Significance
Skin pigmentation reflects strong local adaptation to latitude after humans migrated around the globe. In the Northern Hemisphere, the gene SLC24A5 plays a key role in the genetic basis of light skin pigmentation, where a nonsynonymous mutation in the gene has swept to fixation in contemporary Europeans. Although considered European-specific, we find this mutation at an unexpectedly high frequency in light-skinned KhoeSan from South Africa, far exceeding the European gene flow during colonial migration. Using haplotype analysis and comprehensive demographic modeling including positive selection, we show that this is an example of surprisingly strong adaptation of a recently introduced allele, via back-to-Africa migration, which occurred less than 2,000 y ago.
Keywords: pigmentation, adaptation, SLC24A5, KhoeSan, Africa
Abstract
Skin pigmentation is under strong directional selection in northern European and Asian populations. The indigenous KhoeSan populations of far southern Africa have lighter skin than other sub-Saharan African populations, potentially reflecting local adaptation to a region of Africa with reduced UV radiation. Here, we demonstrate that a canonical Eurasian skin pigmentation gene, SLC24A5, was introduced to southern Africa via recent migration and experienced strong adaptive evolution in the KhoeSan. To reconstruct the evolution of skin pigmentation, we collected phenotypes from over 400 ≠Khomani San and Nama individuals and high-throughput sequenced candidate pigmentation genes. The derived causal allele in SLC24A5, p.Ala111Thr, significantly lightens basal skin pigmentation in the KhoeSan and explains 8 to 15% of phenotypic variance in these populations. The frequency of this allele (33 to 53%) is far greater than expected from colonial period European gene flow; however, the most common derived haplotype is identical among European, eastern African, and KhoeSan individuals. Using four-population demographic simulations with selection, we show that the allele was introduced into the KhoeSan only 2,000 y ago via a back-to-Africa migration and then experienced a selective sweep (s = 0.04 to 0.05 in ≠Khomani and Nama). The SLC24A5 locus is both a rare example of intense, ongoing adaptation in very recent human history, as well as an adaptive gene flow at a pigmentation locus in humans.
Light skin pigmentation has evolved independently in Europeans and East Asians living at northern latitudes (1). Reduced eumelanin count and density facilitates UVB penetration (a subspectrum of UV light) of the skin to promote sufficient cutaneous synthesis of vitamin D (2). The KhoeSan of far southern Africa also possess relatively light skin compared with other sub-Saharan Africans, e.g., ∼50% lighter than equatorial Ghanaians (3, 4). The KhoeSan form the earliest divergence among modern human lineages, branching off from other populations as early as 100 kya to 150 kya (5–8). This raises the possibility that the light to intermediate skin of KhoeSan observed today reflects the ancestral state of the phenotype and light skin pigmentation in Eurasians evolved from standing genetic variation within southern Africa (9).
While the KhoeSan are highly divergent from all other populations, this does not mean they have been isolated completely for the past 100,000 y. Three distinct migrations into southern Africa during the last 2,500 y have impacted the distribution of the ethnicities in the region, and to varying degrees the genetic ancestry of the KhoeSan. The earliest migration is from an eastern African pastoralist population who brought sheep and goat, and possibly cattle, into southern Africa by 2,000 y ago (10, 11); this event is attested by archaeological remains of domestic caprids in Namibia and South Africa (12). Genetic data have not pinpointed the precise source of the gene flow, but the Ethiopian Amhara or another group who themselves are substantially admixed with Near Eastern people has been proposed (13, 14). The second major migration brought Bantu-speaking populations practicing agriculture into southern Africa from central/western Africa, where they are now the demographic majority. Bantu-speaking populations have dark skin pigmentation derived from their equatorial origin (15). The dating of Bantu expansion into southern Africa varies between 1,500 y and 500 y ago, depending on the precise location within southern Africa. Finally, the Dutch East India Company established a refreshment station at present-day Cape Town in 1652; subsequently, Dutch and other European colonists expanded northward from the Cape into the Karoo and southern Kalahari, particularly after 1740 (16).
We recently demonstrated that pigmentation variation in two KhoeSan populations from South Africa is associated with SLC24A5, by performing a genome-wide association analysis in ∼450 individuals quantitatively measured for eumelanin reflectance (ref. 3; see also ref. 17). Variation at rs1426654 is significantly associated with baseline skin pigmentation (P = 9.8e-9). This single locus explains 15.3% of the total variance of baseline skin pigmentation in KhoeSan (Materials and Methods). SLC24A5 is among the top signals in positive selection scans of Europeans and Ethiopians (5–8, 18, 19), and has long been recognized to play a role in pigmentation pathways. The nonsynonymous p.Ala111Thr mutation (G → A) at rs1426654 causes a melanin-reduced phenotype in zebrafish (20). In humans, the derived allele has swept to fixation or is present at high frequency in many European, Near Eastern, and South Asian populations (1, 21, 22); rs1426654 has also been associated with skin pigmentation differences in admixed populations who have recent European ancestors (i.e., African Americans) and within South Asians (1, 2, 23–26). In contrast, the absence of the derived allele in East Asians and most sub-Saharan African populations led to the hypothesis that this mutation originated in Europe, or, potentially, the Near East within the past 10 kya to 35 kya (Table 1) (1).
