Significance
Redox switching driven by [4Fe4S] cluster cofactors modulates DNA binding affinity in proteins, providing a rapid, efficient method of substrate binding and dissociation. Our study establishes an essential redox switch with an aromatic pathway through the yeast DNA primase; a single-residue mutation at position 397 along this redox pathway causes [4Fe4S] cluster degradation and is lethal in yeast.
Keywords: DNA replication, DNA charge transport, iron–sulfur proteins
Abstract
Eukaryotic DNA primases contain a [4Fe4S] cluster in the C-terminal domain of the p58 subunit (p58C) that affects substrate affinity but is not required for catalysis. We show that, in yeast primase, the cluster serves as a DNA-mediated redox switch governing DNA binding, just as in human primase. Despite a different structural arrangement of tyrosines to facilitate electron transfer between the DNA substrate and [4Fe4S] cluster, in yeast, mutation of tyrosines Y395 and Y397 alters the same electron transfer chemistry and redox switch. Mutation of conserved tyrosine 395 diminishes the extent of p58C participation in normal redox-switching reactions, whereas mutation of conserved tyrosine 397 causes oxidative cluster degradation to the [3Fe4S]+ species during p58C redox signaling. Switching between oxidized and reduced states in the presence of the Y397 mutations thus puts primase [4Fe4S] cluster integrity and function at risk. Consistent with these observations, we find that yeast tolerate mutations to Y395 in p58C, but the single-residue mutation Y397L in p58C is lethal. Our data thus show that a constellation of tyrosines for protein-DNA electron transfer mediates the redox switch in eukaryotic primases and is required for primase function in vivo.
DNA replication requires the coordinated activity of several polymerase enzymes (1). DNA primase begins replication on the ssDNA template, synthesizing a short (8–12 nt) RNA primer before handing off the primed template to DNA polymerase α. Primase contains an RNA polymerase subunit (p48) and a regulatory subunit (p58) (2). Primase (3, 4) and many other, if not all, replicative polymerases contain at least one [4Fe4S] cluster (5, 6), a metal cofactor associated with biological redox chemistry (7). Given the high metabolic expense associated with incorporation of a cluster into one of these proteins (8), this cofactor is presumed to play a functional role. Interestingly, the clusters in human DNA primase (9) and yeast DNA polymerase δ (10) have been demonstrated to perform DNA-mediated redox chemistry.
The [4Fe4S] cluster in eukaryotic primases is essential for primer synthesis on the ssDNA template in vitro (3, 4) and in cells (11). The cluster is located in the C-terminal domain of the regulatory subunit, p58C, which is flexibly tethered to the N-terminal domain (12, 13) and can bind DNA independently of the rest of the primase enzyme (14, 15). In human DNA primase, the cluster acts as a redox switch, modulating the interaction of p58C with its substrate (9). Binding of the DNA polyanion shifts the [4Fe4S] cluster redox potential, so that the [4Fe4S]3+ protein is bound tightly, while the [4Fe4S]2+ protein is bound more loosely. Endonuclease III in Escherichia coli similarly undergoes an ∼500-fold increase in DNA binding affinity upon oxidation to the [4Fe4S]3+ state from the resting [4Fe4S]2+ state (16, 17).
We have proposed that the redox switch in human primase may facilitate binding and substrate handoff, through DNA-mediated charge transport (9). In our model, active primase is in the [4Fe4S]3+ state, coupled into the π-stacked bases for redox signaling. The p48 RNA polymerase domain and the p58C [4Fe4S] cluster domain are in contact with the nascent RNA/DNA. When the RNA primer reaches the appropriate length (8–14 nt), we propose that polymerase α, in the [4Fe4S]2+state, contacts the RNA/DNA, becoming activated toward oxidation. Polymerase α sends an electron through the RNA/DNA duplex to reduce primase to the [4Fe4S]2+ state, and in so doing is converted to the [4Fe4S]3+ state. Primase dissociates, and polymerase α is now tightly bound, effectively executing a primer handoff.
During handoff in the redox-switching model, charge must travel through duplex RNA/DNA and protein matrix (9, 16, 17). Biochemical and structural evidence shows that p58C contacts DNA through basic arginine and lysine residues (13, 18, 19), positioning the cluster 25–30 Å from the substrate. Charge must travel from the DNA to the cluster, through a pathway within p58C. Charge-transfer pathways in proteins often comprise aromatic residues, which have comparatively low ionization energies and can transfer charge (20, 21) through single-step tunneling or electron hopping. In human primase, for example, the redox switch is mediated by conserved tyrosine residues, Y309, Y345, and Y347 in the p58C domain. Substitution of these residues with phenylalanine produces redox-deficient mutants, defective for initiation of primer synthesis. Redox pathways through several proteins (22, 23) have been characterized using Tyr → Phe mutations to perturb sites along the electron-hopping pathway.
In yeast (Saccharomyces cerevisiae) primase, residues Y395 and Y397 are conserved and orthologous to Y345 and Y347 in human primase (9). Although positioned differently than their human orthologs within the respective p58C crystal structures, they are spaced ∼10–15 Å apart, and could feasibly comprise a redox pathway (20, 21). We hypothesized that yeast and human p58C have similar redox-switch mechanisms, despite their different tyrosine constellations. Establishing that a yeast primase redox pathway exists also provides an opportunity to assay the viability of yeast cells expressing redox-deficient primase variants.
Results
Redox Switch Through a Tyrosine Pathway in Yeast p58C.
