While mice lacking APE1 throughout neurogenesis were viable with little discernible phenotype at birth, rapid and pronounced brain-wide degenerative changes associated with DNA damage were observed immediately after birth leading to early death. Unexpectedly, Ape1Nes-cre mice appeared hypothermic with persistent shivering associated with the loss of thermoregulatory serotonergic neurons. We found that APE1 is critical for the selective regulation of Fos1-induced hippocampal immediate early gene expression. Finally, loss of APE1 in combination with p53 inactivation resulted in a profound susceptibility to brain tumors, including medulloblastoma and glioblastoma, implicating oxidative DNA lesions as an etiologic agent in these diseases. Our study reveals APE1 as a major suppressor of deleterious oxidative DNA damage and uncovers specific and broad pathogenic consequences of respiratory oxygenation in the postnatal nervous system.
The development and functioning of an organism requires constant genome maintenance (1). This is achieved by multiple DNA-repair pathways, which prevent the accumulation of DNA lesions that can cause severe pathology (1–4). One source of endogenous DNA damage is reactive oxygen species (ROS), generated as by-products of mitochondrial respiration and cellular metabolism. These DNA lesions include damaged bases, abasic sites, and DNA nicks, which are resolved primarily by the base excision repair (BER) pathway (5–7).
BER involves sequential steps in repairing base modifications. To initiate BER, DNA glycosylases recognize and remove different types of base damage, generating apurinic or apyrimidinic (AP) sites (8, 9). AP sites can also be generated directly via spontaneous hydrolysis or through the action of ROS. AP endonuclease 1 (APE1; also known as “APEX1” or redox effector factor 1, “REF1”) recognizes AP sites and cleaves the phosphodiester backbone 5′ to the lesion, giving rise to a DNA single-strand break (7, 10). Although there are two potential class II AP endonucleases in mammals, APE1 is considered the primary AP endonuclease, as APE2 has a very weak endonuclease activity, potentially functioning specifically in antibody diversity in the immune system (11). The scaffold protein XRCC1 (X-ray repair cross-complementing protein 1) helps assemble the enzymatic BER machinery necessary to process DNA single-strand breaks, such as polynucleotide kinase-phosphatase (PNKP), aprataxin (APTX), and tyrosyl-DNA phosphodiesterase-1 (TDP1), which modify noncompatible DNA ends for polymerization or ligation. DNA polymerase β and either DNA ligase 3 (in complex with XRCC1) or ligase 1 function to fill the gap and seal the remaining processed nick (5, 7, 12–14).
Since neurons are highly metabolically active and have high oxygen consumption (15–17), efficient DNA repair of ROS damage is especially important in the nervous system. This is underscored by inherited defects in DNA single-strand break repair enzymes that severely impact the nervous system and cause neurodegeneration (1, 13, 18–20). While several components of the BER pathway have been shown to be critical for the nervous system (2, 4, 12, 21), the exact contribution of APE1, a core participant in classic BER, to the homeostasis of this tissue has not been determined.
APE1 is an ∼35-kDa protein that is composed of a large, conserved endonuclease domain and a unique N-terminal extension not found in its bacterial counterpart, Escherichia coli exonuclease III (12, 22–26). In addition to its powerful AP endonuclease function, APE1 has been shown to exhibit weaker 3′-repair activities on damaged DNA termini through a single enzymatic active site (12, 22–26). Moreover, through its so-called “N-terminal REF1 function” mediated via critical cysteine residues, APE1 can activate/modulate important transcription factors, such as AP-1, Egr1, HIF-1α, p53, and NF-κB (27–30). Therefore, in addition to participating directly in DNA-repair processes, APE1 can potentially regulate transcription, influencing biological events such as stress responses and inflammation.
Despite the critical role of APE1 in cellular DNA repair (10), defining the molecular roles of this protein in vivo has been difficult. Indeed, germline deletion of Ape1 in mice leads to embryonic lethality around E5 (31, 32). Using a tissue-specific gene-deletion approach, we show here that APE1 is essential for postnatal neurodevelopment and genomic integrity in mature neurons. We also found that APE1 is a tumor suppressor and can modulate select AP-1–induced gene expression. These findings show how maintenance of genome stability and stress responses can prevent oxidative stress-induced pathology.
Results
APE1 Is Dispensable for Embryonic Neurogenesis but Is Essential for Postnatal Neurogenesis and Homeostasis.
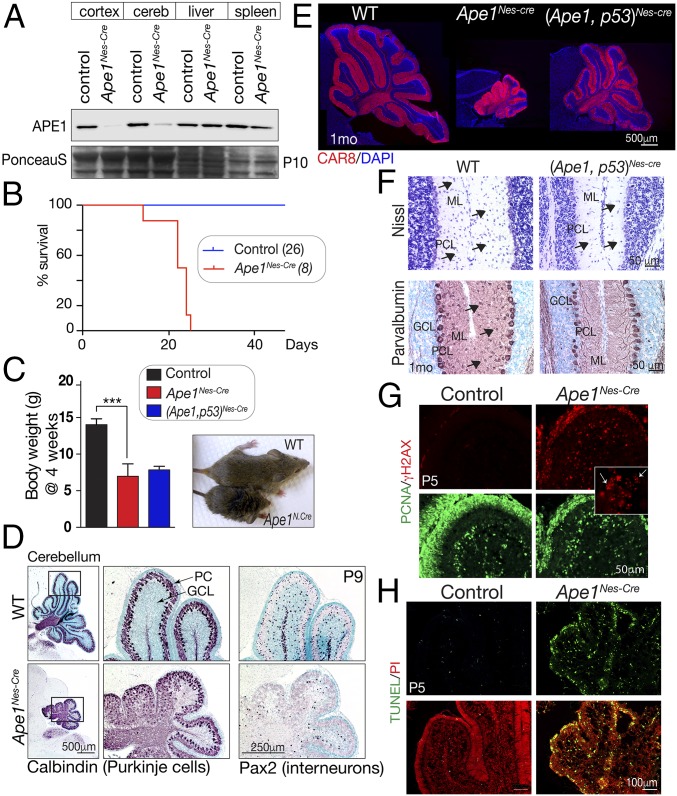
To determine the physiologic contributions of APE1, we generated a conditional Ape1 murine model by flanking exons 2 and 3 of the genomic locus with LoxP sites (SI Appendix, Fig. S1). This strategy generates an Ape1 allele that produces an out-of-frame transcript after cre-mediated exon deletion. We initially generated mice with embryo-wide APE1 inactivation using Sox2-cre, restricting cre expression to the embryo, with no expression in the placenta (as occurs during germline deletion). This resulted in early lethality, as all Ape1Sox2-cre embryos were found dead around E9–E10. Thus, similar to reports of germline lethality for deletion of Ape1 (31, 32) and like germline deletion of other key components of BER such as Xrcc1 (33), we find APE1 is essential for epiblast-derived embryogenesis. In contrast, when Ape1 was deleted throughout neural development using Nestin-cre, we found that embryonic neurogenesis proceeded normally. We analyzed neural development at E12.5 and E15.5 but failed to find any differences in overall tissue histology, cell proliferation, DNA damage, or cell death (SI Appendix, Fig. S1). Notably, and consistent with established cre expression via the Nestin promoter (34, 35), efficient APE1 deletion occurred throughout the nervous system from E12 as determined by immunohistochemistry (SI Appendix, Fig. S1C). Thus, embryonic neural development occurred despite the absence of APE1 during neurogenesis. Concordantly, Ape1Nes-cre mice were born at Mendelian ratios and were initially indistinguishable from their littermates. Western blots further confirmed neural-specific loss of APE1 (Fig. 1A). However, from around postnatal day (P)5 onwards, Ape1Nes-cre mice showed a dramatic decline in vigor, exhibited stunted growth, and developed ataxia, with all mutants dying by 3 wk of age (Fig. 1B). At this stage, Ape1Nes-cre mice were 50% reduced in body weight compared with littermate controls (Fig. 1C). Generation of Ape1Nes-cre mice in which p53 was also inactivated failed to rescue the lethality (Fig. 1C). As will be discussed later, a factor contributing to the early lethality after APE1 inactivation appeared to be hypothermia, which could be mitigated by raising the ambient temperature via use of a heating pad. In the datasets below some tissues were used at ages past 3 wk due to the extension of viability (to ∼2 mo of age) by increased ambient temperature.
Fig. 1.