Table 1.
tMRCA estimates of rs1426654*A
| Source | Method | Haplotype sample size (N) | tMRCA,* kya |
| This study | ρ† | KhoeSan (357) | 32 (±7) |
| Homozygous KhoeSan (148) | 22 (±5) | ||
| CEU (198) | 15 (±3) | ||
| LWK (15) | 101 (±20) | ||
| This study | starmrca | KhoeSan (860) | 22 [17 to 28] |
| Homozygous KhoeSan (462) | 16 [12 to 20] | ||
| CEU (198)‡ | 13 [7 to 33] | ||
| LWK (198) | 27 [17 to 37] | ||
| Beleza et al. (21) | ABC (microsatellites) | Europeans | 11 [1 to 55] |
| Nakagome et al. (44) | ABC | Europeans | 35 [25 to 52] |
Generation time is assumed to be 30 y. Uncertainty in parameter estimates are indicated as (±SE) or [95% C.I.].
Only haplotypes that carry the derived allele are included.
LWK haplotypes that carry the ancestral allele at rs1426654 are used as background haplotypes.
starmrca, starmrca software package (Materials and Methods and ref. 28).
Results
Frequency Distribution of SLC24A5*A.
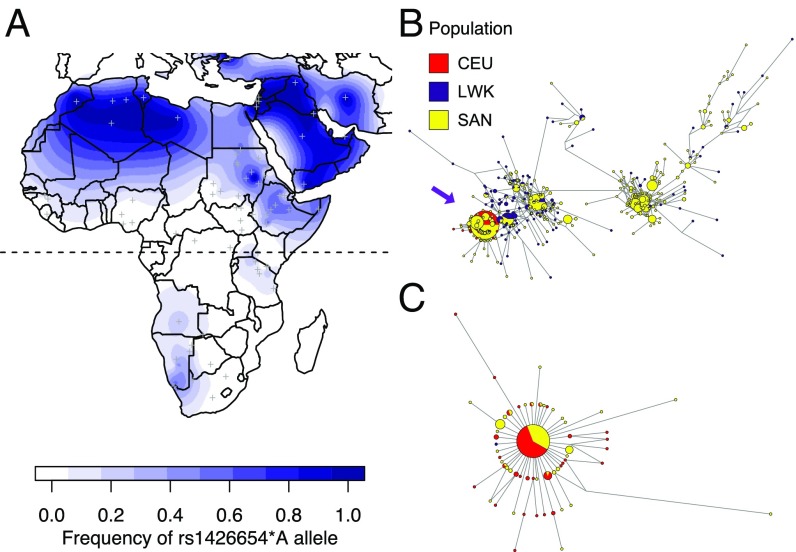
To reconstruct the evolutionary history of SLC24A5 in Africa, we sequenced the entirety of SLC24A5 (31.7 kb) to high coverage (>20×) from samples in two KhoeSan communities from South Africa: the ≠Khomani San (n = 269) and Nama (n = 161). Among the 430 individuals, the derived allele (rs1426654*A) is present at a high frequency of 32.5% in ≠Khomani and 53.5% in Nama. Queries of rs1426654 in publicly available datasets show that the allele is absent or at a very low frequency in other populations from western, central, and southern Africa (Fig. 1A and SI Appendix, Table S1). Other populations with a moderate to high frequency of the allele outside Europe have all experienced gene flow from the Near East or Europe during the Holocene, including those in northern and eastern Africa, and South Asia (17, 22).
Fig. 1.
Frequency of and haplotype structure of the SLC24A5 rs1426654 allele. (A) Frequency map of the derived rs1426654*A allele in Africa and the Near East. Asterisks denote the locations of the ≠Khomani San and Nama samples in this study. Other populations are represented by gray cross-hatches. Color gradients correspond to the binned frequency spectrum (SI Appendix, Table S1). The equator is indicated as the dashed line. (B and C) Haplotype networks were constructed by a median-joining algorithm. Colors denote Europeans (CEU, red), Bantu-speaking Luhya (LWK, purple), and KhoeSan (SAN, yellow). Node size is proportional to number of shared haplotypes, and branch length reflects number of mutations between nodes. Arrow points to the cluster of derived haplotypes. Haplotype network of SLC24A5 is for (B) all individuals carrying either the ancestral or derived allele and (C) individuals who are homozygous derived at rs1426654.
Haplotype Network of SLC24A5.