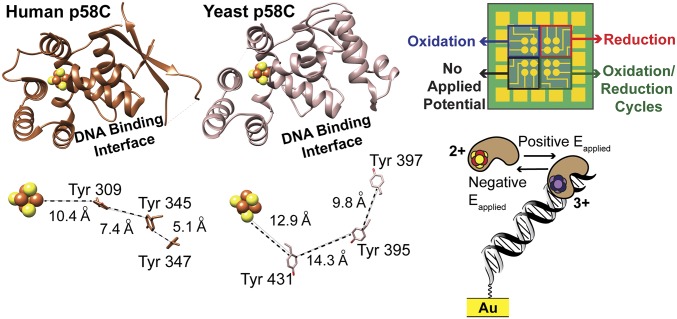
Human and yeast p58C have similar overall structures (Fig. 1) with a backbone root mean squared deviation (rmsd) of only 0.82 Å. Both proteins bind DNA with micromolar affinity (14, 15). Yeast and human p58C each contain a [4Fe4S] cluster buried within the protein, ∼25 Å from the DNA binding interface (14, 15) (Fig. 1). Despite having only 40% identical sequences, yeast and human p58C both have multiple conserved tyrosines between the cluster and the DNA binding interface. In human p58C, these tyrosines shuttle charge through the protein to mediate the redox switch (9) (Fig. 1).
Fig. 1.
Yeast and human p58C structures can both support a redox switch. (Left) Comparison of p58C structures (Upper) and conserved tyrosines (Lower) near the [4Fe4S] cluster in yeast p58C (light brown) and human p58C (dark brown). Structures rendered using human p58C [Protein Data Bank (PDB) ID code 3L9Q] (15) and yeast p58C (PDB ID code 6DI6, SI Appendix, Table S3) structures. (Right) Diagram of the multiplexed DNA electrochemistry platform (Upper) and a cartoon depicting the change in DNA binding associated with redox switching (Lower). Human p58C image adapted from ref. 15. Multiplex chip and yeast p58C images from ref. 9. Reprinted with permission from AAAS.
We sought to determine whether the yeast p58C [4Fe4S] cluster has a redox-switching capability similar to that of human p58C. We identified several tyrosine residues conserved in yeast p58C, which are within feasible charge-transport distance of the yeast p58C cluster or DNA-binding interface, as possible mediators of a redox switch. Residues Y395 and Y397 in yeast p58C (Fig. 1), orthologous to Y345 and Y347, respectively, in the human p58C redox pathway, are prime candidates for a redox role (9). Distances between tyrosine centroids are larger on average in the yeast protein crystal structure (12.9–15.3 Å) (14) than in the human protein crystal structure (5.1–10.4 Å) (15), but fall securely within the distance for feasible microsecond electron transfer. As the distance between centroids, rather than the relative positions of tyrosines, is the primary factor determining the feasibility of charge transfer through protein (21), we predicted that both pathways could mediate redox switching.
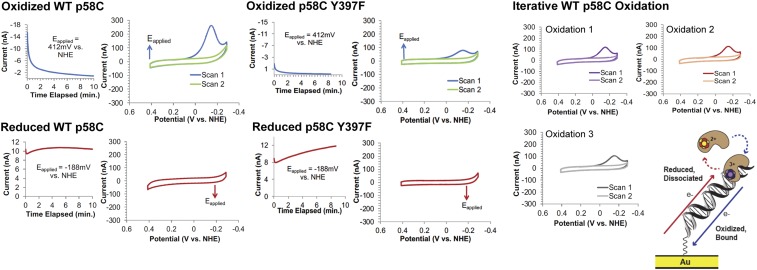
To test whether a change in the oxidation state of the [4Fe4S] cluster in yeast p58C affects DNA binding, we compared anaerobically the electrochemical behavior of oxidized [4Fe4S]3+ p58C and reduced [4Fe4S]2+ p58C on multiplexed DNA electrodes (Fig. 1), modified with a 20-nt DNA duplex substrate containing a 3-nt, 5′- ssDNA overhang (SI Appendix, Table S1) identical to the substrate used to study human p58C (9). Purified WT yeast p58C, in the absence of an applied potential, displays no redox signal in cyclic voltammetry (CV) (SI Appendix, Fig. S1). This form of the protein is thus not coupled to the DNA base pairs for redox signaling. WT p58C was then electrochemically oxidized by applying a positive potential (412 or 512 mV vs. normal hydrogen electrode (NHE); see SI Appendix, Table S2) or electrochemically reduced by applying a negative potential (−188 mV vs. NHE) to the electrode surface. Bulk electrolysis was performed on individual electrodes for a total of 8.3 min, to optimize the yield of electrochemically converted protein. Bulk oxidation of a p58C sample, compared with a buffer control, is shown in SI Appendix, Fig. S2.
Oxidized or reduced p58C samples were scanned by CV immediately after bulk electrolysis. The electrochemically oxidized sample displays a large cathodic peak near −130 to −150 mV vs. NHE in Tris storage buffer (20 mM Tris, pH 7.2, 75 mM NaCl). After a single scan to negative, reducing potentials, the signal disappears, indicating a loss of coupling between the [4Fe4S] cluster and the DNA bases. (Fig. 2). Electrochemically reduced p58C, conversely, does not display any signal after bulk electrolysis. This result suggests that oxidized [4Fe4S]3+ yeast p58C is tightly bound to DNA and electronically coupled into the bases for redox signaling, but reduced [4Fe4S]2+ yeast p58C is loosely associated and not coupled to the DNA for signaling.
Fig. 2.
The yeast p58C redox switch is mediated by conserved tyrosines. (Left) Bulk electrolysis and CV of WT yeast p58C. (Center) Bulk electrolysis and CV of p58C Y397F. (Right) Iterative electrochemical oxidation regenerates a CV signal on DNA. Cartoon (Bottom Right) Illustration of the electrochemically reversible redox switch in yeast p58C. All scans were performed anaerobically, on 30 μM [4Fe4S] p58C (WT, Y395F, or Y397F), 20 mM Tris, pH 7.2, 75 mM NaCl, 100-mV/s scan rate for CV. Illustration from ref. 9. Reprinted with permission from AAAS.