Inactivation of APE1 throughout the nervous system. (A) Western blot analysis of various tissues shows that APE1 (∼35 kDa) is deleted in neural tissue (cerebellum and cortex), while normal protein levels are seen in the liver and spleen. Residual APE1 in the Nes-Cre mice is due to blood/lymphocytes, as no protein is detected by immunohistochemistry in perfused tissue. (B) Without intervention, mice with Ape1 deleted in the nervous system (Ape1Nes-cre) survive only to 3 wk of age. (C) Comparative size of control and Ape1Nes-cre animals at 4 wk of age shows the mutants fail to thrive, and substantial weight reduction is apparent. Inactivation of p53 does not rescue this Ape1Nes-cre phenotype. ***P < 0.0002. (D) P9 development of the cerebellum is markedly affected in the Ape1Nes-cre animals, with a disorganization of Purkinje cells (calbindin immunostaining) due to a loss of granule neurons. Interneurons are also lost, as indicated by Pax2 immunostaining. GCL, granule cell layer; PC, Purkinje cells. (E) Mice in which both Ape1 and p53 have been deleted show partial rescue of the cerebellar phenotype. Substantial postnatal cerebellar folia development is found in the (Ape1;p53)Nes-cre tissue, as shown using CAR8 immunostaining to identify Purkinje cells. The WT image is a two-panel composite to allow presentation of the full cerebellum. (F) Interneurons are markedly reduced in the mutant (Ape1;p53)Nes-cre cerebellum, indicating that while loss of p53 rescues a large fraction of granule neurons, stellate and basket interneurons of the molecular layer are not rescued, as indicated by Nissl and parvalbumin staining. GCL, granule cell layer; ML, molecular layer; PC, Purkinje cells. (G) DNA damage is widespread in the developing cerebellum as shown using γH2AX. The Inset shows the typical γH2AX puncta of DNA damage rather than the pan-γH2AX present in apoptotic cells. (H) The loss of granule neurons occurs via apoptosis as the early P5 postnatal cerebellum contains many TUNEL+ cells.
Examination of the P9 Ape1Nes-cre brain showed a pronounced defect in cerebellar development (Fig. 1D). Because the cerebellum completes development in the 3 wk following birth, this suggests that cerebellar decline in the Ape1Nes-cre mice occurred soon after birth. A major contributor to the observed cerebellar defects was the widespread loss of granule neurons as shown using calbindin immunostaining to identify the disorganized Purkinje cells after granule neuron loss (Fig. 1D). In addition to granule neurons, interneurons were also decreased in the Ape1Nes-cre cerebellum as shown by reduced Pax2 immunostaining, a specific marker for these cerebellar neurons (Fig. 1D). This finding is consistent with the general hypersensitivity of interneurons to genotoxic stress (36, 37). However, coincident inactivation of Ape1 and p53 resulted in a substantial rescue of cerebellar cell loss, generating a granule neuron layer reminiscent of the WT tissue, as seen by the laminar organization of Purkinje cells identified by carbonic anhydrase-related protein 8 (CAR8) immunostaining (Fig. 1E). While the overall phenotype of the (Ape1;p53)Nes-cre cerebellum resembled the WT, clear defects were present, as interneuron numbers in the molecular layer were reduced and those present lacked mature differentiation markers such as parvalbumin (Fig. 1F). These features contrast what has been seen upon inactivation of other DNA-repair factors, such as components of DNA double-strand break repair (e.g., DNA ligase IV), where Nes-cre deletion results in midgestational lethality but p53 inactivation restores viability and relatively normal brain histology (38, 39).
To determine the cause of granule neuron loss, we examined DNA damage via γH2AX and apoptosis using TUNEL. By P5, there were abundant γH2AX+ and TUNEL+ cells throughout the proliferating cell nuclear antigen (PCNA)-positive external granule layer (EGL), which contains the granule neuron progenitors (Fig. 1 G and H). While apoptotic cells can also be γH2AX PCNA+, small nuclear puncta indicative of DNA damage foci were abundant throughout the EGL (Fig. 1G, Inset). Thus, widespread apoptosis of granule neurons, a population known to be susceptible to DNA damage (40, 41), accounts for the reduced size of the Ape1Nes-cre cerebellum (Fig. 1H). Additionally, the substantial rescue by p53 is consistent with DNA damage-induced p53-dependent apoptosis as a causal event in the granule neuron loss and the small cerebellum (Fig. 1E). These data implicate DNA damage-induced apoptosis as precipitating events resulting in cerebellar atrophy in the Ape1Nes-cre mice.
APE1 Is Required for Cortical Homeostasis.
To further establish the neural impact of APE1 loss, we analyzed cortical development. The cortex is a laminar structure consisting of six layers containing specific neuronal classes that provide functional specialization (42, 43). Perturbation of neural development from compromised genome stability can alter the arrangement and content of these six cortical layers (34, 44). While there was no observable effect upon neural development in the embryo, the Ape1Nes-cre cortex became markedly reduced in size after birth (Fig. 2A). To understand the basis for the smaller Ape1Nes-cre cortex, we examined this brain region at various times from early postnates thru 2 mo of age. We found a striking persistence of DNA damage (indicated by γH2AX) initially throughout the cortex which then became more restricted to the upper cortical layers (layers II–IV) by P10 (Fig. 2 B and C and SI Appendix, Fig. S2). Other regions of the Ape1Nes-cre brain, including the thalamus and hippocampus, also showed widespread γH2AX formation (SI Appendix, Fig. S3). To determine the basis for reduced cortical size in the mutant, we examined if the chronic DNA damage coincided with apoptosis. As shown using TUNEL, apoptosis was present throughout the mutant cortex, with sporadic cell death occurring at the regions of γH2AX immunostaining, accounting for the progressive postnatal cell loss and cortical size reduction by 2 mo of age (Fig. 2D).
Fig. 2.
Cortical development shows disruption of upper layers and oligodendrocytes after APE1 loss. (A) The cortex (CTX) and corpus callosum (CC) are reduced in thickness, as is the hippocampal formation (HC) in the Ape1Nes-cre brain. (B) DNA damage resulting from APE1 loss causes elevated γH2AX immunostaining and is more apparent in the upper-layer neurons (I–IV) of the cortex, while the lower-layer neurons (IV–VI) show little γH2AX immunoreactivity. White arrowheads indicate γH2AX cells that are apoptotic, while yellow arrows show the smaller γH2AX puncta indicative of DNA damage. (C) The illustration summarizes the change in the distribution of γH2AX/DNA damage as the cortex develops; darker shades of red indicate more γH2AX. (D) Apoptosis identified using TUNEL shows continual cellular attrition in the P7 and 6-wk-old Ape1Nes-cre brain. (E, Upper) Specific cortical layer markers for upper-layer neurons (Satb2 and Brn2) are absent in the Ape1Nes-cre cortex compared with WT controls. Tbr1, which primarily localizes to lower-layer neurons, is similar in the control and mutant tissue. (Lower) The graphs show quantitation of the loss of Satb2 and Brn2 in the Ape1Nes-cre cortex compared with control cortex. ****P < 0.0001; ***P = 0.0002; n.s., not significant. (F) Myelin production indicated by CNPase immunostaining is markedly reduced in the corpus callosum (CC) of Ape1Nes-cre mice. (G) Loss of oligodendrocytes is indicated by Sox10 immunostaining. The Ape1Nes-cre cortex also has increased gliosis, as shown by GFAP immunostaining, suggesting a general perturbation of neural homeostasis.
We confirmed that APE1 loss targeted neurons in layers II–IV using cell-type–specific markers. Immunostaining for Satb2 and Brn2 [markers specific for the upper layers of the cortex (45, 46)] revealed a nearly complete loss of Satb2-expressing cells, and an ∼70% reduction of Brn2+ cells (Fig. 2E) coinciding with the presence of γH2AX in these layers. However, lower layers (V/VI), which are identified using Tbr1 immunolabeling, show a similar number of neurons in the WT and mutant animals. These data indicate that while early (embryonic) neocortical development is unaffected by APE1 loss, the Ape1Nes-cre cortex undergoes substantial neuronal attrition postnatally, and this phenotype is particularly exacerbated in the upper cortical layers as the cortex undergoes functional compartmentalization and maturation (47, 48).
Given the high levels of γH2AX present in the Ape1Nes-cre brain and because perturbed neural homeostasis involving neurodegeneration can be associated with neuroinflammation (49–51), we used Iba1 immunostaining to determine if inflammation was present after APE1 loss. Iba1 immunostaining was indeed present broadly throughout the cortex, showing that neuroinflammation occurred throughout the Ape1Nes-cre brain (SI Appendix, Fig. S4). Additionally, MAP2 was reduced in the Ape1Nes-cre tissue, also suggesting compromised neural homeostasis (SI Appendix, Fig. S4). These data further illustrate the substantial burden the nervous system faces after birth when APE1 function is compromised.