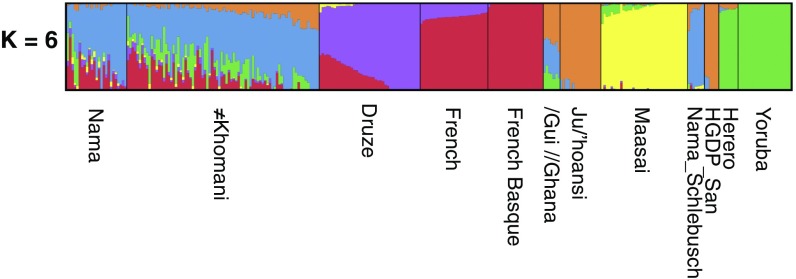
We examined the haplotype structure at SLC24A5 to determine whether the derived allele is identical by descent among populations. While historically isolated, the KhoeSan have received recent gene flow from other populations, including pastoralists from eastern Africa about 2 kya (10, 20, 23), Bantu-speaking agropastoralists from eastern Africa around 500 y ago, and Europeans (including Dutch, French, and German immigrants) about 300 y ago during the colonial era (27). The ≠Khomani San and Nama genomes we investigated derive, on average, 12% and 17%, respectively, of their ancestry from Europeans and 13% and 2%, respectively, from Bantu speakers (the remaining ancestry is primarily KhoeSan; Fig. 2). We obtained comparative SLC24A5 haplotypes from 99 individuals with Northern and Western European ancestry (CEU) and 99 Luhya Bantu speakers from Kenya (LWK) from the 1000 Genomes Project to represent potential source populations for the derived allele. A median-joining haplotype network based on all individuals exhibits extreme variation across the ancestral haplotypes, which are particularly diverse among the KhoeSan (Fig. 1B). However, haplotypes with the derived allele are primarily clustered together (SI Appendix, Fig. S1). We further restricted the network to haplotypes that are homozygous derived at rs1426654 (Fig. 1C), to minimize phasing uncertainty. The network of the homozygous derived haplotypes displays a starburst pattern, where most European and KhoeSan haplotypes in the central node are identical to each other across the entire 31-kb sequence, and other low-frequency haplotypes differ by only one or two mutations from the predominant haplotype.
Fig. 2.
Estimates of ancestry proportions per individual. Ancestry proportions are inferred from ADMIXTURE at k = 6, plotted for the major mode and displaying unrelated individuals common across the 19 running groups; see Materials and Methods. Estimates are obtained with 316,820 genome-wide biallelic markers.
Estimate of Time to the Most Recent Common Ancestor.
Based on the remarkable haplotype similarity among individuals carrying the derived allele across Europe, eastern Africa, and southern Africa, we estimated the age of the derived allele within each of these populations. We dated the time to the most recent common ancestor (tMRCA) of rs1426654*A via two different methods (Table 1). First, we calculated the tMRCA from the haplotype network-based ρ statistic (Materials and Method). We estimated an age of 32 kya (SE is ±7 kya) for all derived haplotypes in KhoeSan. We then restricted the analysis to haplotypes from homozygous derived individuals to avoid possible phase error, which can artificially increase allele age estimates by integrating ancestral sequence onto derived haplotypes. This approach resulted in a more recent tMRCA of 23 kya (±5 kya) for rs1426654*A. European haplotypes have a tMRCA of 15 kya (±3 kya). In contrast, the estimate from the 15 Luhya derived haplotypes was as old as 101 kya (±20 kya).
To explicitly account for recombination between the haplotypes carrying the ancestral allele at rs1426654 (background haplotypes) and derived haplotypes, we adopted a hidden Markov model that takes local recombination into account (SI Appendix, Fig. S3) and is valid for selective sweeps (28). Using both derived and background haplotypes in KhoeSan, we estimated a tMRCA of 22 kya (95% CI: 17 kya to 28 kya) for all individuals and 16 kya (95% CI: 12 kya to 20 kya) for individuals with the AA homozygote. In comparison, Europeans have a slightly younger tMRCA = 13 kya (using background haplotypes from LWK), although the CI range largely overlaps with the KhoeSan (95% CI: 7 kya to 33 kya). The Luhya have the oldest tMRCA = 27 kya (95% CI: 17 kya to 37 kya).
Evidence for Positive Selection.
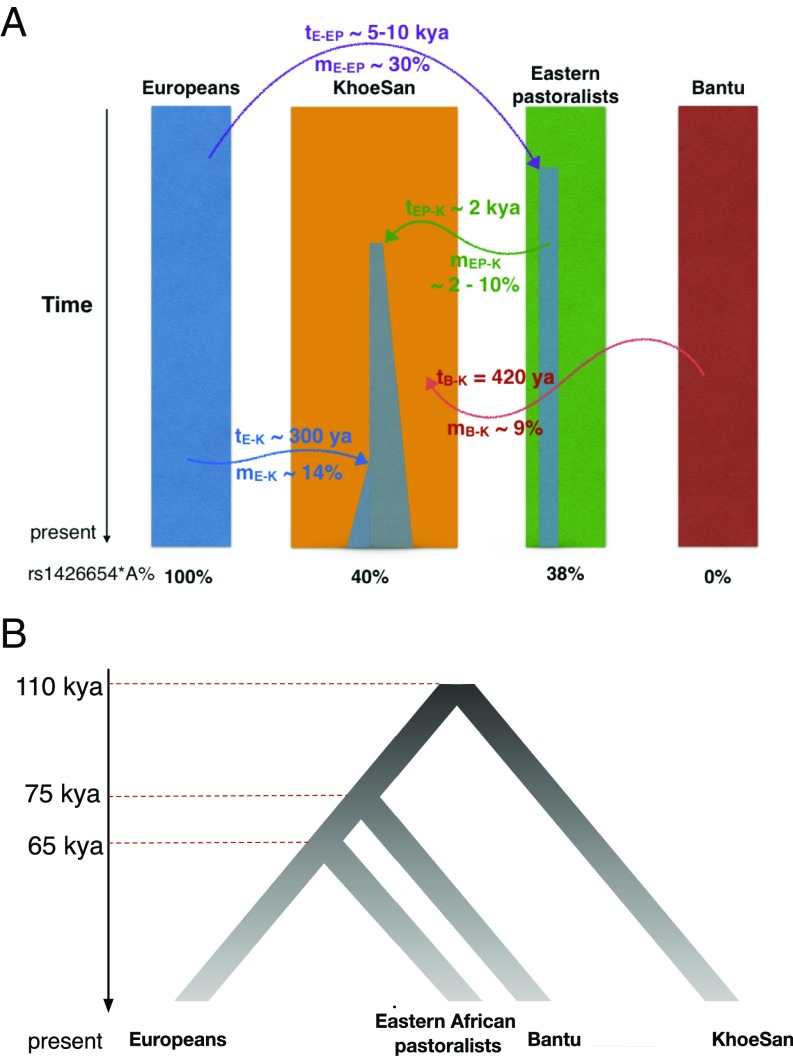
The strong resemblance between the KhoeSan and European haplotypes carrying the derived rs1426654*A allele suggests a common origin. However, the tMRCA estimate for the allele is far younger in both the KhoeSan and Europeans than the estimate from the population divergence between them (Fig. 3). We therefore hypothesized that rs1426654*A allele in the KhoeSan derives from a migration event. Apart from the direct European gene flow to KhoeSan during colonial time, genetic and archeological evidence indicates that the eastern Africans also had contact with ancient west Eurasians from the Near East before their migration to southern Africa, which left detectable traces in current eastern African genomes (13, 29, 30) (SI Appendix, Supplementary Text). Current eastern African populations, such as the pastoralist Maasai from Kenya, carry this allele at 30 to 40% (Fig. 1). Therefore, based on known population history, the two most likely sources of the derived rs1426654*A in KhoeSan are (i) very recent European gene flow within the last 300 y or (ii) eastern African pastoralists who introduced domestic stock into southern Africa within the past 2 ky.
