Significance
The posttranslational modification of nucleosomes is implicated in the regulation of gene expression and chromatin packaging in all eukaryotes. In this study, we investigate the function of histone H4 lysine 16 (H4K16) and its acetylation in Drosophila by generating strains in which lysine 16 is mutated to arginine, glutamine, or alanine. The main conclusion of our paper is that even though H4K16 acetylation was reported to be a critical regulator of chromatin folding in vitro and has therefore been assumed to affect many different nuclear processes, its essential function in Drosophila is in one process: X-chromosome dosage compensation in males.
Keywords: chromatin, histone modification, H4K16 acetylation, dosage compensation complex, Drosophila
Abstract
Acetylation of histone H4 at lysine 16 (H4K16) modulates nucleosome–nucleosome interactions and directly affects nucleosome binding by certain proteins. In Drosophila, H4K16 acetylation by the dosage compensation complex subunit Mof is linked to increased transcription of genes on the single X chromosome in males. Here, we analyzed Drosophila containing different H4K16 mutations or lacking Mof protein. An H4K16A mutation causes embryonic lethality in both sexes, whereas an H4K16R mutation permits females to develop into adults but causes lethality in males. The acetyl-mimic mutation H4K16Q permits both females and males to develop into adults. Complementary analyses reveal that males lacking maternally deposited and zygotically expressed Mof protein arrest development during gastrulation, whereas females of the same genotype develop into adults. Together, this demonstrates the causative role of H4K16 acetylation by Mof for dosage compensation in Drosophila and uncovers a previously unrecognized requirement for this process already during the onset of zygotic gene transcription.
In eukaryotic chromosomes, the genomic DNA exists in the form of chromatin with the nucleosome as its basic building block. Nucleosomes consist of an octamer assembly of the four core histone proteins H2A, H2B, H3, and H4, around which 147 bp of DNA are wrapped in two-helical turns. Every nucleosome is separated from its neighboring nucleosomes by a stretch of linker DNA that, on average, is between 20 and 40 base pairs long. This organization of genomic DNA into arrays of nucleosomes affects almost all processes that occur on DNA. Consequently, a wide repertoire of proteins that modulate chromatin structure has evolved in eukaryotes. These proteins fall into two broad categories: those that make chromatin more accessible and those that make it more compact. At the molecular level, these proteins include factors that remodel nucleosomes, enzymes that add or remove posttranslational modifications on histone proteins, or proteins that bind to these modifications.
Among the different posttranslational histone modifications, acetylation of histone H4 at lysine 16 (H4K16ac) has long been of particular interest because of its roles in chromatin fiber folding, gene silencing in yeast, and dosage compensation in Drosophila. The importance of H4K16 in chromatin folding emerged from structural studies on nucleosomes that showed that a region of the N-terminal tail of H4 encompassing K16 interacts with the acidic patch formed by H2A and H2B residues on the octamer surface of a neighboring nucleosome (1, 2). This direct nucleosome–nucleosome contact is critical for compaction of nucleosome arrays in solution in vitro and, importantly, compaction is disrupted if H4K16 is acetylated (3, 4). In the case of gene silencing in yeast, the specific requirement for H4K16 was first uncovered through genetic studies that showed that yeast cells with H4K16 point mutations are viable and proliferate normally but show defective silencing at the mating type loci (5–7). Subsequent biochemical and structural studies then established that deacetylation of H4K16ac by the SIR protein silencing complex and binding of the complex to nucleosomes with deacetylated H4K16 are key for creating a transcriptionally inactive chromatin structure at the mating type loci and at telomeres (8, 9). In the case of dosage compensation in Drosophila, finally, it was the observation that H4K16ac is specifically enriched on the single X chromosome in males that first indicated that this modification might be linked to X-chromosome dosage compensation (10). Analyses of the dosage compensation regulatory proteins first identified through genetic approaches then revealed that the acetyltransferase Mof, a subunit of the dosage compensation complex (DCC), hyperacetylates H4K16 on the X chromosome in males, and this established a functional connection between H4K16ac and increased transcription of X-chromosomal genes (11–13). Taken together, these studies therefore shaped the view that nonacetylated H4K16 generates compact and transcriptionally repressed chromatin, whereas acetylation of H4K16 provides a mechanism for generating less compact and transcriptionally active chromatin. In both biological settings—that is, for silencing of mating type loci by SIR protein complexes in yeast and for X-chromosome dosage compensation by the DCC in Drosophila—it is the site-specific targeting of these protein complexes to the relevant genomic locations that provides the basis for localized deacetylation and acetylation of H4K16, respectively (reviewed in refs. 14 and 15). For example, while H4K16 acetylation in Drosophila is present at the 5′ transcription start sites of active genes on all chromosomes in both sexes, it is the targeted binding of the dosage compensation complex to X-linked genes in males that results in the specific enrichment of this modification on X-linked gene bodies in this sex (16).
What are the consequences of mutating H4K16 in higher organisms? Here, we generated Drosophila in which H4K16 was substituted by amino acids with different chemical properties. Depending on the amino acid substitution, these mutations cause embryonic lethality in both sexes, male-specific lethality, or permit the mutant animals to develop into adults. Importantly, these studies reveal a stringent requirement for H4K16 acetylation specifically in males but not in females, suggesting that dosage compensation is the main process that critically requires this modification. Moreover, we show that males that lack maternally deposited Mof protein arrest development during gastrulation, whereas females develop into adults. This suggests that H4K16 acetylation by Mof may be critical for a previously unexplained phase of dosage compensation reported to occur during the onset of zygotic gene transcription (17).