APE1 Suppresses Loss of Oligodendrocytes.
Other substantial and wide-ranging neural defects were associated with APE1 loss. In particular, APE1 inactivation resulted in a marked loss of myelination in oligodendrocyte-rich regions throughout the brain. Using key myelin/oligodendrocyte markers, including 2′,3′-cyclic nucleotide 3′-phosphohydrolase (CNPase), myelin basic protein (MBP), and myelin proteolipid protein (PLP), we found widespread loss of myelin throughout white-matter areas of the brain (Fig. 2F and SI Appendix, Fig. S5). Disruption of other BER factors such as PNKP and XRCC1 also impacts myelin-producing oligodendrocytes (37, 52), but myelination, as gauged using MBP immunohistochemistry, was comparatively more affected in the Ape1Nes-cre brain (SI Appendix, Fig. S5B). Moreover, coincident loss of p53 did not correct the myelin defect (SI Appendix, Fig. S5C). This myelin defect was apparent as early as P10 (Fig. 2F) and persisted to adulthood (SI Appendix, Fig. S5C).
We determined if the loss of myelination in the Ape1Nes-cre brain reflects a loss of oligodendrocytes or only the ability to produce myelin. We used SOX10, which identifies oligodendrocytes at various stages of maturation (53), and found that there was a widespread loss of this nuclear marker similar to the loss of myelin, confirming that myelination defects result from the loss of oligodendrocytes rather than from a specific defect in myelination (Fig. 2G). We also used Olig2 to identify oligodendrocyte progenitors and examined early postnatal tissues and found that at this stage there was still a loss of oligodendrocytes (SI Appendix, Fig. S5A). However, progenitor reduction was milder throughout both the cortex and cerebellum, suggesting that the lesions suppressed by APE1 promote a gradual but selective attrition of oligodendrocytes.
Repair of Oxidative DNA Lesions Requires APE1 in Neural Cells.
Given the pathogenic impact of APE1 loss in the nervous system, we determined the genotoxin susceptibility of Ape1−/− primary cortical astrocytes and what types of DNA lesions remained unrepaired. Immunostaining of Ape1Nes-cre astrocytes showed an absence of APE1, a phenotype confirmed by Western blot analysis (Fig. 3A and SI Appendix, Fig. S6A). Additionally, immunoreactivity to GFAP confirmed astrocyte identity (Fig. 3A). Ape1-null astrocytes grew well from establishment and an initial round of passaging, although replication capacity was reduced after passage three (Fig. 3B), in contrast with reports of APE1 loss being incompatible with cellular proliferation (10, 22). The decline in proliferation capacity of the APE1-null (Ape1Nes-cre) cells may be due to checkpoint activation by the chronic accumulation of DNA damage, as high levels of endogenous γH2AX occur in these cells (Fig. 3C). We then determined the relative sensitivity of APE1-null cells compared with BER-deficient XRCC1 cells (each harboring co-inactivation of p53 to abrogate senescence) and appropriate control cells. APE1-null astrocytes were hypersensitive to genotoxins that generate oxidative DNA damage (H2O2), alkylation lesions (methyl methanesulphonate; MMS), or DNA cross-links (cisplatin) compared with control cells (Fig. 3D). In the case of cisplatin, Ape1Nes-cre cells were hypersensitive in comparison with WT or XRCC1-mutant cells (Fig. 3D). In contrast, both APE1-null and XRCC1-null cells were only moderately sensitive to ionizing radiation, compared with the extreme hypersensitivity characteristic of BRCA2-null cells (SI Appendix, Fig. S6B).
Fig. 3.
Loss of APE1 is compatible with cell proliferation but leads to defective DNA repair and sensitizes to select genotoxins. (A) Immunocytochemistry shows APE1 is undetectable in the Ape1Nes-cre-derived primary astrocytes. (B) Ape1−/− astrocytes initially proliferate at nearly normal growth rates, although from passage 3 onwards they grow less well than controls. (C, Upper) Cultured Ape1−/− astrocytes progressively accumulate γH2AX. (Lower) Quantitation of astrocytes containing γH2AX foci (>10 foci per cell; 100 cells were counted per experiment). **P = 0.007. (D) Ape1−/− astrocytes are hypersensitive to H2O2, MMS, and cisplatin. Xrcc1−/− was used as a control for these treatments and was comparably sensitive to Ape1−/−, although APE1 loss resulted in a specific sensitivity to cisplatin. (E, Upper) AP endonuclease activity utilizing an oligomer substrate containing an AP site shows that while WT whole-cell extract (WCE) contains robust endonuclease activity that cleaves the 18mer substrate, Ape1−/− WCE inefficiently processes the oligomer containing the AP site. (Lower) The percent of AP site cleavage. In these experiments, Ape1Cre embryonic fibroblasts were treated with tamoxifen to activate cre-mediated deletion and create Ape1−/− fibroblasts. (F) Endogenous cellular AP sites in primary astrocytes [identified using the aldehyde-reactive probe O-(biotinylcarbazoylmethyl) hydroxylamine) accumulated in the Ape1−/− cells], while both WT controls and Xrcc1−/− cells were competent in removing AP sites. H2O2 produced by epigallocatechin gallate (EGCG) treatment served as control agent to monitor the formation of endogenous AP sites. (G) The inability of APE1-deficient primary astrocytes to cleave at abasic sites was reflected as an accumulation of DNA damage in the alkaline comet assay after Ape1−/− cells were treated with genotoxic agents, including MMS (***P < 0.003), H2O2 (***P < 0.003), and irradiation (****P < 0.002). Xrcc1Nes-cre cells are used as a control for defective BER. Unt, untreated.
Since, in addition to APE1, there is another AP endonuclease, APE2 (54, 55), we determined the total AP endonuclease efficiency of WT and APE1-null cell extracts using a DNA duplex modified by an AP site analog, tetrahydrofuran (THF) (Fig. 3E). While WT cells clearly displayed the ability to cleave the oligo from the 18mer substrate to a 9mer product, this activity was largely lacking in the APE1-null cells (Fig. 3E). To evaluate the occurrence of spontaneous endogenous DNA damage, we measured the levels of AP sites in WT and APE1-null cells using an established technique involving an aldehyde-reactive probe, O-(biotinylcarbazoylmethyl) hydroxylamine. As a positive control, oxidative lesions induced by H2O2 that is produced by epigallocatechin gallate treatment showed abundant aldehyde-reactive signal, as did APE1-null astrocytes; WT controls and XRCC1-null cells, in which intact APE1 activity presumably leads to the elimination of the aldehyde reactivity, showed little AP-site staining (Fig. 3F). Thus, APE1 is the major AP endonuclease activity in mammals, with apparently little contribution by APE2, as is consistent with this enzyme possibly being restricted to the hematopoietic system (54).
Finally, we undertook DNA-repair analysis using an alkaline comet assay to compare the relative recovery of DNA integrity after damage in control and APE1-null cells. APE1-null cells showed a significant difference in the accumulation of DNA damage compared with controls, similar in magnitude to that seen with the loss of XRCC1, indicating an important role for APE1 in processing DNA lesions, such as AP sites and strand breaks, as part of BER (Fig. 3G). Combined with the observation that loss of APE1 leads to hypersensitivity to oxidative and alkylation damage (Fig. 3D), the data indicate that APE1 fulfills an indispensable role in the repair of DNA lesions to maintain genome stability.
APE1 Is Critical for Thermoregulation via Protection of Serotonergic Neurons.
While Ape1Nes-cre mice survived to around 3 wk of age (weaning age), we noticed an obvious shivering in these animals, and found that viability could be extended when their cages were placed on a heating pad. After doing this, survival of Ape1Nes-cre mice was extended to around 2 mo of age. The shivering phenotype and the need for a heating pad for enhanced viability of Ape1Nes-cre mice suggested a thermoregulatory defect was compromising survival.
To further investigate the basis for this phenotype, we used temperature transponders (a dermally implanted IPTT-300 device) to measure body temperature to determine if the shivering was due to low body temperature or an aspect of sensory signaling. We evaluated body temperature while the mice were on the heating pad (set to 37 °C) and also 90 min after the mice were removed from the heating pad and maintained at 20 °C. We collected the temperature at two different time points (8 AM and 12 PM) but could not find a significant difference between body temperature measurements in Ape1Nes-cre mice and controls, suggesting that the shivering was a perturbed intrinsic neurologic response associated with thermal perception/regulation.