Human p58C can be electrochemically converted from the oxidized, tightly bound state to the reduced, loosely associated state (9). Iterative oxidations on a DNA electrode indicate a similar behavior in yeast p58C (Fig. 2). Oxidation of yeast p58C to the [4Fe4S]3+ state produces a large, reductive CV signal, indicating conversion to the resting (3, 4) [4Fe4S]2+ state. The signal disappears in the second CV scan, but a second oxidation regenerates the reductive peak. A single electron transfer reaction thus facilitates conversion between two forms of p58C with dramatically different DNA-binding and redox-signaling properties.
We next investigated whether the observed yeast p58C redox switch is dependent on a pathway of conserved tyrosine residues, as in the human protein. We constructed yeast p58C variants with mutations at Y395 or Y397. As these residues are conserved orthologs to components of the human primase redox pathway, they were prime candidates for the yeast primase redox pathway. All generated p58C variants (Y395F, Y397F, Y395L, and Y397L) load the [4Fe4S] cluster comparably to WT, as assessed by the ratio of absorption at 410 nm/280 nm in UV-visible spectroscopy (SI Appendix, Fig. S3). Fluorescence anisotropy assays on WT and mutant p58C (SI Appendix, Fig. S4) show that all variants bind the substrate with virtually the same low micromolar affinity. This result suggests that the same number of p58C molecules are bound to the DNA on the electrode surface before electrochemical oxidation, when the sample is present largely in the [4Fe4S]2+ state. Since the redox pathway through p58C is the conduit through which cluster oxidation occurs, we expect that fewer mutants versus WT are bound tightly to DNA in the [4Fe4S]3+ state after bulk oxidation, if these tyrosines mediate the redox switch in yeast primase.
Analysis of charge transfer in the reductive peak (QCV, SI Appendix, Table S2 and Figure S5) for WT yeast p58C, p58C Y395F (SI Appendix, Table S2), Y397F (Fig. 2), Y397L (Fig. 3), and Y395L (SI Appendix, Fig. S7) demonstrates that the mutants are consistently redox-deficient relative to WT p58C. It is interesting to observe however that the p58C variants, particularly the Tyr → Phe mutants, retain partial redox-switching ability. Phenylalanine residues are generally less capable of mediating electron transfer through protein than tyrosines, due to their higher ionization energy (20) and inability to form discrete cation radicals; phenylalanines, however, can mediate electron transfer, although less efficiently through tunneling (22), partially inhibiting charge transfer in some protein systems (23, 24).
Fig. 3.
Tyrosine to leucine p58C mutants are redox-deficient. (Left) Bulk electrolysis and CV of WT yeast p58C. (Right) Bulk electrolysis and CV of yeast p58C Y397L. The mutant displays decreased redox activity in CV. All scans were performed anaerobically, on 57 μM [4Fe4S] p58C (WT or Y397L), 20 mM Hepes, pH 7.2, 75 mM NaCl, 100-mV/s scan rate for CV.
Tyrosine Mutations Do Not Change Yeast p58C Structure.
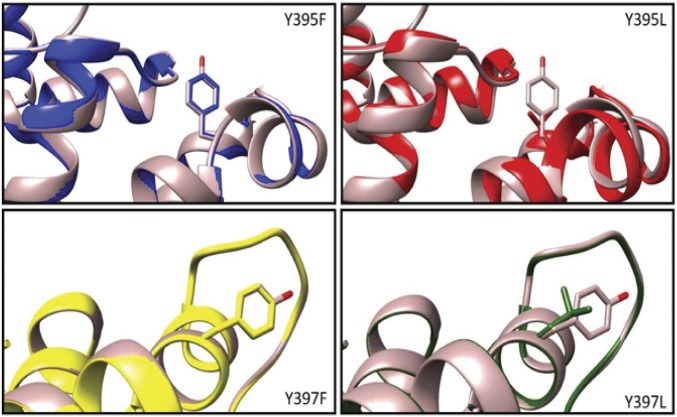
X-ray crystal structures of the WT and mutant yeast p58C demonstrate that no changes in the overall structure of the protein are caused by the tyrosine mutations (Fig. 4 and SI Appendix, Table S3). Overlays of WT yeast p58C and p58C Y395F, Y397F, Y395L, and Y397L, all of which have an rmsd less than 0.5 Å from the WT, underscore the structural similarity. The F395 and F397 aromatic rings in Y395F and Y397F, respectively, are oriented in almost exactly the same position as the aromatic rings in the tyrosines of the WT protein, as had been seen previously with human p58C Y345F and Y347F mutants (9). Notably, the more drastic substitution of the tyrosine aromatic ring with the leucine aliphatic chain also has very little effect on the structure, including the orientation of side chains.
Fig. 4.
Protein structure is preserved upon mutation of Y395 and Y397 in yeast p58C. The WT yeast p58C (brown) crystal structure is overlaid with each redox-deficient mutant. Mutation of tyrosines to aromatic phenylalanine or aliphatic leucine minimally perturbs protein structure.
The similar WT and mutant yeast p58C structures and substrate affinities corroborate that the difference in redox signal between WT and mutant p58C is a result of differences in the redox-switching ability and not the ability to interact with a DNA substrate. All p58C variants were crystallized, moreover, with high [4Fe4S] cluster loading in the protein sample, supported by the UV-visible spectra (SI Appendix, Fig. S3). Overall, we observe that the p58C mutant proteins are less effective in participating in redox signaling on DNA relative to WT, when equal amounts of [4Fe4S] protein (determined spectroscopically) are initially deposited on the Au electrode. Different electrochemical behavior of these variants is thus a consequence of different chemical/electronic properties.
Reversible, Nucleotide Triphosphate-Dependent Redox Activity in Yeast p58C.