Results
H4K16 Point Mutants Reveal Essential Functions of This Residue in Metazoans.
We generated Drosophila in which H4K16 was mutated to arginine, glutamine, or alanine and shall refer to these animals as H4K16R, H4K16Q, and H4K16A mutants, respectively. Each of these amino acid substitutions changes the chemical character of residue 16 in H4 in a different way. In the H4K16R protein, the long aliphatic side chain and the positively charged head that are characteristic of lysine are retained but, unlike lysine, the arginine cannot be acetylated, and H4 therefore constitutively contains a positive charge at residue 16. In the H4K16Q protein, the long aliphatic side chain with a polar head group of glutamine has chemical properties similar to an acetylated lysine and can therefore be regarded as a constitutive acetyl-mimic substitution. In the H4K16A protein, finally, the substitution with the short apolar side chain of alanine causes the most drastic change to the chemical properties of the side chain at this position.
The following genetic strategy was used to produce H4K16R, H4K16Q, or H4K16A mutants. We generated animals that were homozygous for genomic deletions that remove all wild-type H4 gene copies and instead contained transgenes that expressed either wild-type or the above-mentioned H4 mutant proteins under the control of their native promoters. It is important to note that the Drosophila genome contains two types of histone H4 genes. First, there are the canonical histone H4 genes that are expressed during S phase, and these are all contained in a single large locus called HisC (18). HisC comprises 23 repeats of the so-called histone gene unit (HisGU), each of which harbors one copy of the genes for the core histones H2A, H2B, H3, H4, and for the linker histone H1. Second, the Drosophila genome also contains histone H4 replacement (His4r), a single copy gene that encodes an H4 protein with the same amino acid sequence as canonical H4 but is expressed also outside of S phase (19). Because H4 and His4r are identical proteins and therefore subject to the same modifications also at K16, it was essential to create strains with deletions lacking both the canonical H4 genes and the His4r gene. We first generated a His4r∆ deletion allele (SI Appendix, Fig. S1). His4r∆ homozygous animals were viable and fertile and could be maintained as a healthy strain. We next introduced the His4r∆ mutation into a strain carrying a deletion of the entire HisC cluster (HisC∆) (18) to generate HisC∆ His4r∆ mutant animals. Previous studies showed that HisC∆ homozygotes arrest development at the blastoderm stage after exhaustion of the pool of maternally deposited histones but that transgene cassettes providing 12 copies of a wild-type HisGU fragment (12xHisGU) rescue HisC∆ homozygotes into viable adults (18). We found that the wild-type 12xHisGU cassette also rescued HisC∆ His4r∆ homozygotes into viable adults (Fig. 1). Below, we shall refer to individuals with this genotype as H4wt animals (Fig. 1) because they served as control for the H4K16R, H4K16Q, and H4K16A mutant animals that we generated by introducing 12xHisGUH4K16R, 12xHisGUH4K16Q, or 12xHisGUH4K16A mutant cassettes, respectively, into HisC∆ His4r∆ homozygotes. Below, the phenotypes of these H4K16R, H4K16Q, or H4K16A mutant animals are discussed in turn.
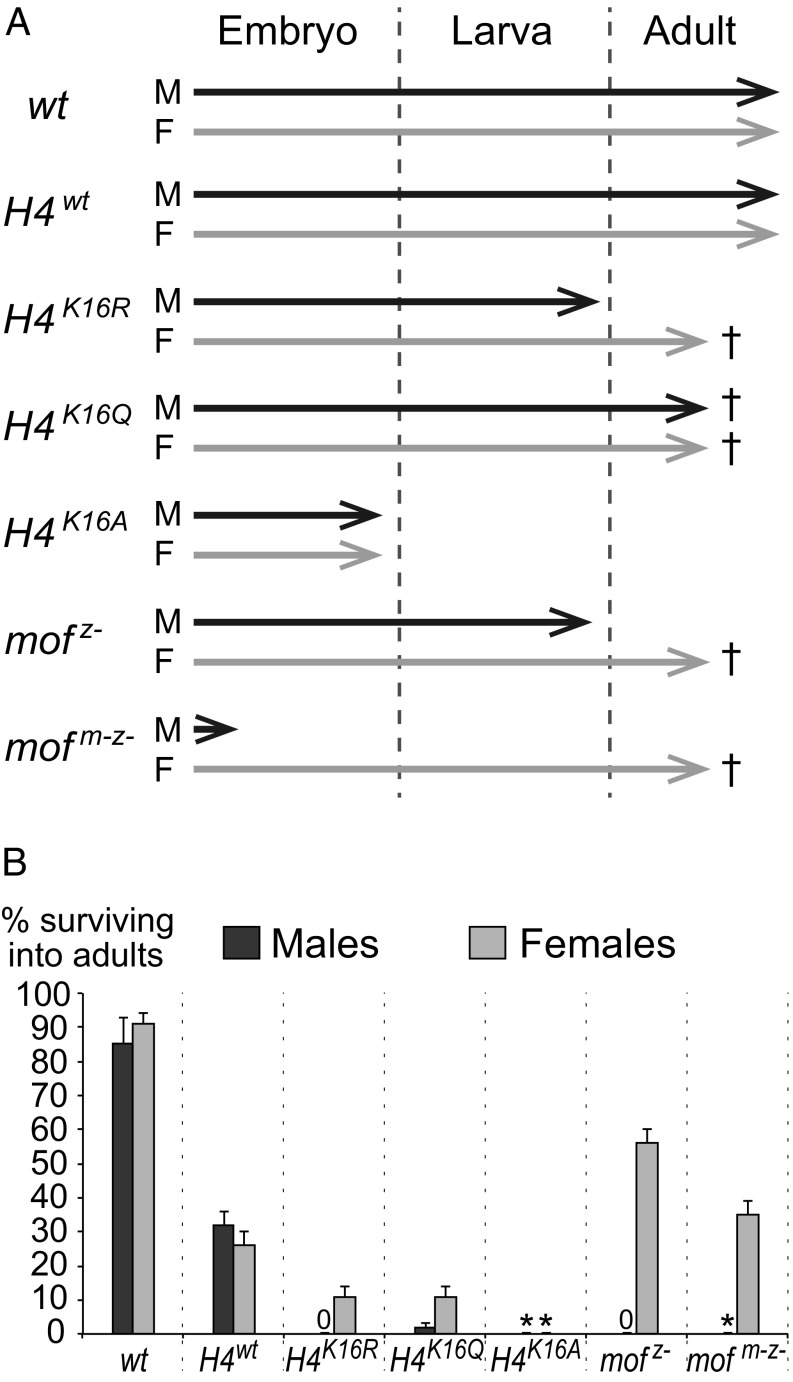
Fig. 1.