Among the neural circuits controlling thermoregulation, several Raphe nuclei, including the rostral medullary Raphe nuclei (RMR) and the dorsal Raphe nuclei (DR) located throughout the brainstem (Fig. 4A), are important for serotonin (5-HT) synthesis and modulation of the cold response (56). The RMR is rich in serotonergic neurons and vesicular glutamate transporter 3 (VGluT3)-positive nonserotonergic neurons, and these populations work together to modulate the response to cold (57–62). 5-HT is a neurotransmitter important for thermoregulation (60, 63), and multiple reports demonstrate that mice lacking central nervous system 5-HT fail to preserve physiologic body temperature when exposed to cold (58, 62, 64–67). We initially used Nissl staining to gauge overall histology in the RMR and DR and found a generalized reduction of cell numbers in the mutant tissue (Fig. 4B, arrows). Additionally, tryptophan hydroxylase 2 (TPH2), a rate-limiting enzyme in serotonin synthesis, 5-HT neurons were markedly decreased (50%) in the Ape1Nes-cre DR and RMR compared with controls (Fig. 4 C and D). Colocalization of TPH2+ neurons with NeuN in the DR further shows the loss of serotonergic neurons in Ape1Nes-cre tissue (Fig. 4E). A similar reduction in TPH2+ neurons was also observed in the Ape1Nes-cre RMR (Fig. 4F).
Fig. 4.
Thermoregulation in APE1 mice is associated with a loss of serotonergic neurons. (A) A cartoon depiction of the thermoregulatory DR and RMR nuclei. (B) Nissl staining of the DR and RMR shows decreased cellularity in the Ape1Nes-cre tissue compared with control. Arrows indicate areas of decreased cellularity. Aq, cerebral aqueduct. (C) Identification of serotonergic neurons in the DR nuclei using TPH2 shows a decrease in TPH2+ neurons in Ape1Nes-cre tissue compared with control tissue; arrows indicate TPH2+ cells. (D) TPH2+ neurons are reduced in number in Ape1Nes-cre tissue compared with control in both the DR (∼25% reduction) and the RMR (50% reduction). ***P < 0.001. (E and F) Colocalization of TPH2 and NeuN immunostaining in the DR (E) and the RMR (F) shows that reduction of TPH2 is due to the loss of neurons. (G) Reduced numbers of vGluT3 neurons (arrows) are also found in Ape1Nes-cre tissue compared with control tissue.
As VGluT3+ nonserotonergic neurons are involved in the response to cold stress (61), we assessed this neuronal population in the RMR as well. We found that the majority of VGluT3+ neurons, which do not overlap with the TPH2+ serotonergic neurons, were markedly decreased in the Ape1Nes-cre brain (Fig. 4G, arrows). These data suggest that the loss of serotonergic (TPH2+) and glutamatergic (VGluT3+) neurons in the Ape1Nes-cre DR/RMR impacts the regulation of cold stress, causing a hypothermic phenotype and contributing to the uniform early lethality characteristic of the Ape1Nes-cre-mutant mice.
APE1 Facilitates Kainic Acid-Induced Expression of Immediate Early Genes.
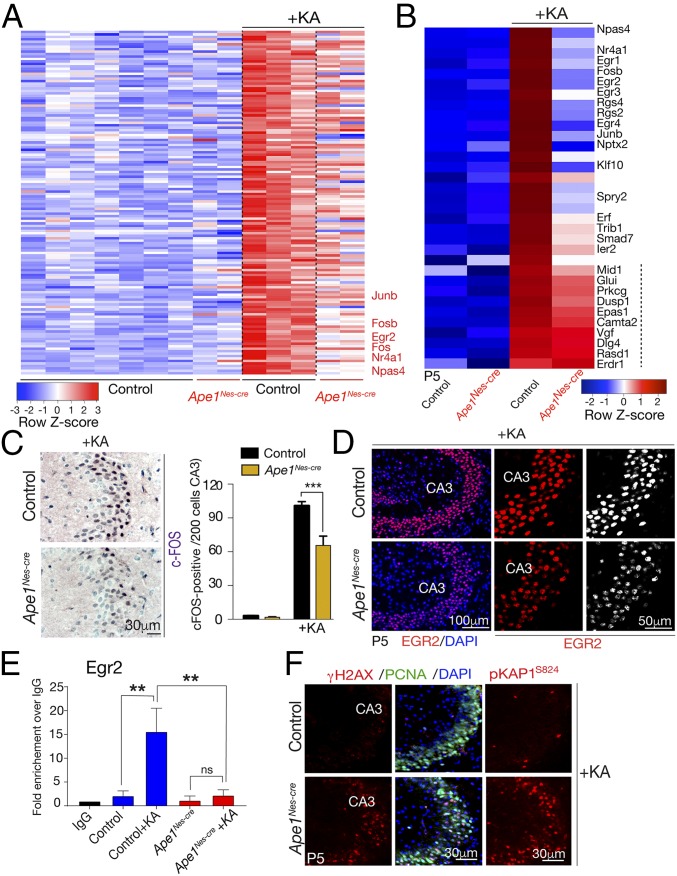
APE1 has been directly implicated in the regulation of gene expression, a function primarily related to its ability to control the redox status and DNA-binding activity of several transcription factors, such as the FOS/JUN heterodimeric complex, activator protein-1 (AP-1) (30). This redox function is considered separate from its DNA-repair activity (12, 24, 25, 68, 69). To investigate the REF1 function of APE1 in a physiologic setting, we used an established paradigm of gene-expression activation via administration of the glutamic acid analog kainic acid (KA) to induce excitatory stimulation of the hippocampus (70, 71), which directly activates the AP-1 complex (30, 68, 72, 73). In this paradigm, KA stimulates a broad gene-expression program in the hippocampus, including activation of immediate early genes (IEGs), a defined group of genes that are rapidly activated in response to stimuli (74, 75). Therefore, we treated P5 WT and Ape1Nes-cre mice with KA and after 2 h assessed gene expression using Affymetrix GeneChips. We initially looked at overall gene expression in response to KA and found a >1.4-fold increase (P < 0.05) in 345 unique genes in WT tissue, while Ape1Nes-cre tissue showed a >1.4-fold increase in only 120 of these genes, with a more modest increase (>1.2) in an additional 120 unique genes (SI Appendix, Fig. S7). There also were 464 genes that were down-regulated in WT tissue, with only 411 of these showing some degree (<1.2-fold change) of decreased expression in Ape1Nes-cre tissues; these genes are involved in various cellular signaling pathways (SI Appendix, Table S1). The identity of the up-regulated genes according to Gene Ontology (GO) groupings listed gene sets associated with synapse function, cell membranes, and cellular junctions (SI Appendix, Table S1). The synapse function GO group is consistent with KA activating NMDA receptors via an excitotoxic stimulatory mode (70, 76). The down-regulated genes were less clearly linked to KA treatment and included G protein-coupled receptors and genes associated with vomeronasal/olfactory function. However, the expression of most genes (over 90% of expressed genes in WT; 32,984 of 33,793 main probes) was unaffected by KA, as is consistent with the selective stimulatory mode of action of this agent.
Significantly and despite the general KA-induced up-regulation of gene expression, some clear differences were identified between control and Ape1Nes-cre tissue. In particular, we found that a select group of 45 genes that were up-regulated in the control were essentially unchanged in the Ape1Nes-cre tissue: 15 genes showed no change (<1.0), and 30 genes were changed by less than 1.2-fold. Examination of these genes revealed a subset of IEGs, including Nptx2, Rsg2/4, and Egr2/4, that were markedly up-regulated after KA treatment in control tissue but not in Ape1Nes-cre tissue (Fig. 5 A and B). This group of genes included Fos and Junb, key components of the AP-1 transcription factor complex, which is known to regulate IEG expression (77, 78). Thus, a subset of IEGs appears to be unresponsive in Ape1Nes-cre tissue, and within this gene set was a smaller subset (Npas4, Nr4a1, Egr1/3, Ptgs2, and others) showing only slight responsiveness but not reaching the levels of up-regulation as seen in control tissue.
Fig. 5.