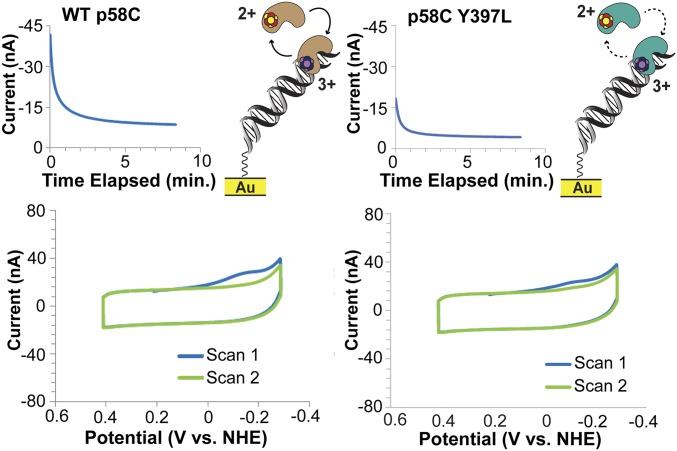
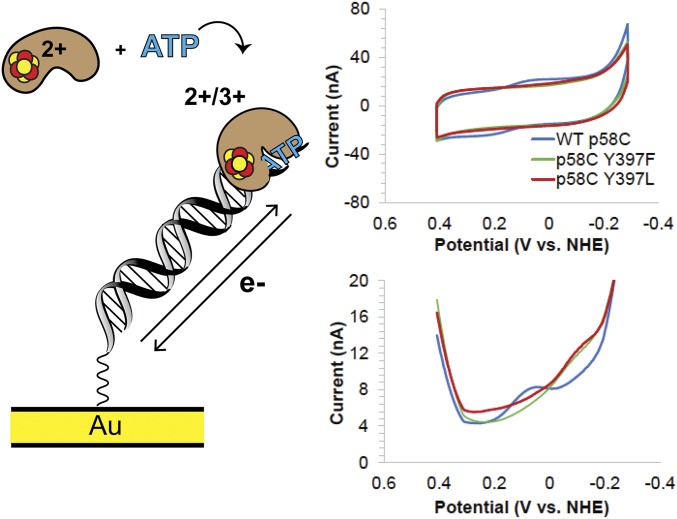
Yeast p58C displays a semireversible, nucleotide triphosphate (NTP)-dependent redox signal on DNA under anaerobic conditions, centered at 149 ± 14 mV vs. NHE, in the presence of 1.25 mM ATP. This potential is within the biological redox potential range (25) and comparable to the potential values observed for other DNA-processing [4Fe4S] proteins, suggesting that active primase may signal other DNA-bound [4Fe4S] proteins during replication (10, 26–28). The signal observed in CV is 71 ± 10% reversible, displaying an average charge-transfer value of 8.7 ± 4 nC in the reductive peak and 5.8 ± 2 nC in the oxidative peak. The larger cathodic wave in CV is consistent with the oxidized [4Fe4S]3+ protein having a higher binding affinity and being more strongly coupled to the DNA bases for redox activity than the reduced [4Fe4S]2+ protein. The reductive peak corresponds to the electrochemical conversion of [4Fe4S]3+ p58C to [4Fe4S]2+ p58C, and the oxidative peak is a measurement of the reverse process. The primase redox-switch model predicts that more oxidized [4Fe4S]3+ protein is bound in the p58C:DNA:NTP complex than reduced [4Fe4S]2+ protein. The data corroborate this model and suggest that NTP binding induces redox switching in yeast p58C.
To assess whether this reversible signal depends on the p58C charge-transfer pathway, we measured the NTP-dependent signals of yeast p58C Y395 and Y397 mutants. Unlike WT yeast p58C, no mutants generated a signal detectable by CV in the presence of 1.25 mM ATP. We therefore used square-wave voltammetry (SWV), an electrochemical technique which can reliably detect smaller signals (29) to aid in characterizing mutant NTP-dependent redox activity. We observe a single reductive peak in SWV at −86 ± 13 mV vs. NHE and −90 ± 4 mV vs. NHE, for Y397F and Y397L, respectively, on a DNA electrode in the presence of 1.25 mM ATP (Fig. 5). This signal appears at a potential distinct in SWV from the semireversible signal for the WT p58C (+85 ± 9 mV vs. NHE), reflecting the presence of a different species (Fig. 5).
Fig. 5.
Reversible, NTP-dependent redox switching in yeast p58C. (Left) Cartoon depicts DNA- and NTP-bound yeast p58C cycling between [4Fe4S]3+ and [4Fe4S]2+ states. (Right) CV scans of WT yeast p58C (blue), p58C Y397F (green), and p58C Y397L (red) in the presence of 1.25 mM ATP. WT p58C displays a signal at 149 ± 14 mV vs. NHE. Reductive SWV of WT and mutant p58C. A small signal near −80 to −90 mV vs. NHE appears for Y397F and Y397L. All scans were performed anaerobically, on 57 μM [4Fe4S] p58C variant and 1.25 mM ATP in 20 mM Hepes, pH 7.2, 75 mM NaCl, 100-mV/s scan rate for CV, 15-Hz frequency, 25-mV amplitude, 60-mV/s scan rate for SWV. Potentials reported are mean ±SD of at least three trials. The increase in current at −0.3 V vs. NHE appears in the buffer scan and is p58C-independent.
We next tested whether the p58C mutants displayed any redox activity in the presence of 2.5 mM ATP. The Y395L mutant displayed no measurable redox signal in the presence of 1.25 or 2.5 mM ATP (SI Appendix, Fig. S8). The Y395F variant displays some reversible charge transport centered at 162 ± 6 mV vs. NHE in the presence of 2.5 mM ATP, although the charge transport (3 ± 1 nC in the cathodic peak, 2 ± 0.6 nC in the anodic peak) is diminished relative to WT with 1.25 mM ATP (SI Appendix, Fig. S9). In the presence of 2.5 mM ATP, Y397F displays an irreversible peak at a similar potential to the peak observed with 1.25 mM ATP (SI Appendix, Fig. S10) in some but not all scans. The Y397L variant displays minimal reversible redox activity in CV (SI Appendix, Fig. S11) centered at 140 ± 17 mV vs. NHE, with charge-transport values of 1.7 ± 1 nC in the cathodic peak and 1.5 ± 1 nC in the anodic peak. The NTP-dependent redox activity of the tyrosine mutants is thus consistently diminished relative to WT p58C, suggesting the primary redox pathway through Y395 and Y397 is important for redox signaling when bound to all necessary substrates for primer synthesis.