Differential viability of Drosophila with H4R16, H4Q16, or H4A16 chromatin support a specific role for H4K16 acetylation in dosage compensation. (A) Graphic representation illustrating the stage of developmental arrest of H4K16R, H4K16Q, H4K16A, and mof2 mutants. Detailed information about each genotype and how it was generated is presented in SI Appendix, Table S1. In the case of H4K16R mutants, males did not develop beyond the third larval instar. In the case of H4K16A mutants, males and females all arrest development at the end of embryogenesis and no larvae were obtained. In the case of mof2 m−z− mutants, only female and no male larvae were obtained; male embryos were analyzed as described in SI Appendix, Fig. S3. mof2 z− males develop to the end of the third larval instar, as previously reported (20). Crosses indicate that adults with these genotypes had a reduced life span and typically died within a few days after eclosion; moreover, these animals were generally weak and we therefore could not assess their fertility. (B) Quantitative analysis of the capacity of the different H4K16 and mof2 mutant genotypes to develop into viable adults. For every genotype that developed beyond embryogenesis, six independent batches with 50 first instar larvae were collected in each case and reared separately in vials. Histograms represent the mean ± SD of the percentages of larvae in individual vials that developed into viable adults. Asterisks show genotypes that arrest development during embryogenesis (A). The diminished viability of H4wt adult animals likely reflects incomplete rescue of HisC∆ His4r∆ homozygotes by the 12xHisGUwt transgene cassettes. It should be noted that H4wt adult males and females showed normal lifespan and were fertile.
H4K16R mutants showed sex-specific lethality. In particular, H4K16R mutant males survived up to the end of larval development and then died but, remarkably, their H4K16R mutant sibling females developed into adults that showed no obvious morphological defects but survived only up to 7 d after eclosion (Fig. 1, see figure legend for details). In females, chromatin that exclusively contains H4R16 nucleosomes therefore permits normal gene regulation and progression of development up to the adult stage. The lethality of males with H4R16 chromatin on the other hand suggests that this chromatin fails to support dosage compensation.
H4K16Q mutants differed from H4K16R mutants in that not only females but also males developed into adults (Fig. 1). While the fraction of surviving H4K16Q adult males was smaller compared with females, neither of the sexes showed any detectable morphological defects, but the animals survived only up to 7 d after eclosion (Fig. 1, see figure legend for details). Chromatin containing only H4Q16 nucleosomes is therefore also able to support normal gene regulation and progression of development.
H4K16A mutants, finally, were strikingly different in that both males and females arrested development by the end of embryogenesis (Fig. 1). The embryonic cuticles of these animals showed no obvious morphological defects and looked comparable to wild-type embryos (SI Appendix, Fig. S2).
Taken together, the comparison of these three different H4 mutants yields two main conclusions. First, the presence of an amino acid containing a long aliphatic side chain with a polar group at the end (i.e., either K, R, or Q) permits normal progression of development, whereas the short apolar side chain of alanine fails to do so. It may appear surprising that Drosophila in which all nucleosomes are positively charged (i.e., H4R16) or in which all nucleosomes are uncharged (i.e., H4Q16) both contain chromatin that is functional to support normal development. We note, however, that both H4K16R and H4K16Q mutant adults had a short lifespan (Fig. 1A), suggesting that presence of a lysine at this position is essential for normal adult viability. The second main conclusion comes from the finding that only H4Q16 chromatin supports the survival of males into adults. The most straightforward explanation of this result is that the glutamine substitution mimics acetylation of H4K16 and thereby permits dosage compensation to occur and allow at least a fraction of males to survive into adults.
H4K16 Acetylation by Mof Early in Embryogenesis Is Critical for Dosage Compensation in Males but Dispensable for Development in Females.