APE1 regulates a subset of IEGs in response to excitotoxic stimulation. (A) A heatmap depicting microarray expression analysis shows that after KA treatment a subset of genes that are markedly up-regulated in the control hippocampal formation are reduced in expression in Ape1Nes-cre tissue. Genes listed in red text are IEGs (a full gene set list is presented in SI Appendix, Table S1). The color bar indicates relative gene-expression levels depicted in the heatmap. (B) Analysis of selected IEGs shows that many of these are decreased in Ape1Nes-cre tissue compared with control tissue. Genes displayed were based on differences in expression between WT tissue after KA treatment and Ape1Nes-cre tissue after KA treatment, with the smallest difference on the bottom and the largest difference at the top. In comparison with IEGs, other genes (marked by the dashed line) show similar expression in control tissue after KA treatment. (C, Left) Immunohistochemical identification of c-FOS after KA treatment shows reduced up-regulation in the Ape1Nes-cre hippocampal region compared with control tissue. (Right) The graph shows the quantification of c-FOS+ cells (of 200 counted cells), showing an ∼35% reduction in levels; ***P < 0.001. (D) Immunostaining indicates the reduction of EGR2 after KA stimulation in the Ape1Nes-cre hippocampal region compared with the control tissue. CA3, cornu ammonis 3 region of the hippocampal formation. (E) ChIP shows reduced AP-1 (JunB) occupancy at the Egr2 promoter in Ape1Nes-cre tissue 1.5 h after KA exposure. n.s., not significant, **P < 0.001. (F) KA treatment induces extensive DNA-strand breaks in the hippocampus as determined using γH2AX and phosphorylation of KAP1 (at serine 824).
We further assessed the gene-expression data using immunohistochemistry to examine levels of FOS and EGR2 after KA treatment. Consistent with the gene-expression data, immunohistochemistry showed a lower level of both proteins after KA stimulation in Ape1Nes-cre tissue than in control tissue, particularly in the hippocampal CA3 region (Fig. 5 C and D). As APE1 redox activity stimulates AP-1 binding to promoters (12, 24, 25, 68, 69), we used ChIP to determine levels of AP-1 bound to IEG promoters in representative genes identified above, including Egr2, Npas4, and JunB. After KA treatment, elevated binding of AP-1 at these IEG promoters was observed in WT tissue, while this binding was markedly reduced in Ape1Nes-cre tissue (Fig. 5E and SI Appendix, Fig. S8). These data indicate that the failure to up-regulate the expression of these genes in APE1-null tissue is probably due to the lack of AP-1 binding to gene promoters and the subsequent stimulation of transcription.
Given the differences in gene expression between the control and Ape1Nes-cre tissue and because a primary function of APE1 is suppressing the accumulation of oxidative DNA damage, we determined levels of DNA damage after KA administration using immunolocalization of γH2AX and pKAP1. As shown in Fig. 5F, there is a marked increase in immunostaining for both markers of DNA-strand breaks in the mutant tissue, while in control tissue no DNA damage above background levels was observed. Notably, both the γH2AX and pKAP1 signals are apparent, suggesting the type of damage is DNA double-strand breaks, likely as a result of the continuing progenitor cell proliferation in this region at P5 (see PCNA signal in Fig. 5F). This indicates that KA treatment induces substantial DNA damage; this damage is effectively suppressed in WT tissue, whereas the loss of repair via APE1 markedly exacerbates the level of DNA breaks. This elevated DNA damage also coincides with attenuation of FOS1 induction, which results in a consequent decrease in IEG expression. Importantly, while these data support the view that APE1 is a regulator of AP-1 activity, the DNA damage coinciding with reduced gene expression may also potentially contribute to a failure of AP-1 binding to gene promoters after excitotoxicity.
APE1 Is a Tumor Suppressor in the Brain.
Persistent endogenous DNA damage resulting from the disruption of DNA repair in conjunction with p53 inactivation can result in early-onset brain tumor development (37, 79, 80). Therefore, we aged (Ape1;p53)Nes-cre mice and followed the pathogenic outcomes of APE1 deletion. Strikingly, all (Ape1;p53)Nes-cre mice rapidly developed medulloblastoma brain tumors by 2–3 mo of age (Fig. 6A). These tumors were highly infiltrative, typically located in the molecular layer of the cerebellum, and in most cases were very large, almost enveloping this tissue (Fig. 6B). Notably, the strong propensity for the loss of APE1 to result in the accumulation of DNA damage that drives oncogenic events is reflected by a somewhat similar latency of tumor development in Ape1Nes-cre animals when p53 is heterozygous (Fig. 6A). We also followed a cohort of p53-homozygous mutant mice with one mutant Ape1 allele [e.g., (Ape1+/L;p53L/L)Nes-cre; L = LoxP, + = WT], and these mice also developed tumors, although with a much longer latency than the homozygous Ape1 mutant (SI Appendix, Fig. S9 A and B). We found that while tumor latency was extended in the (Ape1+/L;p53L/L)Nes-cre mice, around 70% of this genotype still succumbed to brain tumors, mostly gliomas (SI Appendix, Fig. S9). These tumors did not show detectable levels of APE1, as determined by immunohistochemistry, indicating that inactivation of the remaining Ape1 allele is likely causative for tumorigenesis (SI Appendix, Fig. S9D).
Fig. 6.
APE1 is a tumor suppressor. (A) All mice with dual APE1 and p53 inactivation [(Ape1;p53)Nes-cre] and many that were p53-heterozygous [i.e., (Ape1L/L;p53L/+)Nes-cre] developed brain tumors at 1–3 mo of age. Control mice were Ape1 WT, p53Nes-cre. (B) Histology of a representative (Ape1;p53)Nes-cre and (Ape1L/L;p53+/L)Nes-cre medulloblastoma is shown. (Upper) H&E and Nissl staining indicates typical tumor presence in the molecular layer of the cerebellum. (Lower) Immunostaining for PCNA indicates tumor cell proliferation. (C) Immunostaining for MATH1 indicates that the medulloblastoma is a sonic hedgehog-driven tumor derived from granule neuron precursors. P5 developing cerebellum shows normal MATH1 levels in immature granule neurons in the molecular layer. Corresponding medulloblastoma sections indicate widespread MATH1 immunopositivity in tumor tissue. GC, granule cells; MB, medulloblastoma. (D) Recurrent cytogenetic abnormalities are present in the tumors, including loss of Ptch1 and loss of a copy of Chr13 (which contains Ptch1) and MycN amplification. Mixt, the N-myc amplification was heterogeneous and not identified in all tumor cells. (E) Cytogenetic analysis utilizing FISH indicates the loss of Patched-1 and in some cases chromosome 13 (arrow). (F) Amplification of MycN is also observed in many medulloblastomas and often appears as an homogeneously stained region in the chromatin (arrows). The FISH images represent two of the five tumors listed in the table in D.
Medulloblastoma in (Ape1;p53)Nes-cre mice was characterized by Patched-1 (Ptch1) inactivation, which was often linked to chromosome 13 loss (this chromosome contains Ptch1) (Fig. 6D). Elevated MycN expression and gene amplification via double minute chromosomes or homogeneously staining regions were also common features in APE1-null medulloblastomas (Fig. 6D). In addition, high expression levels of MATH1 (also known as “ATOH1”) were observed in these tumors, similar to the P5 EGL, underscoring that the medulloblastoma arose from granule neuron precursors due to deregulated sonic hedgehog signaling stemming from Ptch1 inactivation (Fig. 6C). Thus, inactivation of Ptch1 promotes persistent sonic hedgehog signaling, which, together with oncogene activation (e.g., MycN amplification), causes deregulated proliferation, a situation characteristic of other mouse medulloblastomas originating from combined genome instability and loss of functional p53 (79). The high granule neuron progenitor numbers in the cerebellar EGL likely present an abundant target population for early transformation to medulloblastoma. In contrast, in Ape1-heterozygous [(Ape1+/L;p53L/L)Nes-cre] brain tissue, glioma formation predominates (SI Appendix, Fig. S9), because after 3 wk of age (the period of postnatal cerebellar development) the cerebellum no longer contains the susceptible granule neuron progenitors.
Discussion
Defects in maintaining genome stability can impact multiple organ systems and are associated with a variety of human diseases. While all DNA-repair pathways are broadly required for normal neural developmental, germline defects in components of BER have been specifically linked to neurologic disease, remarkably without an obvious impact elsewhere in the body (21). Using conditional inactivation of Ape1 in the mouse brain, we find that this central BER factor is critical for the maintenance of the postnatal nervous system. In particular, our findings reveal that APE1 is expendable for embryonic neurogenesis, but after birth and coincident with respiration and commencement of oxidative metabolism, APE1 fulfills a critical and indispensable role in genome maintenance in the nervous system. We also observed that the loss of APE1 affected specific neural cell populations, including myelin-producing oligodendrocytes and serotonergic neurons in the Raphe nuclei, leading to premature lethality associated with thermoregulatory defects. Additionally, the enigmatic role ascribed to APE1 as a modulator of gene expression/transcription was apparent in the setting of excitotoxic stimulation of the hippocampus via KA, implicating the REF1 function of the protein in regulating AP-1 transcription factor binding. Last, our data indicate that APE1 is a potent tumor suppressor, and that multiple brain tumor types can arise from the loss of this enzyme, supporting a role for oxidative DNA lesions as initiating events in tumorigenesis.