This irreversible reductive peak between −80 and −90 mV vs. NHE likely corresponds to the [3Fe4S]+ degradation product of the [4Fe4S] cofactor (30). This irreversible secondary reductive peak is actually observed in WT yeast p58C, alongside the primary wave, in the presence of a large excess of ATP (SI Appendix, Fig. S12). A signal in the reductive wave at comparable potentials was recently observed electrochemically and spectroscopically in a cancer-causing variant of human base excision repair protein MUTYH, when exposed to atmospheric oxygen (31). The [3Fe4S]+ degradation product is a consequence of oxidation from the resting [4Fe4S]2+ state to the [4Fe4S]3+ state (26, 31) in a mutant with a destabilized metal cofactor. The [3Fe4S]+ product can also occur as a result of oxidation in an aerobic atmosphere during purification or sample preparation (6, 26). Since the [3Fe4S]+/0 reduction peak occurs in p58C Y397F/Y397L in the presence of NTPs and in the absence of oxygen, however, these mutants likely undergo some electron transfer when bound to DNA and NTPs, anionic substrates expected to shift the potential of the [4Fe4S] cluster (25, 32). Since the compromised redox pathway inhibits reversible cycling between the [4Fe4S]2+ and [4Fe4S]3+ states, redox signaling in these p58C variants can lead to a “trapped” high-energy [4Fe4S]3+ species which cannot easily be reduced back to the [4Fe4S]2+ state; the cluster then becomes unstable and degrades. Efficient redox switching is essential for regulation of activity, as well as [4Fe4S] cluster stability.
The Yeast p58C Redox Switch Is Necessary for Viability.
After characterizing the redox switch in yeast DNA primase, we sought to investigate the consequences of this chemistry in yeast cells. Both subunits of yeast primase are essential; partially defective alleles compromise DNA synthesis and cell growth (33, 34). Liu and Huang (11) additionally have shown that mutations of the cysteine residues ligating the p58C [4Fe4S] cluster cause growth defects in cells, suggesting the importance of the primase cluster for viability. We have shown that mutations at p58 residues Y395 or Y397 along the redox pathway impair the Fe–S redox switch in vitro and hypothesized that these mutations would compromise cell growth. Since we observe the formation of a putative [3Fe4S]+ species in the presence of DNA and NTPs electrochemically for the Y397 mutants, we were interested in potential differences between Y395 and Y397 tyrosine pathway mutants. Single-site mutations were introduced into the chromosomal gene encoding the p58 subunit of S. cerevisiae DNA primase (PRI2, ref. 34) at positions Y395 and Y397 under control of the endogenous promoter. We incorporated Y395F, Y395L, Y397F, or Y397L mutations into the yeast genome to investigate the biological effects of both mutations at different loci along the charge-transfer pathway and mutations to aromatic phenylalanine versus aliphatic leucine. As these redox-pathway mutations do not affect [4Fe4S] cluster loading or DNA binding of p58C in vitro, we could specifically assess the effects of the cluster redox switch on cellular fitness.
We investigated whether the Y395F and Y397F mutations in p58C, which retain some redox switching on DNA electrochemically (Fig. 2 and SI Appendix, Fig. S5), affect cellular fitness. Haploid strains expressing the pri2Y395F and pri2Y397F alleles were viable at 30 °C and grew comparably to the parental PRI2 strain (SI Appendix, Fig. S13). The Tyr → Phe mutants retain some redox-switching activity in the electrochemical scans, and the viability of cells is consistent with a partially but not completely inhibited redox pathway in these variants. Phenylalanine has an aromatic side chain, and although the ionization energy of this residue is higher than that of tyrosine (20), phenylalanine is still found disproportionately in oxidoreductase enzymes, suggesting that it can aid in redox pathways through protein (21).
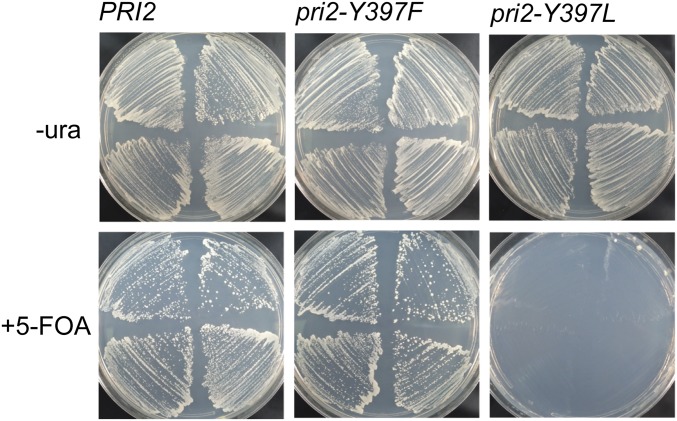
We next investigated the effects of the pri2Y395L and pri2Y397L mutations on yeast viability. As aliphatic leucine more strongly abrogates the redox pathway through p58C (Fig. 3 and SI Appendix, Fig. S7), we expected to observe more severe phenotypes with Tyr → Leu mutations in PRI2 than with Tyr → Phe mutations. We constructed a haploid pri2Y395L strain and observed, as with the Tyr → Phe variants, no growth defect relative to WT PRI2 strains. In contrast, we were unable to construct a haploid strain containing the pri2Y397L allele. To confirm lethality of pri2Y397L, we transformed the WT haploid strain with a URA3-marked PRI2 complementing plasmid and subsequently integrated either WT or mutant PRI2 alleles at the endogenous locus. Complemented strains expressing WT PRI2, pri2Y397F, or pri2Y397L grew well on media lacking uracil, as expected. However, when plated on media containing 5-fluoroorotic acid (5-FOA), only the pri2Y397L strain failed to generate colonies (Fig. 6). Since the protein product of the URA3 gene converts 5-FOA to toxic 5-fluorouracil, this result confirms that cells expressing the lethal pri2Y397L allele cannot tolerate loss of the URA3-marked PRI2 complementing plasmid. Phenotypes of the pri2Y395L and pri2Y397L mutants were confirmed by tetrad analysis (SI Appendix, Fig. S14A).