The sex-specific developmental defects of H4 mutants prompted us to compare their phenotypes with those of mof mutants. Previous studies had focused on the analysis of mof zygotic mutant animals (mof z−) (11, 20). These studies showed that mof z− males develop up to the late larval stage and arrest development shortly before pupation (20). Here, we investigated the phenotype of animals that lacked not only zygotically expressed but also maternally deposited Mof protein (Mof m−z−). Using mof2, a presumed null allele that lacks acetyltransferase activity (20, 21), we generated mof2 m−z− animals from females with mof2 mutant germ cells. Interestingly, mof2 m−z− mutant males arrested development shortly after gastrulation, whereas their mof2 m−z− mutant female siblings developed into viable adults (Fig. 1 and SI Appendix, Fig. S3 and Table S1). Like mof2 z− mutant females (16, 21), mof2 m−z− mutant females had a short lifespan and exhibited very low fertility (Fig. 1). We next generated mof2 m−z+ males that we obtained as the progeny from mothers with mof2 mutant germ cells and fathers that provided a wild-type copy of the mof+ gene (SI Appendix, Table S1). Such mof2 m−z+ males also failed to develop into adults. This shows that zygotic expression of Mof protein is insufficient to compensate for defects caused by the lack of maternally deposited Mof protein during the earliest stages of embryogenesis. In summary, these analyses reveal that, in males, H4K16 acetylation by Mof is critically required already during the onset of zygotic gene transcription and that in its absence, males are unable to develop past the gastrulation stage. Furthermore, these data demonstrate that H4K16 acetylation by Mof is fully dispensable for development of female zygotes into adults.
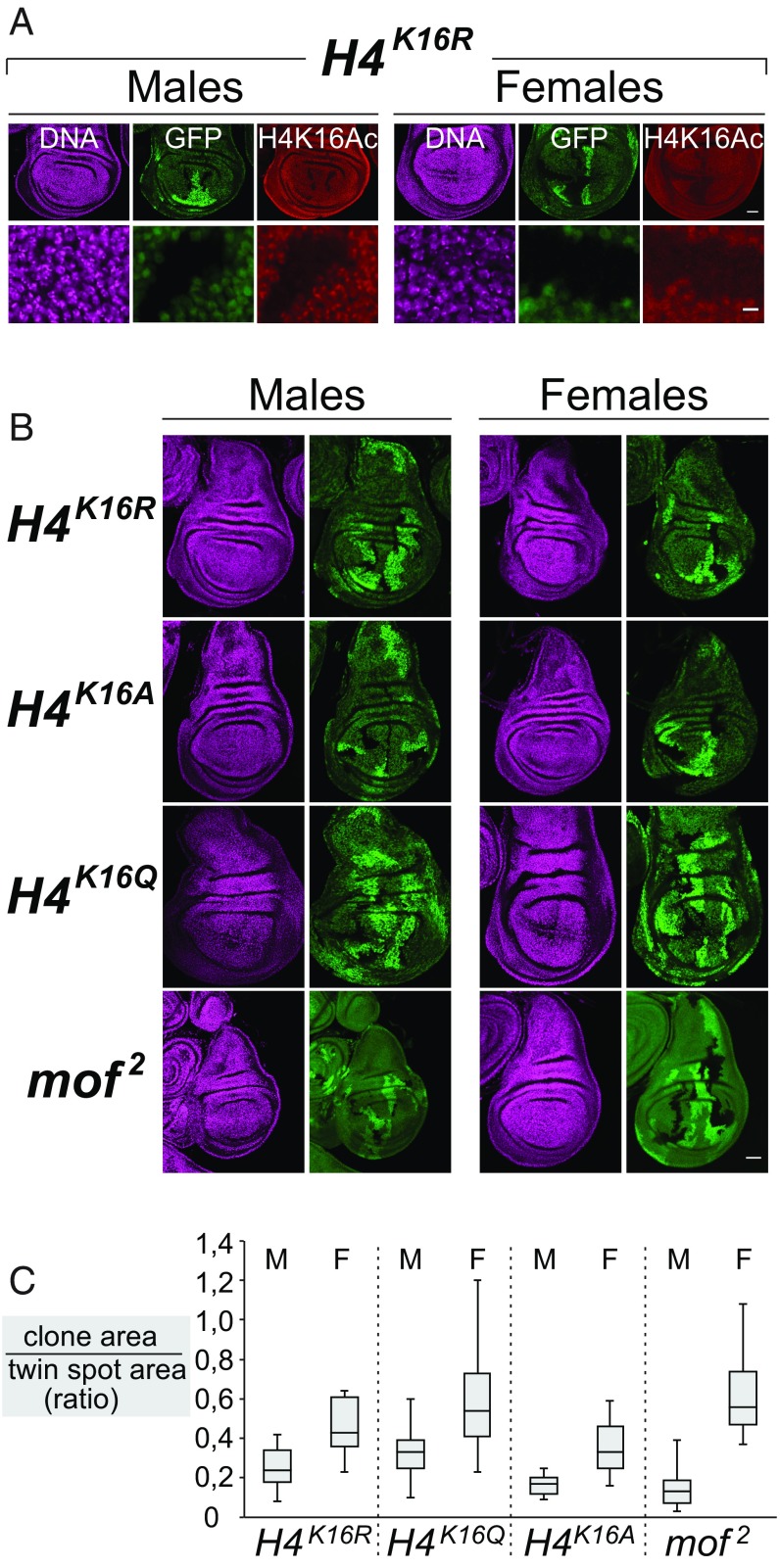
Is Mof the principal H4K16 acetyltransferase in Drosophila? Previous analyses in mof mutants led Conrad et al. (21) to propose that Mof creates most H4K16 acetylation in both males and females, whereas studies by Gelbart et al. (16) and Feller et al. (22) found that H4K16ac levels in mof mutant females are only about twofold reduced compared with wild type. Here, we analyzed imaginal discs with clones of mof2 mutant cells to directly compare H4K16ac levels in wild-type and mof2 mutant cells in the same tissue. While mof2 mutant cells in males showed a drastic reduction of H4K16ac signal, mof2 mutant cells in females still contained substantial levels of H4K16ac and the signal appeared only mildly reduced compared with wild-type cells (SI Appendix, Fig. S4A). Similarly, H4K16ac levels also appeared only mildly reduced in imaginal discs from mof2 m−z− mutant female larvae that had never contained any Mof activity (SI Appendix, Fig. S4B). These observations are consistent with the findings from Gelbart et al. (16) and Feller et al. (22) and support the view that a considerable fraction of total H4K16 acetylation in females is generated by acetyltransferases other than Mof. Considering that females with H4R16 chromatin that cannot be acetylated also develop into adults (Fig. 1), the role of this Mof-independent H4K16 acetylation in females currently remains enigmatic. We conclude that the essential function of H4K16 acetylation in Drosophila is for male-specific X-chromosome dosage compensation.