Our findings suggest that the etiology of BER-related syndromes and their dramatic effects on the nervous system probably stem from the high oxygen utilization of the brain that leads to frequent oxidative DNA lesions (1). In this regard, APE1 is key for suppressing the accumulation of oxidative DNA damage, being an essential factor in BER after both direct formation of AP sites and damaged base removal by one of several DNA glycosylases (7, 9, 81). The shift to mitochondrial respiration markedly impacts neonatal physiology, including the nervous system (82). Indeed, while the Ape1Nes-cre mouse exhibited no obvious phenotype during embryonic neural development, postnatal neurogenesis in the cerebellum was severely disrupted. The profound cerebellar tissue disruption coincided not only with the shift from placental to respiratory oxygenation but also with the extensive DNA replication during postnatal neurogenesis. Elsewhere, throughout the Ape1Nes-cre nervous system, there was gradual and widespread cellular attrition. Similar to the profound Ape1Nes-cre phenotype, neural inactivation of DNA ligase 3, which is essential for mtDNA replication, also had little embryonic effect but led to widespread apoptosis in the postnatal nervous system (14). Additionally, inactivation of XRCC1 resulted in the failure of postnatal cerebellar interneuron proliferation and maturation, but not embryonic progenitor expansion, via p53-dependent cell-cycle arrest (37). Thus, when respiration becomes the source of oxygenation in the brain, it is critical to suppress oxidative DNA damage.
Mice appear more tolerant of the loss of DNA-repair factors than humans are, often lacking the corresponding severe phenotypic consequences seen in the respective human genetic disorder such as seen with APTX and TDP1 inactivation (83, 84). Currently, no inherited human neurologic syndromes resulting from APE1 mutations have been reported, possibly indicating the severe outcome of the loss of APE1 function. However, this scenario had been considered the likely reason that defects in another key BER factor, XRCC1, had not been identified in human disease, but a recent report described hypomorphic XRCC1 mutations as causative for ataxia (19, 85).
Interestingly, higher levels of γH2AX foci were observed in Ape1Nes-cre mice within the cortical neurons present in upper layers (II–IV). There is substantial developmental maturation in this region between the second and fourth postnatal weeks, resulting in a 10-fold cortical layer-specific increase in the number of both excitatory and inhibitory synapses (48, 86). Glutamatergic and GABAergic synapses also form in the upper-layer (II/III) cortical pyramidal neurons during the first few weeks after birth (47). The increased γH2AX in the Ape1Nes-cre upper cortical layers could reflect cortical functional compartmentalization associated with higher metabolic/oxidative utilization during this period of rapid synaptogenesis.
We also observed striking defects in oligodendrocytes, where widespread loss of myelination and a reduction in oligodendrocyte number were found after APE1 inactivation. Other BER factors, such as PNKP and XRCC1, are similarly required to prevent myelin and oligodendrocyte defects, further indicating the high sensitivity of this neural cell type to oxidative DNA damage (37, 52). The sensitivity of oligodendrocytes to oxidative DNA damage potentially implicates genotoxic stress as an etiologic agent in multiple sclerosis. In a recent study, a role for APE1 in preserving the survival of neurons and oligodendrocytes after stroke-related damage was also identified. In this work, tamoxifen-induced Ape1 deletion showed that the protein was necessary to reduce both gray- and white-matter tissue damage following ischemic injury (87).
Our findings that serotonergic neuron loss in the Raphe nuclei and thermoregulation defects occur in the Ape1Nes-cre mice point to additional specific vulnerabilities of APE1 deficiency. This unexpected phenotype reveals a previously unknown susceptibility of the serotonergic and VGluT3+ glutamatergic neuronal population to oxidative DNA lesions. While the loss of these neurons is sufficient to explain the thermoregulatory defect, additional issues may contribute to this feature of the Ape1Nes-cre phenotype. The Ape1Nes-cre mice have extensive and varied cell loss, which could affect broader aspects of thermoregulation, such as defects in the thalamus and hypothalamus or neurotransmission. The demyelination that occurs in the Ape1Nes-cre mice might also be involved, since in humans multiple sclerosis is linked to thermoregulation defects (88). Finally, metabolic defects such as hypoglycemia and various endocrine conditions can influence thermoregulation (89). If metabolism impacts thermoregulation in Ape1Nes-cre mice, this could potentially result from neuroendocrine abnormalities (90, 91).
The availability of the Ape1Nes-cre mice provided a means for physiologically testing the role of APE1 in the regulation of gene expression. Toward this end, we used an established paradigm of gene expression involving KA, which is a potent agonist that activates the AMPA subclass of ionotropic glutamate receptors (70, 92, 93). After KA treatment we observed that a subset of genes in the Ape1Nes-cre hippocampus either were not expressed or were expressed at much reduced levels relative to WT tissue. These genes were IEGs (94) or recognized FOS-responsive genes (78). Thus, our data indicate that APE1/REF1 is a necessary factor for FOS-induced gene regulation in a physiologically relevant in vivo setting. In addition, our data indicate that KA treatment induces substantial levels of DNA damage in the hippocampus, damage that is suppressed by APE1. Previous reports similarly linked γH2AX formation to stimulation by KA or glutamate agonists in cultured cortical neurons, indicating that these stimulatory activities promote neural DNA damage (95). IEG expression has recently been functionally linked to DNA-strand breaks caused by topoisomerase activity (96). Consequently, it is possible that APE1 fulfills an important regulatory role in restraining excessive DNA damage associated with transcription, perhaps in concert with the DNA-strand breaks reportedly caused by topoisomerase activity during transcription (96, 97).
Inactivation of APE1 in the nervous system in the setting of defective p53 leads to a range of tumor types. Medulloblastoma occurs when both Ape1 and p53 alleles are inactivated, and gliomas predominate when tumor latency is extended by the initial loss of only one Ape1 or p53 allele. The much longer tumor latency in Ape1-heterozygote mice likely represents the time required for inactivation of the remaining Ape1 allele. Despite APE1’s clear role as a potent tumor suppressor in mice, inactivating mutations have not yet been directly linked to tumorigenesis in humans. In addition, only a few APE1 mutations in human cancer are reported in the human cancer COSMIC database (https://cancer.sanger.ac.uk/cosmic). Nevertheless, the striking tumor predisposition after APE1 inactivation (together with p53 loss) in mice implicates the endogenous DNA lesions suppressed by APE1 as causative in cancer. Possibly, in humans, these lesions, most likely AP sites, are restrained by other/additional DNA-repair factors not present in rodents, or human cells activate cell-death responses more effectively, eliminating the potential for replication-dependent mutagenesis.
In summary, our findings have elaborated important biological roles for APE1 and the prominence of this DNA-repair factor in preventing pathogenic genome damage. It will now be important to connect APE1 and tissue homeostasis more thoroughly in different disease scenarios. Furthermore, delineating APE1 repair and transcriptional regulatory functions is now possible in a physiologic setting, permitting examination of these separable roles in different tissues.
Methods
Generation of the Ape1 Conditional Knockout Mouse.
Exons 2 and 3 of Ape1, including the initiating ATG and the second coding exon, were deleted using recombineering, and chimeric mice were generated. To inactivate Ape1 in the nervous system, Ape1LoxP mice were crossed with Nes-cre [B6.Cg-Tg(Nes-cre)1Kil/J] mice (Jackson Laboratory no. 003771) to generate Ape1Nes-cre mice. Sox2-cre [Tg(Sox2-cre)1Amc] mice (Jackson Laboratory no. 004783) was used to delete Ape1 throughout the embryo to generate an Ape1-null (Ape1−/−) mouse. Cre mice [B6.Cg-Tg(CAG-cre/Esr1)5Amc/J] (Jackson Laboratory no. 004682) were used to generate E14 primary mouse embryonic fibroblasts. The generation of p53LoxP, Xrcc1LoxP, Brca2LoxP mice has been described previously (37, 79). All animal experiments were carried out according to NIH regulations and were approved by the St. Jude’s Children’s Research Hospital Animal Care and Use Committee.
Body temperature measurement.
An IPTT-300 temperature transponder (Bio Medic Data Systems) was used to collect the body temperature. Three mice per genotype were used for this experiment.
Histology and immunodetection.