Fig. 3.
Simulation of KhoeSan demographic history with positive selection. (A) Demographic schematic to introduce the derived SLC24A5 into the KhoeSan under a complex four-population model. An alternative model assumes no contribution from eastern African pastoralists (SI Appendix, Fig. S5). Relative timing and migration rates are noted as t and m, and their footnotes denote the initials of population: E, Europeans; EP, Eastern Pastoralists; B, Bantu; and K, KhoeSan (SI Appendix, Table S2). For simplicity, the frequency in Luhya is set to 0, as they carry a low frequency of the derived allele, which introduces a negligible number of derived copies during migration. (B) Divergence among populations that have contributed gene flow to KhoeSan.
While eastern African ancestry has been detected across several different Khoe and San populations, estimates of this ancestry in present-day individuals rarely exceed 10%. On average, eastern African ancestry in our dataset was 4%. Additionally, the average European admixture is 12 to 17% in the ≠Khomani San and Nama. Using these estimates of global ancestry and the frequency of the allele in the source populations, we would predict a frequency of ∼15% for rs1426654*A under a model of migration and neutral evolution. The observed frequency of 33 to 53% in the ≠Khomani San and Nama is much higher than that expected from admixture alone. This discrepancy between global ancestry and allele frequency, together with the starburst pattern in network of the derived haplotypes in KhoeSan, led us to test a hypothesis of positive selection. Assuming a deterministic model with infinite population size (31), we first estimated the distribution of selection coefficients given the present-day allele frequency for a range of starting allele frequencies and migration times (Fig. 4 and SI Appendix, Fig. S4). If we assume that Europeans introduced the allele during the colonial era 300 y ago and with present European admixture proportions, the selection coefficient would need to be s ≈ 0.16 to reach current allele frequencies, which is unrealistically high—higher than prior positive selection coefficient estimates for most phenotypes (except for infectious diseases) (21, 32–34). If we assume that the allele was introduced from eastern African pastoralists 2 kya, the initial frequency would be low (given that the allele is not fixed in eastern Africa and KhoeSan groups retain little ancestry from this event). Even under this model, however, the selection coefficient would largely range from s ≈ 0.05 to 0.1, assuming a range of East African ancestry between 4% and 10% and rs1426654*A of 30 to 50% in the migrating population (Fig. 4).
Fig. 4.
Estimates of the selection coefficient associated with rs1426654 are exceptionally strong. (A) The contour plot reflects the expected selection coefficient of rs1426654*A under a deterministic model, given a combination of the time of migration (in generations, g) at which the derived allele was introduced (x axis) and the initial allele frequency in the migrants (y axis), and taking the current allele frequency to be 53.5% (i.e., observed in the Nama). (Inset) The selection coefficients under a European era introduction. (B) The posterior distribution of s under East African Source model after ABC.
Full Demographic Model via ABC Analysis with Coalescent Simulations.
The deterministic model in Evidence for Positive Selection, while suggestive of extremely strong recent selection, does not incorporate the effect of genetic drift. We therefore performed model selection on our two alternate hypotheses for source of the SLC24A5 allele by simulating detailed three- or four-population demographic scenarios with selection using realistic effective population sizes (Ne; Fig. 3A and SI Appendix, Fig. S5 and Table S2). The first model (“European Source”) considers recent European gene flow as the initial source of the derived allele, which was followed by positive selection in the KhoeSan over the past 300 y. The second model (“East African Source”) assumes that the allele derives initially from eastern African pastoralists 1 kya to 2 kya, with a second pulse of recent European admixture during the past 300 y; positive selection occurs as soon as the allele arrives in the KhoeSan. We tested the ≠Khomani San and Nama separately to account for fine-scale population structure and the difference in rs1426654*A frequency.
