Significance
T cell-mediated immune responses are compromised in aged individuals, leading to increased morbidity and reduced response to vaccination. Finding new ways to boost T cell immunity in the elderly is key for enhancing their immune competence. In this work, we performed a systematic analysis of proteins and metabolites in young versus aged T cells. Metabolic rewiring occurs in young T cells following stimulation but is dampened in aged T cells. Moreover, we show that aged T cell functions can be enhanced by metabolite addition.
Keywords: metabolism, T cells, mitochondria, one-carbon metabolism, aging
Abstract
T cell-mediated immune responses are compromised in aged individuals, leading to increased morbidity and reduced response to vaccination. While cellular metabolism tightly regulates T cell activation and function, metabolic reprogramming in aged T cells has not been thoroughly studied. Here, we report a systematic analysis of metabolism during young versus aged naïve T cell activation. We observed a decrease in the number and activation of naïve T cells isolated from aged mice. While young T cells demonstrated robust mitochondrial biogenesis and respiration upon activation, aged T cells generated smaller mitochondria with lower respiratory capacity. Using quantitative proteomics, we defined the aged T cell proteome and discovered a specific deficit in the induction of enzymes of one-carbon metabolism. The activation of aged naïve T cells was enhanced by addition of products of one-carbon metabolism (formate and glycine). These studies define mechanisms of skewed metabolic remodeling in aged T cells and provide evidence that modulation of metabolism has the potential to promote immune function in aged individuals.
Immune responses are compromised in the elderly, resulting in increased frequency and severity of infectious diseases and impaired responsiveness to vaccination. In response to infection, T cells rapidly proliferate and differentiate into specialized subpopulations that defend against encountered threats. T lymphocytes are among the immune cell populations most detrimentally affected by aging, displaying compromised effector functions and impaired T cell memory formation (1). A key factor contributing to these defects is depletion of the naïve T cell pool (2, 3) and reduced responsiveness of existing cells (4, 5).
Rewiring of intracellular metabolism is key for T cell activation and effector functions, but the metabolic changes in T cells during aging are not well understood. T cell receptor activation and costimulatory signals increase fuel uptake and metabolism to generate precursors for synthesis of macromolecules, fulfill increased energy demands, and enable stress responses (6). To facilitate this high metabolic demand, activated T cells up-regulate glycolysis, glutaminolysis, mitochondrial biogenesis, and oxidative phosphorylation (OXPHOS) (6–9). Previously, we and others identified mitochondrial one-carbon metabolism as one of the most induced metabolic pathways in early T cell activation (8, 10). One-carbon metabolism utilizes serine to generate one-carbon units for purine and thymidine biosynthesis and contributes to redox homeostasis by generating glycine for glutathione synthesis and NADPH for reduction of oxidized thioredoxin and glutathione (11). One-carbon metabolism also contributes to mitochondrial protein synthesis, supporting OXPHOS (12, 13). T cells with low levels of one-carbon metabolism show impaired proliferation and increased DNA damage, leading to cell death (8). Moreover, serine starvation impairs T cell expansion (14).
Considering the importance of mitochondrial adaptation and metabolism during early T cell activation, we set out to systematically test whether aged T cells (from 20-mo-old mice) demonstrated altered mitochondrial function. Activation of T cells from aged mice induced mitochondrial biogenesis, but yielded smaller mitochondria with lower respiratory capacity compared with young animals. Likewise, analysis of intracellular metabolites by mass spectrometry confirmed an overall reduction in activation-induced metabolism, manifested by decreased levels of metabolites in glycolysis, the pentose phosphate pathway (PPP), and the TCA cycle. Finally, we defined the proteome of aged T cells during activation using quantitative mass spectrometry. This analysis identified a specific defect in the induction of enzymes in mitochondrial one-carbon metabolism. Replenishing one-carbon metabolism reinvigorated the function of aged T cells.
Results
Naïve CD4+ T Cells Are Scarce in Aged Mice and Fail to Properly Activate in Response to Stimulation.