Brains and embryos were collected in either 4% (wt/vol) buffered paraformaldehyde (PFA) or were fixed in 10% buffered formalin (vol/vol). NISSL and H&E stainings were performed according to standard procedures. For immunodetection, sections were subjected to antigen retrieval and were treated overnight with primary antibodies. FITC- and Cy3-conjugated secondary antibodies were used for immunofluorescence. For colorimetric immunodetection, the standard protocol was performed using biotinylated secondary antibody and avidin-biotin complex (Vectastain Elite kit; Vector Laboratories). Apoptosis was measured by TUNEL assay using the ApopTag kit (Millipore) according to the manufacturer’s protocol. At least three independent tissue samples from each genotype were used for all experiments and analyses.
Primary astrocytes and immunofluorescence.
Primary mouse astrocytes were prepared from P3 cortices. Astrocytes were grown on chamber slides and fixed with a 4% PFA solution. Slides were treated with primary antibodies for 3 h and were treated with Alexa-Fluor 488/555-conjugated secondary antibodies for 1 h. All experiments were performed in triplicate.
APE1 Enzymatic Assay.
Primary E14 fibroblasts were isolated from Ape1CreTM mice. Total cell extracts were prepared using 1× RIPA buffer with 1× Halt protease inhibitor mixture (Pierce). AP-site–incision assays were performed using a double-stranded oligonucleotide substrate in which one strand harbored an abasic site analog, THF. Whole-cell extracts and 1 pmol of 5′-[32P]–labeled double-stranded DNA were incubated with the labeled strand harboring THF. Reactions were heated before separation on a 20% polyacrylamide-urea denaturing gel.
AP site detection assay.
Primary astrocytes were grown on CC2-treated glass chamber slides, and the AP-site Assay Kit (ab133076; Abcam) was used according to the manufacturer’s protocol.
Proliferation assay.
Passage 2 primary astrocytes were seeded onto 24-well plates and then were trypsinized and counted every 3 d and reseeded at the same density. Seeding and counting were repeated until passage 6. The experiment was performed in triplicate using multiple clones.
Cell treatments and survival assay.
Passage 3 primary astrocytes were grown on 24-well plates at day 0. On day 1, cells were treated with MMS (Sigma), H2O2 (Thermo Fisher), cis-Diammineplatinum(II) (cisplatin; Sigma), or γ-irradiation. Cells were counted on day 4. The experiment was performed in triplicate using multiple clones.
Alkaline comet assay.
Cells were treated with H2O2 (Thermo Fisher), MMS (Sigma), or γ-irradiation (20 Gy). Comets were analyzed using the Comet Assay IV system (Perceptive Instruments), and at least 100 comets per experiment were counted.
KA treatment.
P5 mice were i.p. injected with KA (K0250; Sigma). After 2 h, hippocampi were collected to generate cryosections for immunofluorescence analysis or for microarray analysis.
Microarray analysis.
RNA from P5 mice hippocampi was analyzed using the Mouse Gene 2.0 ST platform. Fold change (FC) was calculated after normalization using samples from untreated control compared with controls 2 h after KA treatment. Heatmaps were generated in R with normalized, batch-corrected microarray data; row z-scores were utilized for scaling. Probes were selected based on FC cutoff levels (either >1.4 or >1.5) between untreated controls and controls 2 h after KA treatment, using a P value < 0.05 from a Student’s t test.
ChIP.
WT and Ape1Nes-cre P5 mice were injected with KA (K0250; Sigma), and hippocampi were collected 90 min later. Tissue extract was cross-linked with 1% formaldehyde and then washed with ice-cold PBS containing a protease inhibitor mixture (P8340; Sigma). Chromatin was extracted in lysis buffer, washed, and then sheared to generate DNA fragments of 200–500 bp using a Covaris M220 sonicator (Covaris). c-Jun antibody (5 µg; ab31419; Abcam) and Magna ChIP Protein A+G Magnetic Beads (20 µL; 16-663; Millipore) were added to the chromatin and incubated overnight at 4 °C. Beads were washed and immunoprecipitated, and DNA was eluted at 65 °C overnight. Immunoprecipitated DNA was analyzed via quantitative PCR (7500 Real Time PCR System; Applied Biosystems). Primers used for PCR amplification of precipitated DNA are as follows: Egr2, 5′-CTGCCATTAGTAGAGGCTCAGG (forward) and 5′-GTTCTACAGCCGTCATCCTGG (reverse).
Full methodology is presented in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jingfeng Zhao for genotyping and technical help and the Animal Resource Center, the Small Animal Imaging Center, the Cytogenetics Core, and the Transgenic Core Unit for support with mouse work. P.J.M. is supported by NIH Grants NS-37956 and CA-21765, Cancer Center Support Grant P30 CA21765, and the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital. M.S. received support from the Uehara Memorial Foundation of Life Sciences, Japan, and D.M.W. was supported by the Intramural Research Program of the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809682115/-/DCSupplemental.
References
- 1.McKinnon PJ. Genome integrity and disease prevention in the nervous system. Genes Dev. 2017;31:1180–1194. doi: 10.1101/gad.301325.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sykora P, Wilson DM, 3rd, Bohr VA. Base excision repair in the mammalian brain: Implication for age related neurodegeneration. Mech Ageing Dev. 2013;134:440–448. doi: 10.1016/j.mad.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 6.Dizdaroglu M. Oxidatively induced DNA damage: Mechanisms, repair and disease. Cancer Lett. 2012;327:26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dizdaroglu M, Coskun E, Jaruga P. Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat Res. 2017;771:99–127. doi: 10.1016/j.mrrev.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee AJ, Wallace SS. Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? Free Radic Biol Med. 2017;107:170–178. doi: 10.1016/j.freeradbiomed.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 11.Guikema JE, et al. Apurinic/apyrimidinic endonuclease 2 regulates the expansion of germinal centers by protecting against activation-induced cytidine deaminase-independent DNA damage in B cells. J Immunol. 2014;193:931–939. doi: 10.4049/jimmunol.1400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbotts R, Wilson DM., 3rd Coordination of DNA single strand break repair. Free Radic Biol Med. 2017;107:228–244. doi: 10.1016/j.freeradbiomed.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbari M, Morevati M, Croteau D, Bohr VA. The role of DNA base excision repair in brain homeostasis and disease. DNA Repair (Amst) 2015;32:172–179. doi: 10.1016/j.dnarep.2015.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du F, et al. Tightly coupled brain activity and cerebral ATP metabolic rate. Proc Natl Acad Sci USA. 2008;105:6409–6414. doi: 10.1073/pnas.0710766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magistretti PJ, Pellerin L. Cellular bases of brain energy metabolism and their relevance to functional brain imaging: Evidence for a prominent role of astrocytes. Cereb Cortex. 1996;6:50–61. doi: 10.1093/cercor/6.1.50. [DOI] [PubMed] [Google Scholar]
- 17.Raichle ME, Gusnard DA. Appraising the brain’s energy budget. Proc Natl Acad Sci USA. 2002;99:10237–10239. doi: 10.1073/pnas.172399499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 19.Hoch NC, et al. Care4Rare Canada Consortium XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2017;541:87–91. doi: 10.1038/nature20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 21.McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumi T, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi T, Mitra S. Deletion analysis of human AP-endonuclease: Minimum sequence required for the endonuclease activity. Carcinogenesis. 1998;19:525–527. doi: 10.1093/carcin/19.3.525. [DOI] [PubMed] [Google Scholar]
- 24.Luo M, et al. Characterization of the redox activity and disulfide bond formation in apurinic/apyrimidinic endonuclease. Biochemistry. 2012;51:695–705. doi: 10.1021/bi201034z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Wilson DM., 3rd Human apurinic/apyrimidinic endonuclease 1. Antioxid Redox Signal. 2014;20:678–707. doi: 10.1089/ars.2013.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgiadis MM, et al. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 29.Nishi T, et al. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J Biol Chem. 2002;277:44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 30.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig DL, et al. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- 32.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tebbs RS, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y, et al. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nat Neurosci. 2012;15:819–826. doi: 10.1038/nn.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Brown EJ, Chang S, McKinnon PJ. Pot1a prevents telomere dysfunction and ATM-dependent neuronal loss. J Neurosci. 2014;34:7836–7844. doi: 10.1523/JNEUROSCI.4245-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 39.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci USA. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altman J, Bayer SA. Prenatal development of the cerebellar system in the rat. I. Cytogenesis and histogenesis of the deep nuclei and the cortex of the cerebellum. J Comp Neurol. 1978;179:23–48. doi: 10.1002/cne.901790104. [DOI] [PubMed] [Google Scholar]