H4 Point Mutants Highlight Dosage Compensation as the Key Cellular Process Requiring K16 Acetylation.
To compare H4 and mof mutants at the cellular level, we generated clones of H4K16R, H4K16Q, H4K16A, or mof2 mutant cells in wild-type animals (Materials and Methods). We induced such clones in first instar larvae and analyzed them 96 h after induction in third instar larvae. Immunofluorescence labeling of imaginal discs with clones of H4K16R mutant cells showed that 96 h after clone induction, H4K16ac was no longer detectable in these mutant cells (Fig. 2A). However, the size of these H4K16R mutant clones in males was smaller than in females (Fig. 2B, first row and Fig. 2C). This is best quantified by comparing the size of the mutant clones relative to that of their “twin spot” control clones that were generated by the reciprocal recombination event (Fig. 2C). In males, H4K16R mutant clones were smaller than their twin spot control clones, whereas in females, H4K16R mutant clones and twin spot control clones were more similar in size (Fig. 2B, first row and Fig. 2C). Similarly, clones of H4K16Q, H4K16A, or mof2 mutant cells were also smaller in males compared with females (Fig. 2B, second, third, and fourth rows and Fig. 2C). No obvious sex-specific differences in clone size were observed in H4wt clones, consistent with the similar percentage of surviving H4wt males and females (Fig. 1). Interestingly, when we compared the clone sizes between males with different H4 mutant genotypes, we found that H4K16Q mutant clones were on average larger than H4K16R, H4K16A, or mof2 mutant clones (Fig. 2). The most straightforward explanation for these observations is that H4K16R, H4K16A, or mof2 mutant cells in males are more severely compromised for proliferation and/or survival because of defective dosage compensation. On the other hand, H4K16Q mutant cells, containing H4Q16 acetyl-mimic chromatin, proliferate and survive better because dosage compensation is sustained in these cells, in agreement with the observation that males consisting solely of H4K16Q mutant cells can survive into adults (Fig. 1). A final point worth noting is that, in females, H4K16A mutant cells proliferate to form large patches of mutant tissue (Fig. 2B, third row), suggesting that chromatin consisting entirely of H4A16 nucleosomes permits cells to proliferate for multiple generations.
Fig. 2.
Clones of cells with H4R16, H4Q16, or H4A16 chromatin show male-specific growth defects. (A) Imaginal wing disc from male (Left) and female (Right) larvae with clones of H4K16R mutant cells that are marked by the lack of GFP, stained with antibody against H4K16ac, and with Hoechst (DNA) to visualize nuclei. Clones were analyzed 96 h after clone induction. (Scale bar: 30 μm.) Lower row shows representative area of disc with wild-type and H4K16R mutant cells at higher magnification. (Scale bar: 5 μm.) In wild-type cells in males, the brightly labeled H4K16ac spot superimposed on the more uniform H4K16ac signal in every nucleus corresponds to the X-chromosome territory (10); wild-type nuclei in females only show the more uniform H4K16ac signal. Note that H4K16R mutant cells lack H4K16ac signal. (B) Imaginal wing discs from male (Left) and female (Right) larvae with clones of H4K16R (first row), H4K16Q (second row), H4K16A (third row), and mof2 (fourth row) mutant cells, in each case marked by the lack of GFP; Hoechst staining labels all nuclei. Clones were analyzed 96 h after induction. Note that in males, the mutant cell clones are significantly smaller compared with their juxtaposed brightly labeled GFP+/GFP+ twin spot clones induced by the reciprocal recombination event. This smaller size of the mutant cell clones is most notable in the case of H4K16R, H4K16A, and mof2, and less pronounced in the case of H4K16Q. (Scale bar: 30 μm.) (C) Quantitative analysis of the results shown in B, in box plot representation. For each genotype and sex, the total area of GFP-negative mutant clone tissue and of GFP+/GFP+ twin spot clone tissue was measured in individual imaginal discs using ImageJ. The ratio of total clone area to total twin spot area was calculated in individual discs, and for each genotype and sex, the results from 15 imaginal discs were plotted. These results are consistent with males being more sensitive than females to H4K16 mutations in all cases, but also with decreased viability of females with mutant H4 compared with wild type (Fig. 1).