Our approximate Bayesian computation (ABC) simulation approach operates as follows (Materials and Methods): After sampling selection coefficients from a prior, we generate forward-in-time allele frequency trajectories for the focal allele, and we only accept a trajectory if the final simulated allele frequency matches the observed frequency in the Nama or ≠Khomani San. Next, we perform coalescent simulations conditional on a forward allele trajectory using mssel. Finally, we conduct model selection based on three summary statistics (Materials and Methods).
The ABC analysis rejects neutral scenarios (SI Appendix, Supplementary Text and Table S3), consistent with positive selection as suggested from the deterministic model. Further, ABC strongly supports the East African Source model over the European Source model for both populations: ≠Khomani San (posterior probability: 0.876 vs. 0.124, respectively) and Nama (posterior probability: 0.801 vs. 0.199). Posterior predictive checks confirmed that the East African Source is a good fit for the observed data (SI Appendix, Fig. S6). As part of our model selection, we also obtain the posterior distributions of selection coefficients consistent with present-day allele frequencies (Fig. 4B). The estimates on the focal allele in both populations are very strong, with a posterior mode of s = 0.04 (95% CR: 0.03 to 0.12) in the ≠Khomani and s = 0.05 (95% CR: 0.04 to 0.14) in the Nama (SI Appendix, Fig. S7). The posterior selection coefficients obtained under the European Source model are also much higher: s = 0.26 to 0.31 (95% CR: 0.23 to 0.35), and are unlikely to represent a reasonable evolutionary scenario.
We further explored demographic parameters associated with the East African Source model. The modal posterior estimate of the eastern African pastoralist migration time (t) is 990 y ago (95% CR: 930 to 2,910) in the ≠Khomani and 930 y ago (95% CR: 930 to 2,940) in the Nama. The migration rate (m) estimates of eastern African pastoralists remain relatively low: 0.03 (95% CR: 0.02 to 0.1) for both the ≠Khomani and the Nama. The highest probability from the joint posterior distribution for t and m is around 1,200 y ago with a migration rate of 0.03 in both KhoeSan communities, although the posterior surface for t is relatively flat (SI Appendix, Fig. S8).
Discussion
Here, we have demonstrated that the derived rs1426654*A in SLC24A5 in the KhoeSan has a common origin with Europeans, reflected by the similar tMRCA estimates and identical modal haplotypes. This is consistent with the extended haplotype similarity observed between San from Botswana and Europeans (17). We asked whether the common origin of rs1426654*A haplotype could be due to recent migration between Europe and South Africa, or via an indirect route of migration through eastern Africa. A pastoralist migration from eastern Africa to southern Africa 2 kya is well documented from human genetic and archaeological data, and is likely associated with the introduction of domestic sheep and goats into the region. The summary statistics in our demographically explicit ABC approach clearly discriminate between the East African Source and European Source models, and strongly favor the East African Source model (Materials and Methods and SI Appendix, Supplementary Text). We cannot speculate as to the precise geographic origin of this allele, due to the limited sequencing data from eastern African and Near Eastern populations.
Additionally, the allele frequency of rs1426654*A in the KhoeSan exceeds the expected frequency under a neutral, migration-based scenario. Both deterministic modeling and simulation of complex demography suggest strikingly strong positive selection on this allele in far southern Africa (s = 0.04 to 0.05), similar to the strength of its counterpart in Europe at high northern latitudes [s = 0.08 under an additive model (21)]. The modal values of the selection coefficients from our modeling are primarily shaped by the current allele frequency, with minor updates to s from the ABC (SI Appendix, Fig. S7). Our selection coefficients are comparable to or exceed other well-known examples of positive selection, including lactase persistence in Europeans [s = 0.012 (32)] and eastern Africans [s = 0.035 to 0.097 (33)], malaria resistance [s = 0.02 to 0.20 (34)], and other pigmentation genes in Europeans [s = 0.02 to 0.04 (21)]. Furthermore, the selective sweep, albeit incomplete, appears to have occurred in fewer than 1,500 y, making it one of the few examples of selection during very recent human history.
We have shown that the phenotypic consequences of rs1426654*A have a large effect on the pigmentation of present-day KhoeSan; individuals who carry the derived homozygote are 14% lighter than the population average. The biological or cultural advantage of the lighter pigmentation phenotype remains to be tested in southern Africa. The southern tip of Africa receives relatively lower ultraviolet radiation intensity (35), which would serve as the major driving force of selection on the lighter skin pigmentation of the local populations. Additional selection force may have come from a possible diet shift about 2,000 y ago as evidenced in the Cape and coastal regions, as Khoekhoe populations transitioned from vitamin D-enriched marine fish and terrestrial food, likely including animal liver, to pastoralism (36–38). This decrease of dietary intake of vitamin D could have accelerated depigmentation for more photosynthesis of vitamin D, to promote calcium absorption, bone formation and innate immunity. However, as the ≠Khomani San live in the southern Kalahari and many individuals were still practicing a hunter-gatherer subsistence 100 y ago, it is unclear how the transition to pastoralism might have impacted their ancestors. It is particularly interesting to note that the model of evolution at SLC24A5 is remarkably similar to selection for the eastern African lactase persistence allele in the Nama (11). An alternative hypothesis is that sexual selection for light skin pigmentation drove the allele frequency. We find the third hypothesis unlikely, as this sexual preference would make most sense under the European colonialist regime, but we find that that selection most likely began before European arrival (East Africa Source model; SI Appendix). While the biological cause of the selective event merits further investigation, we have demonstrated an unusually rapid case of selection for lighter skin pigmentation based on a recently introduced allele <2,000 y ago, the first case of pigmentation adaptation from migration in humans.