Fig. 6.
Yeast p58 (Pri2) Y397L mutation is lethal. Haploid yeast were transformed with a URA3-marked complementing plasmid expressing PRI2. The indicated PRI2 alleles (PRI2 WT, Y397F, or Y397L) were introduced by integration at the chromosomal locus (Materials and Methods). After verification of the genotype, individual transformants were streaked on media lacking (Upper) or containing (Lower) 5-FOA to select for cells that have lost the complementing plasmid.
While the pri2Y397L mutation has no discernible effect on the protein structure in vitro (Fig. 4), we wanted to confirm that the mutant is expressed and associates with the primase complex in yeast. If so, we would predict that the Y397L mutation in PRI2 impairs yeast growth when overexpressed in an otherwise WT background, competing with the normal protein for incorporation into the primase complex. Conditional overexpression of the Y397L variant, but not of WT PRI2, causes severe growth defects (SI Appendix, Fig. S14B). The Y397L mutation thus generates a p58 protein that can associate with primase, but is severely impaired in vivo.
The severe phenotype in the pri2Y397L strain is consistent with the effect of the Y397L mutation on p58C redox signaling activity (Figs. 3 and 5). This mutant displays minimal redox switching on DNA, and the signal in the presence of DNA and NTPs suggests that redox signaling leads to oxidative degradation of the cluster in p58C Y397L. Small-molecule reactive oxygen species such as hydrogen peroxide can damage the [4Fe4S] cluster directly, causing degradation to the [3Fe4S]+ form (30), or indirectly through DNA charge transport. We have observed, for example, that guanine radicals in oxidized duplex DNA induce redox switching in a bound [4Fe4S] Endonuclease III protein (35). WT p58C can more easily cycle between the [4Fe4S]2+ and [4Fe4S]3+ states than the tyrosine mutants when bound to both DNA and NTPs, mimicking the active primase. Y395F and Y395L variants appear to lose redox-switching activity, but are not oxidatively degrading on the DNA electrode. The Y397F and Y397L mutants, conversely, appear to be oxidized from the [4Fe4S]2+ state to the [4Fe4S]3+ state, but they are not easily reduced back to the [4Fe4S]2+ state, causing the reductive peak likely associated with the irreversible [3Fe4S]+/0 couple on DNA (31) to appear. We observe therefore that the combination of greater redox attenuation overall in Tyr → Leu variants (Fig. 3 and SI Appendix, Fig. S7), and the oxidative degradation observed in Y397F/L variants (Fig. 5), dysregulate primase activity to a point for which cellular machinery cannot compensate.
Discussion
Reversible, redox-driven switches control DNA binding affinity in [4Fe4S] repair and replication enzymes, facilitating rapid binding and dissociation (9, 25, 32). The oxidized [4Fe4S]3+ bacterial glycosylase Endonuclease III, for example, binds the DNA polyanion 550-fold more tightly than the reduced [4Fe4S]2+ Endonuclease III. Reversible cluster oxidation and reduction facilitates DNA-mediated redox signaling between Endonuclease III and other [4Fe4S] repair proteins in the first steps of locating oxidative DNA damage (32). The [4Fe4S] enzyme yeast DNA polymerase δ is DNA-bound and active in the [4Fe4S]2+ state, when associated with proliferating cell nuclear antigen (PCNA) (36). When oxidized to the [4Fe4S]3+ state, however, PCNA-associated polymerase δ binds DNA even more tightly, stalling replication (10). This change in binding may allow polymerase δ to sense and respond to oxidative stress. Similar redox switching chemistry thus regulates diverse, specialized DNA-processing [4Fe4S] enzymes.
Here we establish a redox switch, driven by a change in [4Fe4S] cluster oxidation state, that regulates DNA binding and redox signaling in eukaryotic DNA primase. This switch regulates primer synthesis, but not catalytic activity, in human primase (9). Structural and biochemical evidence suggests that primase adopts a compact configuration during activity, with both the p48 (RNA polymerase) subunit and the p58C [4Fe4S] domain contacting the ∼8–14-nt RNA/DNA duplex substrate (13, 18, 19). Redox pathways comprising conserved tyrosines spaced 10–15 Å apart, shuttle charge approximately ∼25–30 Å from the DNA binding interface to the [4Fe4S] cluster. Aromatic tyrosines spaced ≤15 Å apart can facilitate microsecond electron transfer (20, 21) in protein, possibly through formation of hopping intermediates. Despite previous arguments that structural differences in yeast and human primase preclude a general redox role for these residues (37), the electrochemical and biological data unequivocally demonstrate that the electron-transfer pathway is conserved. Electrochemical attenuation of the p58C redox signal on DNA and lethality in yeast due to a single-residue redox-pathway mutation suggest a more significant role than the contributions of these tyrosines to any network of p58C/substrate hydrogen bonds (13).