In Males, Cells with H4K16 Mutant Chromatin or Lacking Mof Protein Differentiate to Form Tissues with Normal Morphology.
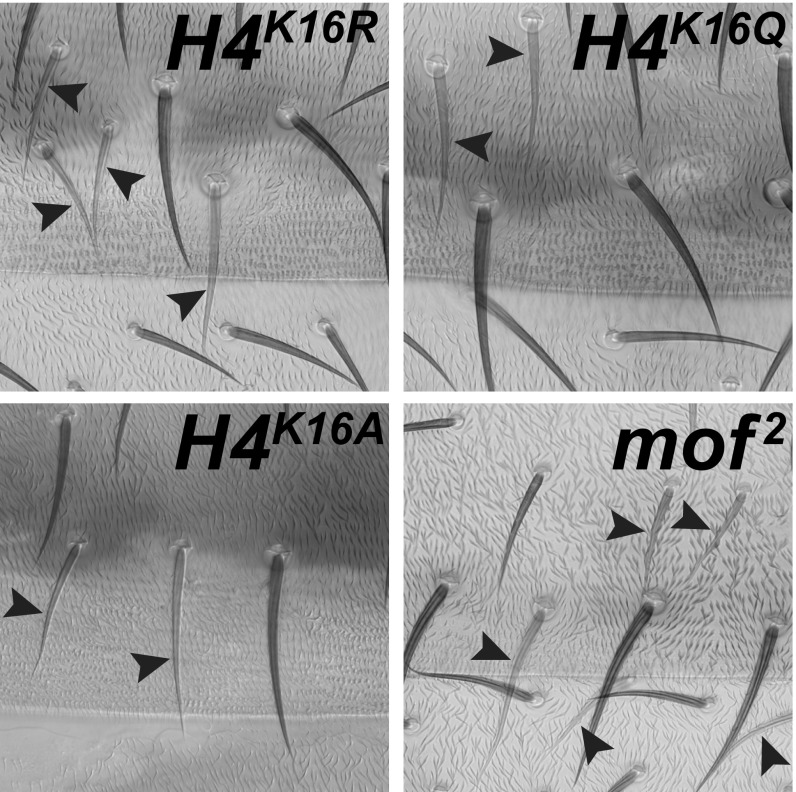
The results described above show that growth of H4K16R, H4K16A, and mof2 mutant cell clones in male larvae is impaired. We next investigated whether such mutant cells are able to differentiate. We induced clones of H4K16R, H4K16A, mof2, or also H4K16Q mutant cells in male larvae and tested whether they differentiate to form epithelial structures in adults. As shown in Fig. 3, mutant cells of all four genotypes formed epidermal structures with sensory bristles that looked indistinguishable from neighboring wild-type tissue. In conclusion, even though cells that contain H4R16 or H4A16 chromatin, or that lack the Mof acetyltransferase, are all impaired in proliferation in males, they nevertheless have the capacity to undergo differentiation and build tissues with apparent wild-type morphology.
Fig. 3.
In males, cells with H4R16 or H4A16 chromatin or lacking Mof protein retain the capacity to differentiate normally. Portions of abdominal segment A2 from adult males with clones of H4K16R, H4K16Q, H4K16A, or mof2 mutant cells. In each case, mutant clone cells are marked by the yellow mutation, and bristles in the mutant clone tissue therefore are more lightly pigmented than bristles in the neighboring wild-type tissue. Note that the morphology of the mutant bristles (arrowheads) and associated tissue is in all four cases comparable to the neighboring wild-type tissue. H4K16Q mutant clones were analyzed for comparison and as control, recall that males consisting solely of H4K16Q mutant cells can survive into adults with wild-type morphology (Fig. 1).
Discussion
Mutational analyses of histone amino acid residues that are subject to posttranslational modifications provide a direct approach for probing the physiological role of these residues and their modification. Here, we investigated the function of H4K16 and its acetylation in Drosophila by generating animals in which all nucleosomes in their chromatin were altered to constitutively carry a positively charged H4R16, an acetyl-mimic H4Q16, or a short apolar H4A16 substitution. These three types of chromatin changes have different physiological consequences that lead to the following main conclusions. First, H4R16 and H4Q16 chromatin both support development of female zygotes into adults. This suggests that, in females, modulation of H4K16 by acetylation is a priori not essential for the regulation of gene expression and the chromatin folding that occurs during development of the zygote. Second, unlike in females, only H4Q16 but not H4R16 chromatin supports development of male embryos into adults. This difference between males and females directly supports the critical role of H4K16 acetylation for dosage compensation in males. Third, cells with H4A16 chromatin are viable, proliferate, and can differentiate to form normal tissues in both males and females, but animals that entirely consist of cells with H4A16 chromatin arrest development at the end of embryogenesis. This lethality contrasts with the viability of animals with H4K16, H4R16, or H4Q16 chromatin and suggests that presence of a long aliphatic side chain with a polar group (i.e., either K, R, or Q) at residue 16 is more important for H4 function than the ability to regulate the charge of this residue by acetylation. A fourth main conclusion of this work comes from the finding that males that completely lack Mof protein (i.e., mof m−z− males) arrest development during gastrulation, whereas females of the same genotype develop into morphologically normal adults. This uncovers a previously unknown critical requirement of Mof acetyltransferase activity in males, already during the onset of zygotic gene transcription. In the following sections, we discuss the results reported here in the context of our current understanding of the role of H4K16 and its acetylation.