Materials and Methods
Ethics Statement and Sample Collection.
As described previously (3, 9, 27), sampling of the ≠Khomani San took place in the Northern Cape of South Africa in the southern Kalahari Desert region (near Upington and neighboring villages) in 2006, 2010, 2011, 2013, and 2015. Sampling of the Nama took place in the Richtersveld in 2014 and 2015. Institutional review board (IRB) approval was obtained from Stanford University, Stony Brook University, and the University of Stellenbosch, South Africa. The ≠Khomani San N|u-speaking individuals, Nama individuals, local community leaders, traditional leaders, nonprofit organizations, and a legal counselor were all consulted regarding the aims of the research before collection of DNA (9). Research was conducted with the permission of the Working Group of Indigenous Minorities in Southern Africa and, subsequently, the South African San Council. All individuals gave signed written and verbal consent, with a witness present, before participating. Individuals collected in 2006 were reconsented under an updated protocol. Ethnographic interviews of all individuals were conducted, including questions about age, language, place of birth, and ethnic group of the individual and of his/her mother, maternal grandparents, father, and paternal grandparents. DNA was obtained via saliva, collected using Oragene saliva collection kits (DNAGenotek).
Sequencing of SLC24A5 Region and Variant Calling.
We captured the entirety of SLC24A5 (31.7 kb) for 453 individuals from two KhoeSan communities: the ≠Khomani San (n = 269) and Nama (n = 184). As described in ref. 3, the complete 31.7 kb of SLC24A5 (chr15:48403169 to 48434869 based on GRCh37) was enriched by using NimbleGen SeqCap EZ Choice Enrichment Kit, then sequenced with Illumina NextSeq. Sequenced data were then processed through a standard pipeline informed by the 1000 Genome Project. Briefly, we aligned reads to the hg19 reference genome using bwa-mem 0.7.10. We then sorted bam files and marked duplicate reads with Picard v1.92. We next ran RealignerTargetCreator, IndelRealigner, BaseRecalibrator, PrintReads, HaplotypeCaller, GenotypeGVCFs, and VariantRecalibrator, and ApplyRecalibration with GATK (v3.2.2). Quality control of the sequence is described in SI Appendix, SI Materials and Methods.
Phasing Haplotypes and Its Visualization.
We used 99 samples of unrelated Utah residents with northern and western European ancestry (CEU) and 99 samples of unrelated Luhya in Webuye, Kenya (LWK) from 1000 Genome Project, after the variants were recalled together during the targeted sequencing step, and pooled with 453 KhoeSan samples for the 31.7-kb region. We then phased the region from the three populations using SHAPEIT2 (v2.r778) (39). Pedigrees from both KhoeSan communities, inferred from the self-reported ethnographic information, were added to improve phasing.
We filled in ancestral/derived allele information at each locus based on the 1000 Genomes Project. Visualization of haplotypes, annotated with ancestral and derived allele at each position, was plotted using R package Adegenet (40).
Haplotype Network Construction.
We included haplotypes of 99 CEU, 99 LWK from the phasing step, and 430 KhoeSan individuals who do not carry Damara or Bosluis Baster ancestries as indicated in their ethnographic report. The networks of all samples, haplotypes carrying the derived allele, and samples that are homozygous derived in each population separately and together were constructed under a median-joining multistate algorithm using NETWORK 4.5 (41) (www.fluxus-engineering.com/sharenet.htm).
Age Estimate of tMRCA Using Rho.
Rho (ρ), the average number of sites differing between a set of haplotypes and their specified common ancestor (42), was calculated in NETWORK 4.5. Assuming no natural selection involved, a ρ-based age estimate of the common ancestor of a haplotype network follows the equation
| [1] |
where μ is mutation rate per locus per generation, L is the haplotype length, and g is the generation time (30 y).
Age Estimate of tMRCA of the Derived Allele.
To test the ancestral haplotype carrying the derived allele and the timing of its appearance, we adopted a hidden Markov model that exploits patterns of recombination between selected and background haplotypes, implemented in R package startmrca (28). We calculated a uniform recombination rate of SLC24A5 region to be 6e-10 per site per generation, using an African American recombination map (43). The mutation rate is 2e-8 per site per generation, with a generation time of 30 y. The Monte Carlo Markov chain was set to run for 5,000 iterations, with a maximum number of individuals to include as the selected and reference panels to be 100 and 40 each time. Replicate chains were run five times, and the one with the highest posterior probability was reported as the age estimate.