The primary redox pathway affecting the yeast p58C [4Fe4S] cluster redox switch likely involves Y395 and Y397. When these conserved residues are mutated, two possible events may occur. First, the mutant residue at position 395 or 397 may transfer charge through single-step tunneling, as opposed to electron hopping onto the tyrosine. Additionally, the mutation may route the electron through another primary hopping pathway, less efficient than the WT, within p58C. Multiple potential pathways through p58C, involving conserved residues Y431 and Y352 for example, are available to transfer charge between bound DNA and the cluster.
Our model for redox-driven primer handoff is compatible with the time scale of primer synthesis. Studies of calf thymus primase (38) suggest that primer synthesis is quite slow, with a first-order rate constant of 0.0027 s−1. Picosecond DNA-mediated electron transfer (39), microsecond electron transfer through primase/polymerase-α protein matrix (21), and microsecond/millisecond conformational changes in the polymerase-α active site (40) would all occur securely within this time. The aerobically measured binding constants of eukaryotic DNA primase and polymerase α (∼100 nM–1 μM) (9, 14, 41) interestingly correspond to the upper limit of the estimated primase/polymerase-α concentration in the yeast nucleus, based on protein copy number (42) and yeast cell volume (43) estimates. Tighter protein-DNA binding than these numbers indicate, which redox switching provides a means to access, may be necessary to efficiently execute replication.
Redox signaling in eukaryotic DNA primase is mediated by an aromatic pathway in the primase [4Fe4S] domain. Mutation of a conserved aromatic tyrosine at position 395 attenuates redox-switching activity driven by the primase [4Fe4S] cluster. Mutation of conserved tyrosine 397 attenuates redox switching and leads to oxidative degradation during redox signaling on DNA. The Y397L mutation abrogates the aromatic pathway and leads to cluster degradation during the redox switch, conferring lethality in yeast. Tyrosines 395 and 397 are not located near the primase catalytic site, yet a mutation at a position between the [4Fe4S] cluster and DNA-binding domain affects the regulatory redox switch and can prohibit cell growth. These observations support our proposal that the conserved redox chemistry of the [4Fe4S] cluster in DNA primase plays a central role in coordinating the initial steps of eukaryotic DNA priming.
Materials and Methods
DNA Modified Electrode Assembly/Preparation.
DNA and multiplexed chips were prepared and cleaned as described in SI Appendix. Assembled chips were transported into an anaerobic glove bag (Coy Products) and washed with deoxygenated protein storage buffer.
Sample Preparation for Electrochemistry.
Yeast p58C and tyrosine variants were overexpressed and purified as described in SI Appendix. Tyrosine mutagenesis and characterization of p58C variants is described in SI Appendix. Samples were buffer exchanged if necessary through a procedure detailed in SI Appendix. Samples were deposited onto multiplex chip quadrants, then bulk solution was added to a final volume of 200–300 μL.
WT/Mutant p58C Electrochemistry.
All electrochemistry was performed using a CHI620D potentiostat and 16-channel multiplexer (CH Instruments), in an anaerobic glove chamber. Details of measurements under specific conditions can be found in SI Appendix.
X-Ray Crystallography.
The protein was dialyzed into 20 mM Hepes (pH 6.8), 2 mM DTT, and 75 mM NaCl and concentrated to ∼5 mg/mL. Details of crystal formation and data collection/analysis can be found in SI Appendix.
Yeast Strain Construction.
PRI2 mutations were introduced into haploid strain YKF201 (MATa trp1 leu2 ura3 his7) by two-step gene replacement (44). Detail of this procedure and the procedure used to overexpress PRI2 alleles can be found in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to the NIH for Operating Grants R01 GM126904 (to J.K.B.), R35 GM40120 (to W.J.C.), and R01 GM123292 (to K.L.F.), and Training Grants T32 GM07616 (to E.O.) and T32 GM08230 (to L.E.S.). We received additional support from the Moore Foundation (J.K.B.) and a Ralph M. Parsons Fellowship (to E.O.). Diffraction data were collected at the Advanced Photon Source, operated by the US Department of Energy (Contract DE-AC02-06CH11357).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes: WT p58C (Saccharomyces cerevisiae), 6DI6; Y395F (S. cerevisiae), 6DTV; Y395L (S. cerevisiae), 6DU0; Y397F (S. cerevisiae), 6DTZ; and Y397L (S. cerevisiae), 6DI2].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810715115/-/DCSupplemental.