Differences in H4K16R, H4K16Q, and H4K16A Mutant Phenotype Severity in Flies Versus Yeast Reflect Organism-Specific Chromatin Functions of H4K16.
In yeast and flies, the comparison of the severities of the phenotypes caused by different amino acid substitutions at H4K16 highlights how the two organisms have evolved to use this conserved residue and its modification in different ways. In yeast, H4K16ac is present genome-wide and SIR silencing is the key physiological process that requires H4K16, in its deacetylated state (5–7). Yeast cells with H4K16R, H4K16Q, or H4K16A mutations are viable but they show defective SIR silencing. Silencing is much more strongly impaired in H4K16Q or H4K16A mutants than in H4K16R mutants (5–7). This is because SIR3 protein binding to deacetylated H4K16 (9), a prerequisite for silencing, is probably less severely impaired by the arginine substitution than by the alanine or glutamine substitutions. In Drosophila, the phenotypic differences between H4K16R, H4K16Q, and H4K16A mutants suggest that H4K16 is associated with two other, distinct physiological functions that are critical for the organism. The male-specific lethality of H4K16R mutants and the restoration of male viability in H4K16Q mutants demonstrate that dosage compensation is one essential process that critically requires the acetylated form of H4K16. A reduction of internucleosomal contacts by H4K16ac (3) to generate chromatin that is more conducive to gene transcription on the male X chromosome currently is the simplest mechanistic explanation for how H4K16 acetylation enables dosage compensation. The observation that an H4K16A mutation causes lethality in both sexes suggests that, unlike in yeast, a long aliphatic side chain at this residue is essential for H4 function in Drosophila. It is currently not known why Drosophila H4K16A mutants die. However, it is important to note that H4K16A mutant cells retain the capacity to proliferate and differentiate and the mutation therefore does not disrupt any fundamental process required for cell survival.
Mutation of His4r Is Critical for Assessing the Consequences of K16 Mutations in Canonical Histone H4.
Previous studies that investigated the function of histone H3 modifications by histone replacement genetics showed that for modifications associated with transcriptionally active chromatin it is essential to remove not only the wild-type copies of the canonical histone genes but to also mutate the histone H3.3 variants (23, 24). The analyses of H4K16R, H4K16Q, and H4K16A mutant phenotypes reported here were all performed in the genetic background of animals lacking His4r, the only histone H4 variant in Drosophila. Importantly, we found that in a His4r+ background, where only the canonical H4 proteins are replaced with mutant H4, the modifiable His4r protein permitted H4K16R His4r+ mutant males and, surprisingly, also H4K16A His4r+ mutant females and males to develop into adults. These animals were therefore not analyzed further. Supporting these observations, a recent study by Armstrong et al. (25) that used a similar strategy for replacing canonical histone H4 with H4K16R also found that H4K16R His4r+ mutant males develop into normal adults. This suggests that, like His3.3 (26, 27), the His4r protein might also preferentially be incorporated into transcriptionally active chromatin and become acetylated by Mof. Although the viable H4K16R His4r+ males have been reported to show a significant reduction of X-linked gene expression (25), a full assessment of transcriptional defects in animals containing only H4R16 nucleosomes would require that such molecular analyses be performed in H4K16R His4r∆ mutant males.
A final point that should be noted here is that during the early stages of embryogenesis, H4K16R, H4K16Q, or H4K16A mutants also still contain maternally deposited wild-type H4 protein that becomes incorporated into chromatin during the preblastoderm mitoses and only eventually becomes fully replaced by mutant H4 proteins during postblastoderm cell divisions. During the earliest stages of embryogenesis it has therefore not been possible to assess the phenotype of animals with chromatin containing exclusively H4R16, H4Q16, or H4A16 nucleosomes. This needs to be kept in mind when considering comparisons between the phenotypes of H4K16 point mutants and mof m–z– mutants.
Mof Acetyltransferase Activity in the Early Embryo Is Essential to Permit Male Development Beyond Gastrulation.
We found that males without Mof protein (i.e., mof m−z− males) arrest development during gastrulation while their female siblings develop into adults. Moreover, mof m–z+ males also fail to develop, demonstrating that zygotic expression of Mof protein is insufficient to rescue male embryos that lacked maternally deposited Mof protein. The most straightforward explanation for these observations is that H4K16 acetylation by Mof is critically required for hypertranscription of X-chromosomal genes that has been reported to occur already during the onset of zygotic gene transcription (17) and that the early developmental arrest of males is a direct consequence of failed dosage compensation.
How does this early requirement for Mof activity at the blastoderm stage relate to our current understanding of the temporal requirement for the DCC for dosage compensation? Previous studies showed that males lacking the DCC subunits Msl-1, Msl-2, Msl-3, or Mle complete embryogenesis and arrest development much later, around the stage of puparium formation (28). For example, Msl-1 protein null mutants (i.e., msl-1 m–z– mutants) die as late third instar larvae, yet Msl-1 directly interacts with Mof to incorporate it into the DCC (29) and is critical for targeting the complex and H4K16ac accumulation on the X chromosome in larvae (30, 31). One possible explanation for the conundrum that the lack of Mof but not that of Msl-1 or other DCC subunits results in lethality during gastrulation could be that during these early stages, H4K16 acetylation by Mof for dosage compensation is not as strictly dependent on the other DCC subunits as during later developmental stages, or that there is redundancy between Msl-1, Msl-2, or Msl-3 for targeting Mof to the X chromosome in the early embryo.