To control for phasing errors, we compared estimates from all 430 KhoeSan individuals with that on a subset of 231 KhoeSan that are homozygous at rs1426654 (including 74 homozygous derived, 157 homozygous ancestral). We also tested 99 Luhya individuals from 1000 Genome project phased with KhoeSan together as a separate run, further excluding one Luhya (NA19404) whose haplotype appeared to have phasing errors as shown in the network. We then tested 99 CEU individuals, together with 85 Luhya who carry homozygous ancestral haplotypes as the reference panel (where the regional recombination rate was calculated from DECODE project).
Estimation of Selection Coefficient Under a Deterministic Model.
We used the model proposed by Ohta and Kimura (31) and similarly applied in Breton et al. (11). It assumes a large selective advantage of the beneficial allele in a randomly mating diploid population, and where the frequency of the focal allele is not extreme (i.e., close to 0 or 1). The frequency at a given generation fits the equation
| [2] |
where is the frequency of the beneficial allele at generation t, is the initial frequency when selection starts (presumably here after the initial migration), and s is the selection coefficient of the allele. Rearranging [2], we get
| [3] |
Variance of Baseline Pigmentation Explained by the Allele.
We estimated the skin pigmentation variance explained by rs1426654 in the KhoeSan by estimating the fractional reduction of the phenotypic variance on this locus, similar to the method in ref. 25. A linear mixed effect model (lme) testing the association between genotypes and baseline skin pigmentation quantified in melanin units (MI) of the same KhoeSan cohort was done in a separate study by Martin et al. (3), where the global European and Bantu ancestries, and an admixture-corrected genetic relationship matrix, were controlled for in the lme regression. Taking the β-effect size of 3.58 MI of rs1426654 obtained from the lme in ref. 3, we calculated a fractional reduction of the phenotype as
| [4] |
where Y stands for the baseline skin pigmentation (as the phenotype), β is the effect of the locus estimated from the lme, and G is the genotype (0, 1, or 2) per individual at the locus. The phenotypic variance in KhoeSan explained by the locus thereby is defined as
| [5] |
Note that this method treats the effect of rs1426654 as fixed instead of random, and does not perform a joint analysis by partitioning the whole genome into loci of interest vs. the rest of the genome as in ref. 3. Therefore, it can inflate the effect from loci linked with rs1426654 or epistatic effects on rs1426654, with the variance explained appearing larger than the method adopted in ref. 3.
Modeling Demographic and Evolutionary Scenarios with Coalescent Simulations Through ABC.
We modeled two possible demographic scenarios with or without positive selection that could give rise to the observed patterns in our KhoeSan SLC24A5 samples. The general outline of these models is presented in Fig. 3 and SI Appendix, Fig. S5, and we describe the details of the four models with their demographic parameters in SI Appendix, SI Materials and Methods. We chose Luhya from Kenya and Maasai from Kenya/Tanzania as representatives for Bantu speakers and eastern African pastoralists, respectively. Parameters used in each model are listed in SI Appendix, Table S2.
We performed coalescent simulations of all models in a two-step framework similar to ref. 44 (SI Appendix, SI Materials and Methods), conditioning on the final frequency of the focal allele in each population from Wright−Fisher forward simulations. Summary statistics of pairwise nucleotide differences (π), number of segregating sites (s), and extended haplotype homozygosity are calculated from simulated sequences and the empirical data in ≠Khomani San and Nama. Model selection and parameter inference were further conducted under an ABC framework implemented in “abc” package (45) (SI Appendix, SI Materials and Methods).
Data and Script Availability.
Data for the Nama are available through dbGap (https://www.ncbi.nlm.nih.gov/gap). Data for the ≠Khomani San are available upon application to the South African San Council.
Mssel simulator, written by Richard R. Hudson, can be found at genapps.uchicago.edu/newlabweb/software.html.
Scripts for multipopulation trajectories that go into mssel based on https://github.com/shigekinakagome/sim_trajectory_3pops and pipelines including adding sequencing errors and calculating summary statistics can be found at https://github.com/menglin44/additional_sim_scripts.
Supplementary Material
Acknowledgments
We thank Joel Smith and Richard Hudson for assistance with computational tools. We thank the ≠Khomani San and Nama communities of South Africa for their generous participation in this project. M.L. and B.M.H. were supported by National Institutes of Health (NIH) Grant R01GM118652. A.R.M. was supported by an NIH Genetics and Developmental Biology Training Program (Grant T32GM007790) and a trainee research grant from the Stanford Center for Computational, Evolutionary, and Human Genomics. This research was partially funded by the South African government through the South African Medical Research Council (E.G.H. and M.M.). This work was also supported by the National Research Foundation of South Africa. The content is solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.G.J. is a guest editor invited by the Editorial Board.
Data deposition: Data for the Nama are available through dbGap, https://www.ncbi.nlm.nih.gov/gap (study ID 31621). Data for the ≠Khomani San are available upon application to the South African San Council (admin@sasi.org.za).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801948115/-/DCSupplemental.