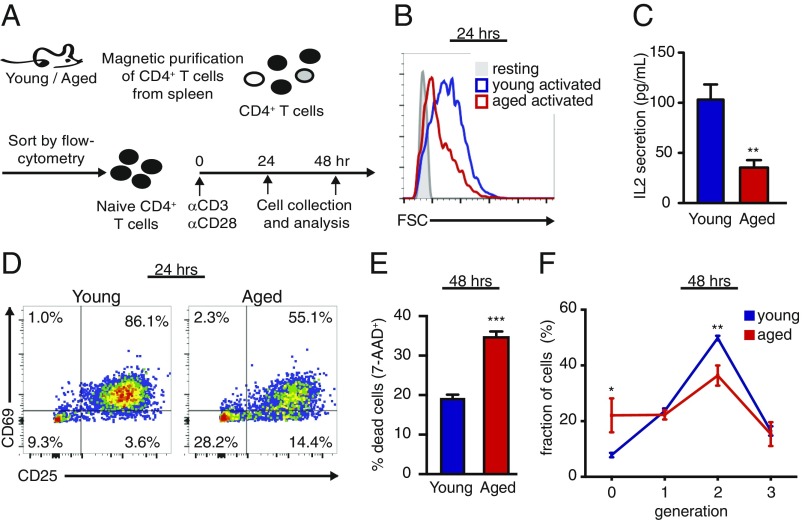
To identify changes in T cell populations during aging, we analyzed T cells from the spleens of young and aged C57BL/6 mice. In agreement with previous studies (1), we identified an overall reduction in T cell frequency and number in the spleen (SI Appendix, Fig. S1A), a ∼2.5-fold increase in Foxp3+ T regulatory cells, and more than fivefold decrease in the percentage of T conventional cells with a naïve phenotype (CD4+CD25−CD62L+CD44lo). To test whether naïve T cells in aged mice retained function, we sorted naïve CD4+ T cells from young and aged mice and activated them ex vivo using plate-bound anti-CD3/anti-CD28 antibodies (Fig. 1A). Naïve CD4+ T cells from aged mice were inferior to young CD4+ T cells in all parameters tested, showing reduced cell growth (Fig. 1B), lower IL-2 secretion (Fig. 1C), reduced expression of early activation markers (Fig. 1D and SI Appendix, Fig. S1B), as well as a 50% increase in cell death at 48 h postactivation (Fig. 1E).
Fig. 1.
Naïve CD4+ T cells in aged mice respond poorly to stimulation. (A) Splenocytes were isolated from young and aged mice, enriched for CD4+ T cells using anti-CD4 magnetic beads, and sorted by flow cytometry to yield a pure naïve CD4+ T cell population (CD4+CD25−CD62L+CD44lo; purity >99%) and stimulated ex vivo using plate-bound anti-CD3/anti-CD28. Cultured cells were harvested at 24 h and analyzed by flow cytometry to assess (B) cell size, (C) IL-2 concentration in culture media, (D) expression of early activation markers: CD69 and CD25, and (E) cell death by 7-AAD incorporation. To assess proliferation, cells were loaded with CFSE and analyzed by flow cytometry at 48 h postactivation. Cell divisions were detected by dilution of the CFSE. (F) Quantitation of cell division, showing the percentage of cells in each generation. *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test comparing young vs. aged T cells). Data are representative of at least two independent experiments.
To assess the proliferative capacity of aged naïve T cells, we labeled naïve CD4+ T cells with carboxyfluorescein succinimidyl ester (CFSE), a cell permeable dye, and assessed CFSE dilution at 48 h postactivation. Aged T cells had an increased percentage of nondividing cells (generation = 0) and a decreased percentage of cells that underwent two cell divisions (generation = 2; Fig. 1F). Thus, we observed a significant contraction of the naïve T cell pool in aged mice, and the existing aged naïve CD4+ T cells exhibited diminished activation and function in response to stimulation.
Metabolism Is Defective in T Cells from Aged Mice.
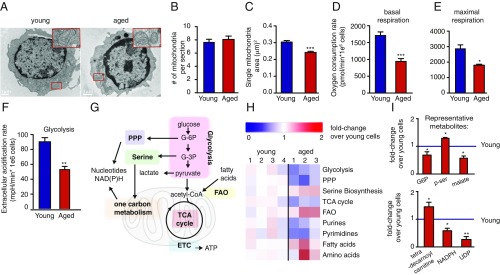
Our recent work identified a program of mitochondrial biogenesis that is initiated upon naïve T cell stimulation and is critical for T cell activation and survival (8). To test whether T cell mitochondrial biogenesis declined with age, naïve CD4+ T cells from young and aged mice were stimulated ex vivo for 24 h, followed by fixation and imaging by electron microscopy (Fig. 2A). While mitochondria numbers were similar (Fig. 2B), the mitochondria in stimulated T cells from aged mice were significantly smaller (Fig. 2C and SI Appendix, Fig. S1 C and D), leading to a reduction in mitochondrial mass.
Fig. 2.