A final point worth noting is that Mof is also present in another protein assembly called the NSL complex (32). NSL was reported to act genome-wide for regulating housekeeping gene transcription in both sexes (33, 34) and several NSL subunits are essential for Drosophila viability (35). The finding that mof m–z– mutant females develop into morphologically normal adults shows that the NSL complex must exert regulatory functions that are essential for viability independently of Mof H4K16 acetyltransferase activity.
Concluding Remarks.
The acetylation of lysine residues in the N termini of histones is generally associated with chromatin that is conducive to gene transcription. Mutational studies in yeast showed that there is substantial functional redundancy between most of the different acetylated lysine residues in the N termini of histone H3 and H4 but that H4K16 has unique effects on transcriptional control (36), with well-defined phenotypic consequences (5–7). Here, we show that in Drosophila the principal function of H4K16 acetylation is X-chromosome dosage compensation in males.
Materials and Methods
Drosophila Stocks and Genetic Analyses.
Genotypes of fly strains and of the animals shown in each figure are listed in SI Appendix, Table S1. SI Appendix, Table S1 also contains information on how mof2 m–z– males and females were generated. Induction and analyses of mitotic cell clones in imaginal discs and in the adult epidermis were performed as described (37). Discs were stained with Hoechst and rabbit anti-H4K16ac antibody (Active Motif no. 39167).
Generation of Histone Transgenes.
Site-directed mutagenesis on pENTR221-HisGU.WT, pENTRL4R1-HisGU.WT, and pENTRR2L3-HisGU.WT (18) was used to convert the AAG codon for H4K16 to CGT (Arg), to CAG (Gln), or GCC (Ala). The final constructs ϕC31-attB-3xHisGU.H4K16R, ϕC31-attB-3xHisGU.H4K16Q, and ϕC31-attB-3xHisGU.H4K16A were generated by Gateway LR recombination of the above vectors. The transgene cassettes were integrated into the attP sites VK33, 68E, or 86Fb, as described (37). Standard meiotic recombination was then used to generate chromosomes carrying appropriate combinations of these transgene insertions and the His4rΔ deletion allele, as listed above.
Generation of His4r∆.
The His4rΔ deletion allele was generated by imprecise excision of the P{EPgy2}His4rEY06726 P-element; this resulted in a deletion from 3R:14,625,816–3R:14,626,410 (Berkeley Drosophila Genome Project, Release 6) that removes the entire His4r coding region.
Supplementary Material
Acknowledgments
We thank Frank Schnorrer for providing the M{attP}ZH-86Fb stock lacking the RFP marker gene and Shigeo Hayashi for the Y-GFP+ stock. This work was supported by the Max Planck Society (J.M.) and by NIH Grant GM126944 (to M.I.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817274115/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 3.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 4.Robinson PJJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: Essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 7.Park EC, Szostak JW. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 9.Armache K-J, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 11.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith ER, et al. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 14.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 15.Lucchesi JC, Kuroda MI. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 2015;7:a019398. doi: 10.1101/cshperspect.a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol. 2009;16:825–832. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lott SE, et al. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9:e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günesdogan U, Jäckle H, Herzig A. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 2010;11:772–776. doi: 10.1038/embor.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhmanova A, Miedema K, Hennig W. Identification and characterization of the Drosophila histone H4 replacement gene. FEBS Lett. 1996;388:219–222. doi: 10.1016/0014-5793(96)00551-0. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Szauter P, Lucchesi JC. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev Genet. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Conrad T, et al. The MOF chromobarrel domain controls genome-wide H4K16 acetylation and spreading of the MSL complex. Dev Cell. 2012;22:610–624. doi: 10.1016/j.devcel.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Feller C, Forné I, Imhof A, Becker PB. Global and specific responses of the histone acetylome to systematic perturbation. Mol Cell. 2015;57:559–571. doi: 10.1016/j.molcel.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Hödl M, Basler K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol. 2012;22:2253–2257. doi: 10.1016/j.cub.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Penke TJR, McKay DJ, Strahl BD, Matera AG, Duronio RJ. Functional redundancy of variant and canonical histone H3 lysine 9 modification in Drosophila. Genetics. 2018;208:229–244. doi: 10.1534/genetics.117.300480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong RL, et al. Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila. Genome Res. 2018;28:1688–1700. doi: 10.1101/gr.239913.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 27.Wirbelauer C, Bell O, Schübeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belote JM, Lucchesi JC. Male-specific lethal mutations of Drosophila melanogaster. Genetics. 1980;96:165–186. doi: 10.1093/genetics/96.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadlec J, et al. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol. 2011;18:142–149. doi: 10.1038/nsmb.1960. [DOI] [PubMed] [Google Scholar]
- 30.Bone JR, et al. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 31.Rastelli L, Richman R, Kuroda MI. The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech Dev. 1995;53:223–233. doi: 10.1016/0925-4773(95)00438-7. [DOI] [PubMed] [Google Scholar]
- 32.Mendjan S, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Feller C, et al. The MOF-containing NSL complex associates globally with housekeeping genes, but activates only a defined subset. Nucleic Acids Res. 2012;40:1509–1522. doi: 10.1093/nar/gkr869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam KC, et al. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8:e1002736. doi: 10.1371/journal.pgen.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raja SJ, et al. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell. 2010;38:827–841. doi: 10.1016/j.molcel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 36.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.