References
- 1.Norton HL, et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 2.Jablonski NG, Chaplin G. Human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin AR, et al. An unexpectedly complex architecture for skin pigmentation in Africans. Cell. 2017;171:1340–1353.e14. doi: 10.1016/j.cell.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durazo-Arvizu RA, et al. 25-hydroxyvitamin D in African-origin populations at varying latitudes challenges the construct of a physiologic norm. Am J Clin Nutr. 2014;100:908–914. doi: 10.3945/ajcn.113.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song S, Sliwerska E, Emery S, Kidd JM. Modeling human population separation history using physically phased genomes. Genetics. 2017;205:385–395. doi: 10.1534/genetics.116.192963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veeramah KR, et al. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol Biol Evol. 2012;29:617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlebusch CM, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henn BM, et al. Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa. Proc Natl Acad Sci USA. 2008;105:10693–10698. doi: 10.1073/pnas.0801184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breton G, et al. Lactase persistence alleles reveal partial East African ancestry of southern African Khoe pastoralists. Curr Biol. 2014;24:852–858. doi: 10.1016/j.cub.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Pleurdeau D, et al. “Of sheep and men”: Earliest direct evidence of caprine domestication in southern Africa at Leopard Cave (Erongo, Namibia) PLoS One. 2012;7:e40340. doi: 10.1371/journal.pone.0040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickrell JK, et al. Ancient west Eurasian ancestry in southern and Eastern Africa. Proc Natl Acad Sci USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlebusch CM, et al. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science. 2017;358:652–655. doi: 10.1126/science.aao6266. [DOI] [PubMed] [Google Scholar]
- 15.Coussens AK, et al. High-dose vitamin D3 reduces deficiency caused by low UVB exposure and limits HIV-1 replication in urban Southern Africans. Proc Natl Acad Sci USA. 2015;112:8052–8057. doi: 10.1073/pnas.1500909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penn N. The Forgotten Frontier: Colonist and Khoisan on the Cape’s Northern Frontier in the 18th Century. Juta and Company Ltd; Lansdowne, S Africa: 2005. [Google Scholar]
- 17.Crawford NG, et al. Loci associated with skin pigmentation identified in African populations. Science. 2017;358:eaan8433. doi: 10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72, erratum (2007) 5:e147. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsetskhladze ZR, et al. Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS One. 2012;7:e47398. doi: 10.1371/journal.pone.0047398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beleza S, et al. The timing of pigmentation lightening in Europeans. Mol Biol Evol. 2013;30:24–35. doi: 10.1093/molbev/mss207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu Mallick C, et al. The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet. 2013;9:e1003912. doi: 10.1371/journal.pgen.1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 24.Quillen EE, et al. OPRM1 and EGFR contribute to skin pigmentation differences between indigenous Americans and Europeans. Hum Genet. 2012;131:1073–1080. doi: 10.1007/s00439-011-1135-1. [DOI] [PubMed] [Google Scholar]
- 25.Beleza S, et al. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 2013;9:e1003372. doi: 10.1371/journal.pgen.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stokowski RP, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81:1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uren C, et al. Fine-scale human population structure in Southern Africa reflects ecogeographic boundaries. Genetics. 2016;204:303–314. doi: 10.1534/genetics.116.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith J, Coop G, Stephens M, Novembre J. Estimating time to the common ancestor for a beneficial allele. Mol Biol Evol. 2018;35:1003–1017. doi: 10.1093/molbev/msy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llorente MG, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science. 2015;350:820–822. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 30.Hodgson JA, Mulligan CJ, Al-Meeri A, Raaum RL. Early back-to-Africa migration into the horn of Africa. PLoS Genet. 2014;10:e1004393. doi: 10.1371/journal.pgen.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta T, Kimura M. The effect of selected linked locus on heterozygosity of neutral alleles (the hitch-hiking effect) Genet Res. 1975;25:313–326. doi: 10.1017/s0016672300015731. [DOI] [PubMed] [Google Scholar]
- 32.Gerbault P, Moret C, Currat M, Sanchez-Mazas A. Impact of selection and demography on the diffusion of lactase persistence. PLoS One. 2009;4:e6369. doi: 10.1371/journal.pone.0006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedrick PW. Population genetics of malaria resistance in humans. Heredity (Edinb) 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaplin G, Jablonski NG. Hemispheric difference in human skin color. Am J Phys Anthropol. 1998;107:221–223, and discussion (1998) 107:223–224. doi: 10.1002/(SICI)1096-8644(199810)107:2<221::AID-AJPA8>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 36.Sealy J, et al. Diet, mobility, and settlement pattern among Holocene hunter-gatherers in southernmost Africa. Curr Anthropol. 2006;47:569–595. [Google Scholar]
- 37.Lee-Thorp JA, Sealy JC, Morris AG. Prehistoric Human Bone. Springer; Berlin: 1993. Isotopic evidence for diets of prehistoric farmers in South Africa; pp. 99–120. [Google Scholar]
- 38.Sealy J, Pfeiffer S. Diet, body size, and landscape use among Holocene people in the Southern Cape, South Africa. Curr Anthropol. 2000;41:642–655. [Google Scholar]
- 39.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 40.Jombart T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 41.Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 42.Forster P, Harding R, Torroni A, Bandelt H-J. Origin and evolution of native American mtDNA variation: A reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 43.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagome S, et al. Estimating the ages of selection signals from different Epochs in human history. Mol Biol Evol. 2016;33:657–669. doi: 10.1093/molbev/msv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Csilléry K, François O, Blum MGB. abc: An R package for approximate Bayesian computation (ABC) Methods Ecol Evol. 2012;3:475–479. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.