The metabolic rewiring driven by T cell stimulation is defective in T cells from aged mice. (A) Representative electron micrograph (EM) images of young and aged T cells that were isolated and stimulated for 24 h, as described in Fig. 1A. (B and C) Quantitation of EM micrographs showing differences in the number (B) and surface area (C) of the mitochondria. Oxygen consumption rate (mitochondrial respiration) and extracellular acidification rate (indicator of glycolysis) measured in T cells from young and aged mice activated for 24 h, using the Seahorse Extracellular Flux Analyzer. (D) Basal respiration. (E) Maximal respiratory capacity was calculated based on the increase in oxygen consumption following treatment with FCCP, a mitochondrial uncoupler. (F) Glycolysis was assessed based on the changes in extracellular acidification rate following treatment with oligomycin, an ATP synthase inhibitor. (G) Major metabolic pathways involved in T cell activation. (H) Average fold change in metabolite levels for central metabolic pathways in activated aged vs. young CD4+ T cells. Fold change compared with average levels in young T cells was calculated. (I) Fold change of representative metabolites in young vs. aged T cells. *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test comparing young vs. aged T cells). Data are representative of at least two independent experiments.
Analysis of oxygen consumption rate (a measure of mitochondrial respiration) in activated naïve CD4+ T cells from young and aged mice revealed a significant reduction in basal respiration (Fig. 2D) and maximal respiratory capacity (Fig. 2E) in aged T cells. Although respiration impairment often induces a compensatory increase in glycolysis, in aged T cells, extracellular acidification rate (a measure of glycolysis) also was decreased (Fig. 2F). Consistent with decreased glycolysis and respiration, levels of central carbon intermediates in glycolysis, the PPP, and the TCA cycle were lower in T cells from aged mice (Fig. 2 G–I). Moreover, purines, pyrimidines, and NADPH were also decreased. In contrast, fatty acids, acyl carnitines, amino acids, and phosphoserine were higher in the aged T cells, perhaps reflecting impaired consumption of these metabolites due to slower respiration and protein synthesis. Thus, upon their activation, naïve CD4+ T cells from aged mice have broad-based defects in central carbon metabolism (Fig. 2 H and I and SI Appendix, Fig. S2).
Quantitative Proteomics Identifies Changes in Mitochondrial Composition Between Young and Aged T Cells.
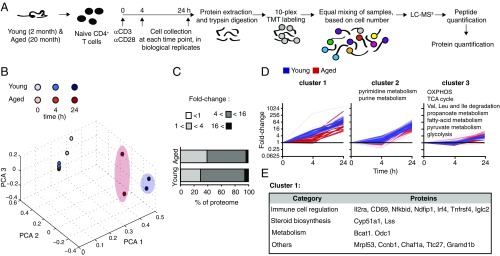
To quantify the proteome of young and aged T cells, we employed tandem mass spectrometry after isobaric peptide tagging (TMT) in naïve versus activated CD4+ T cells from young and aged mice (Fig. 3A) and determined dynamic changes in >3,600 proteins (Dataset S1). According to principal component analysis (PCA), the largest changes in protein composition were induced by activation (PCA1 represents 85.99% of the variance and separates between 4 and 24 h). Protein composition of T cells from young and aged mice was different at 0 and 24 h postactivation (PCA2 represents 12.79% of the variance and separates between young and aged T cells at T = 0 h and T = 24 h). PCA3 (0.5% of the variance) separates all replicates (Fig. 3B). At the 24-h time point, we identified an attenuation of fold-change increase in protein content in aged compared with young T cells (Fig. 3C and SI Appendix, Fig. S3A), in agreement with the reduced size of aged T cells (Fig. 1B) and their accumulation of free amino acids (Fig. 2H).
Fig. 3.
Quantitative proteomics identifies distinct activation-induced changes in the proteome between young and aged T cells. (A) Naïve CD4+ T cells from young and aged mice were activated using plate-bound anti-CD3/anti-CD28, collected, and processed by protein extraction and digestion. We quantified two separate pools of mice for every age group at 4 and 24 h postactivation, and one representative naïve cell sample. The peptides from each of the 10 samples were labeled with a specific TMT label. Peptides were pooled based on cell numbers and analyzed by LC-MS3 to quantify proteins. (B) Principal component analysis. (C) Distribution of fold-change induction in protein content at 24 h postactivation. (D) Proteins were clustered based on magnitude and kinetics of expression in young T cells (blue). The expression of the same cluster of proteins in aged T cells is shown in red. Three representative clusters are shown (see SI Appendix, Fig. S2 for clusters 4–12). Kegg pathway analysis identified the specific metabolic pathways enriched within each cluster. (E) List of the proteins in cluster 1.