References
- 1.Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner BE, et al. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem. 2007;282:33444–33451. doi: 10.1074/jbc.M705826200. [DOI] [PubMed] [Google Scholar]
- 4.Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol. 2007;14:875–877. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuss JO, Tsai CL, Ishida JP, Tainer JA. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim Biophys Acta. 2015;1853:1253–1271. doi: 10.1016/j.bbamcr.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Netz DJA, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees DC, Howard JB. The interface between the biological and inorganic worlds: Iron-sulfur metalloclusters. Science. 2003;300:929–931. doi: 10.1126/science.1083075. [DOI] [PubMed] [Google Scholar]
- 8.Rouault TA. Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nat Rev Mol Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- 9.O’Brien E, et al. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science. 2017;355:eaag1789. doi: 10.1126/science.aag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartels PL, Stodola JL, Burgers PMJ, Barton JK. A redox role for the [4Fe4S] cluster of yeast DNA polymerase δ. J Am Chem Soc. 2017;139:18339–18348. doi: 10.1021/jacs.7b10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, Huang M. Essential role of the iron-sulfur cluster binding domain of the primase regulatory subunit Pri2 in DNA replication initiation. Protein Cell. 2015;6:194–210. doi: 10.1007/s13238-015-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Núñez-Ramírez R, et al. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res. 2011;39:8187–8199. doi: 10.1093/nar/gkr534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranovskiy AG, et al. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J Biol Chem. 2016;291:10006–10020. doi: 10.1074/jbc.M116.717405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauguet L, Klinge S, Perera RL, Maman JD, Pellegrini L. Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One. 2010;5:e10083. doi: 10.1371/journal.pone.0010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaithiyalingam S, Warren EM, Eichman BF, Chazin WJ. Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc Natl Acad Sci USA. 2010;107:13684–13689. doi: 10.1073/pnas.1002009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boal AK, et al. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry. 2005;44:8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 17.Sontz PA, Mui TP, Fuss JO, Tainer JA, Barton JK. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc Natl Acad Sci USA. 2012;109:1856–1861. doi: 10.1073/pnas.1120063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirk BW, Kuchta RD. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: Primase and the family X polymerases share significant sequence homology. Biochemistry. 1999;38:7727–7736. doi: 10.1021/bi990247c. [DOI] [PubMed] [Google Scholar]
- 19.Arezi B, Kirk BW, Copeland WC, Kuchta RD. Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: Implications for the role of the p58 subunit. Biochemistry. 1999;38:12899–12907. doi: 10.1021/bi9908991. [DOI] [PubMed] [Google Scholar]
- 20.Plekan O, Feyer V, Richter R, Coreno M, Prince KC. Valence photoionization and photofragmentation of aromatic amino acids. Mol Phys. 2008;106:1143–1153. [Google Scholar]
- 21.Gray HB, Winkler JR. Electron flow through metalloproteins. Biochim Biophys Acta. 2010;1797:1563–1572. doi: 10.1016/j.bbabio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Liang N, Pielak GJ, Mauk AG, Smith M, Hoffman BM. Yeast cytochrome c with phenylalanine or tyrosine at position 87 transfers electrons to (zinc cytochrome c peroxidase)+ at a rate ten thousand times that of the serine-87 or glycine-87 variants. Proc Natl Acad Sci USA. 1987;84:1249–1252. doi: 10.1073/pnas.84.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukacs A, Eker APM, Byrdin M, Brettel K, Vos MH. Electron hopping through the 15 A triple tryptophan molecular wire in DNA photolyase occurs within 30 ps. J Am Chem Soc. 2008;130:14394–14395. doi: 10.1021/ja805261m. [DOI] [PubMed] [Google Scholar]
- 24.Guergova-Kuras M, Boudreaux B, Joliot A, Joliot P, Redding K. Evidence for two active branches for electron transfer in photosystem I. Proc Natl Acad Sci USA. 2001;98:4437–4442. doi: 10.1073/pnas.081078898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartels PL, et al. Electrochemistry of the [4Fe4S] cluster in base excision repair proteins: Tuning the redox potential with DNA. Langmuir. 2017;33:2523–2530. doi: 10.1021/acs.langmuir.6b04581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boal AK, et al. Redox signaling between DNA repair proteins for efficient lesion detection. Proc Natl Acad Sci USA. 2009;106:15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. J Am Chem Soc. 2011;133:16378–16381. doi: 10.1021/ja207222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grodick MA, Segal HM, Zwang TJ, Barton JK. DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity. J Am Chem Soc. 2014;136:6470–6478. doi: 10.1021/ja501973c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osteryoung JG, Osteryoung RA. Square wave voltammetry. Anal Chem. 1985;57:101A–110A. [Google Scholar]
- 30.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 31.McDonnell KJ, et al. A human MUTYH variant linking colonic polyposis to redox degradation of the [4Fe4S]2+ cluster. Nat Chem. 2018;10:873–880. doi: 10.1038/s41557-018-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tse ECM, Zwang TJ, Barton JK. The oxidation state of [4Fe4S] clusters modulates the DNA-binding affinity of DNA repair proteins. J Am Chem Soc. 2017;139:12784–12792. doi: 10.1021/jacs.7b07230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francesconi S, et al. Mutations in conserved yeast DNA primase domains impair DNA replication in vivo. Proc Natl Acad Sci USA. 1991;88:3877–3881. doi: 10.1073/pnas.88.9.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foiani M, Santocanale C, Plevani P, Lucchini G. A single essential gene, PRI2, encodes the large subunit of DNA primase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3081–3087. doi: 10.1128/mcb.9.7.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yavin E, et al. Protein-DNA charge transport: Redox activation of a DNA repair protein by guanine radical. Proc Natl Acad Sci USA. 2005;102:3546–3551. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson E, Majka J, Burgers PMJ. Structure of DNA polymerase δ from Saccharomyces cerevisiae. J Biol Chem. 2001;276:43824–43828. doi: 10.1074/jbc.M108842200. [DOI] [PubMed] [Google Scholar]
- 37.Baranovskiy AG, et al. Comment on “The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport.”. Science. 2017;357:eaan2396. doi: 10.1126/science.aan2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheaff RJ, Kuchta RD. Mechanism of calf thymus DNA primase: Slow initiation, rapid polymerization, and intelligent termination. Biochemistry. 1993;32:3027–3037. doi: 10.1021/bi00063a014. [DOI] [PubMed] [Google Scholar]
- 39.O’Neill MA, Becker H-C, Wan C, Barton JK, Zewail AH. Ultrafast dynamics in DNA-mediated electron transfer: Base gating and the role of temperature. Angew Chem Int Ed Engl. 2003;42:5896–5900. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 40.Perera RL, et al. Mechanism for priming DNA synthesis by yeast DNA polymerase α. eLife. 2013;2:e00482. doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheaff R, Ilsley D, Kuchta R. Mechanism of DNA polymerase alpha inhibition by aphidicolin. Biochemistry. 1991;30:8590–8597. doi: 10.1021/bi00099a014. [DOI] [PubMed] [Google Scholar]
- 42.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 43.Moran U, Phillips R, Milo R. SnapShot: Key numbers in biology. Cell. 2010;141:1262–1262.e1. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 44.Ji H, Platts MH, Dharamsi LM, Friedman KL. Regulation of telomere length by an N-terminal region of the yeast telomerase reverse transcriptase. Mol Cell Biol. 2005;25:9103–9114. doi: 10.1128/MCB.25.20.9103-9114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.