- 41.Chong MJ, et al. Atm and Bax cooperate in ionizing radiation-induced apoptosis in the central nervous system. Proc Natl Acad Sci USA. 2000;97:889–894. doi: 10.1073/pnas.97.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y, et al. ATR maintains select progenitors during nervous system development. EMBO J. 2012;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, et al. Expression of transcription factor Satb2 in adult mouse brain. Anat Rec (Hoboken) 2013;296:452–461. doi: 10.1002/ar.22656. [DOI] [PubMed] [Google Scholar]
- 47.Virtanen MA, et al. Development of inhibitory synaptic inputs on layer 2/3 pyramidal neurons in the rat medial prefrontal cortex. Brain Struct Funct. 2018;223:1999–2012. doi: 10.1007/s00429-017-1602-0. [DOI] [PubMed] [Google Scholar]
- 48.De Felipe J, Marco P, Fairén A, Jones EG. Inhibitory synaptogenesis in mouse somatosensory cortex. Cereb Cortex. 1997;7:619–634. doi: 10.1093/cercor/7.7.619. [DOI] [PubMed] [Google Scholar]
- 49.Czirr E, Wyss-Coray T. The immunology of neurodegeneration. J Clin Invest. 2012;122:1156–1163. doi: 10.1172/JCI58656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 52.Shimada M, Dumitrache LC, Russell HR, McKinnon PJ. Polynucleotide kinase-phosphatase enables neurogenesis via multiple DNA repair pathways to maintain genome stability. EMBO J. 2015;34:2465–2480. doi: 10.15252/embj.201591363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stolt CC, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guikema JE, et al. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hadi MZ, Wilson DM., 3rd Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen. 2000;36:312–324. [PubMed] [Google Scholar]
- 56.Morrison SF. Central control of body temperature. F1000 Res. 2016;5:880. doi: 10.12688/f1000research.7958.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boulant JA. Role of the preoptic-anterior hypothalamus in thermoregulation and fever. Clin Infect Dis. 2000;31:S157–S161. doi: 10.1086/317521. [DOI] [PubMed] [Google Scholar]
- 58.Martín-Cora FJ, Fornal CA, Metzler CW, Jacobs BL. Single-unit responses of serotonergic medullary and pontine raphe neurons to environmental cooling in freely moving cats. Neuroscience. 2000;98:301–309. doi: 10.1016/s0306-4522(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 59.McAllen RM, et al. Human medullary responses to cooling and rewarming the skin: A functional MRI study. Proc Natl Acad Sci USA. 2006;103:809–813. doi: 10.1073/pnas.0509862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morrison SF, Nakamura K. Central neural pathways for thermoregulation. Front Biosci. 2011;16:74–104. doi: 10.2741/3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura K, et al. Identification of sympathetic premotor neurons in medullary raphe regions mediating fever and other thermoregulatory functions. J Neurosci. 2004;24:5370–5380. doi: 10.1523/JNEUROSCI.1219-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nason MW, Jr, Mason P. Medullary raphe neurons facilitate brown adipose tissue activation. J Neurosci. 2006;26:1190–1198. doi: 10.1523/JNEUROSCI.4707-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barnes NM, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38:1083–1152. doi: 10.1016/s0028-3908(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 64.Alenina N, et al. Growth retardation and altered autonomic control in mice lacking brain serotonin. Proc Natl Acad Sci USA. 2009;106:10332–10337. doi: 10.1073/pnas.0810793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cummings KJ, Li A, Nattie EE. Brainstem serotonin deficiency in the neonatal period: Autonomic dysregulation during mild cold stress. J Physiol. 2011;589:2055–2064. doi: 10.1113/jphysiol.2010.203679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hodges MR, et al. Defects in breathing and thermoregulation in mice with near-complete absence of central serotonin neurons. J Neurosci. 2008;28:2495–2505. doi: 10.1523/JNEUROSCI.4729-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murray NM, Buchanan GF, Richerson GB. Insomnia caused by serotonin depletion is due to hypothermia. Sleep (Basel) 2015;38:1985–1993. doi: 10.5665/sleep.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: Not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sperk G, et al. Kainic acid induced seizures: Neurochemical and histopathological changes. Neuroscience. 1983;10:1301–1315. doi: 10.1016/0306-4522(83)90113-6. [DOI] [PubMed] [Google Scholar]
- 71.Vendrell M, Curran T, Morgan JI. A gene expression approach to mapping the functional maturation of the hippocampus. Brain Res Mol Brain Res. 1998;63:25–34. doi: 10.1016/s0169-328x(98)00248-4. [DOI] [PubMed] [Google Scholar]
- 72.Franza BR, Jr, Rauscher FJ, 3rd, Josephs SF, Curran T. The Fos complex and Fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science. 1988;239:1150–1153. doi: 10.1126/science.2964084. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, et al. c-fos regulates neuronal excitability and survival. Nat Genet. 2002;30:416–420. doi: 10.1038/ng859. [DOI] [PubMed] [Google Scholar]
- 74.Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kasof GM, et al. Kainic acid-induced neuronal death is associated with DNA damage and a unique immediate-early gene response in c-fos-lacZ transgenic rats. J Neurosci. 1995;15:4238–4249. doi: 10.1523/JNEUROSCI.15-06-04238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jochum W, Passegué E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 78.Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: Involvement of the inducible proto-oncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 79.Frappart PO, et al. Recurrent genomic alterations characterize medulloblastoma arising from DNA double-strand break repair deficiency. Proc Natl Acad Sci USA. 2009;106:1880–1885. doi: 10.1073/pnas.0806882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee Y, McKinnon PJ. DNA ligase IV suppresses medulloblastoma formation. Cancer Res. 2002;62:6395–6399. [PubMed] [Google Scholar]
- 81.Jacobs AL, Schär P. DNA glycosylases: In DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hillman NH, Kallapur SG, Jobe AH. Physiology of transition from intrauterine to extrauterine life. Clin Perinatol. 2012;39:769–783. doi: 10.1016/j.clp.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Becherel OJ, et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013;9:e1003435. doi: 10.1371/journal.pgen.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katyal S, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Connor E, et al. Mutations in XRCC1 cause cerebellar ataxia and peripheral neuropathy. J Neurol Neurosurg Psychiatry. 2018;89:1230–1232. doi: 10.1136/jnnp-2017-317581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Micheva KD, Beaulieu C. Quantitative aspects of synaptogenesis in the rat barrel field cortex with special reference to GABA circuitry. J Comp Neurol. 1996;373:340–354. doi: 10.1002/(SICI)1096-9861(19960923)373:3<340::AID-CNE3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 87.Stetler RA, et al. APE1/Ref-1 facilitates recovery of gray and white matter and neurological function after mild stroke injury. Proc Natl Acad Sci USA. 2016;113:E3558–E3567. doi: 10.1073/pnas.1606226113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davis SL, Wilson TE, White AT, Frohman EM. Thermoregulation in multiple sclerosis. J Appl Physiol (1985) 2010;109:1531–1537. doi: 10.1152/japplphysiol.00460.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheshire WP., Jr Thermoregulatory disorders and illness related to heat and cold stress. Auton Neurosci. 2016;196:91–104. doi: 10.1016/j.autneu.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 90.Arancibia S, Rage F, Astier H, Tapia-Arancibia L. Neuroendocrine and autonomous mechanisms underlying thermoregulation in cold environment. Neuroendocrinology. 1996;64:257–267. doi: 10.1159/000127126. [DOI] [PubMed] [Google Scholar]
- 91.Gale CC. Neuroendocrine aspects of thermoregulation. Annu Rev Physiol. 1973;35:391–430. doi: 10.1146/annurev.ph.35.030173.002135. [DOI] [PubMed] [Google Scholar]
- 92.Coyle JT. Neurotoxic action of kainic acid. J Neurochem. 1983;41:1–11. doi: 10.1111/j.1471-4159.1983.tb11808.x. [DOI] [PubMed] [Google Scholar]
- 93.Obrenovitch TP, Urenjak J. Altered glutamatergic transmission in neurological disorders: From high extracellular glutamate to excessive synaptic efficacy. Prog Neurobiol. 1997;51:39–87. doi: 10.1016/s0301-0082(96)00049-4. [DOI] [PubMed] [Google Scholar]
- 94.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 95.Crowe SL, Movsesyan VA, Jorgensen TJ, Kondratyev A. Rapid phosphorylation of histone H2A.X following ionotropic glutamate receptor activation. Eur J Neurosci. 2006;23:2351–2361. doi: 10.1111/j.1460-9568.2006.04768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madabhushi R, et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.