Overall, aged T cells demonstrated a lower induction of proteins upon activation compared with young T cells. A total of 150 proteins were induced at least twofold less in aged T cells (SI Appendix, Fig. S3A and Dataset S2) and included proteins associated with inflammation and immune regulation, such as Vnn1 (15), Nfkbid (16), and foxp4 (17). Interestingly, the majority of these proteins are not well studied in the context of immune cell function and may highlight pathways contributing to immunosenescence. We further identified 40 proteins that were elevated at least twofold more in aged T cells compared with young T cells (SI Appendix, Fig. S3A and Dataset S2), suggesting that the aged T cell phenotypes were not solely due to blunted activation. The proteins most induced in activated aged T cells included Gm16519, a predicted ribosomal protein; Iglc2, an immunoglobulin; and Bicd2, involved in Golgi trafficking (18). While immunoglobulins are generated by B lymphocytes, our proteomic data from sorted T cells did not detect other B cell markers such as CD19, ruling out a general B cell contamination. One possible explanation for detection of Iglc2 is attachment of the antibody to the T cells’ surface, that is not completely excluded by the wash.
To identify functional patterns during activation, proteins were grouped based on the kinetics and magnitude of induction in young cells (Fig. 3D and SI Appendix, Fig. S3B). Cluster 1 contains early and highly induced proteins in young T cells and includes known transcription regulators of T cell activation and early activation markers (Fig. 3E). Induction of proteins in this cluster was delayed in aged cells, and most show no induction at 4 h poststimulation. Pathways associated with nucleic acids were enriched in young T cells within the second most highly induced cluster (cluster 2), whereas proteins associated with OXPHOS, TCA cycle, glycolysis, and fatty acid oxidation (FAO) were enriched within the least induced cluster (cluster 3).
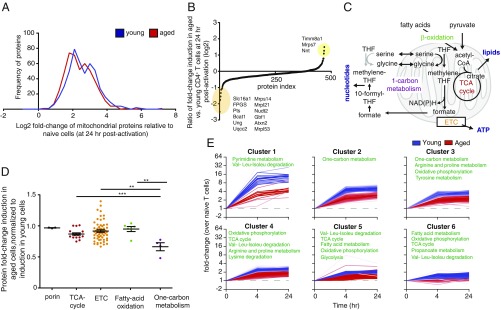
Given the substantial defects in respiration in the aged T cells, we also inspected the mitochondrial proteome. We used the Mitocarta database as a filter to generate a list of 469 mitochondrial proteins detected in our dataset (19). There was a slight decrease in magnitude of fold-change induction in aged compared with young T cells (Fig. 4 A and B), consistent with reduced mitochondrial mass. The top 20 overinduced and underinduced proteins are listed in SI Appendix, Table S1. Analysis of central metabolic pathways in the mitochondria (Fig. 4C) demonstrated that enzymes involved in TCA cycle, electron transport chain (ETC), and FAO were similarly induced in young and aged T cells (Fig. 4D). Strikingly, the least induced metabolic pathway in aged T cells was one-carbon metabolism; enzymes in this pathway were induced ∼35% less in aged T cells compared with young ones (Fig. 4D and SI Appendix, Fig. S4A).
Fig. 4.
Induction of mitochondrial enzymes of one-carbon metabolism is defective in aged T cells. (A) Distribution of mitochondrial proteome induction following CD4+ T cell activation in young (blue) and aged (red) T cells. (B) Log2 ratio of fold-change induction (activated/naïve) in mitochondrial proteome in aged vs. young T cells. Highlighted are proteins with the greatest differences in induction rate between young and aged cells. In yellow: proteins that were induced more in aged cells. In orange: proteins that were induced less in the aged cells. (C) Central metabolic pathways in the mitochondria. (D) Ratio of fold change (fold-change aged vs. fold-change young) of enzymes in central metabolic pathways in the mitochondria. (E) Mitochondrial proteome was clustered following application of the ccRemover algorithm, based on magnitude and kinetics of expression in young T cells (blue). Expression of the same cluster of proteins in aged T cells is shown in red. Kegg pathway analysis identified specific metabolic pathways enriched within each cluster. **P < 0.01, ***P < 0.001 (Student’s t test).
Our analysis of young and aged T cells was performed at 24 h postactivation, before proliferation occurs. To further validate that the observed differences in mitochondrial proteome are not due to differences in cell cycle, we reanalyzed our proteomic dataset after applying the ccRemover algorithm to remove cell cycle effects (20). Mitochondrial proteins were then grouped into clusters based on kinetics and magnitude of activation (Fig. 4E). Consistent with our findings, one-carbon metabolism was enriched within clusters 2 and 3, in which we observed a difference in magnitude between young and aged T cells. This analysis identified enrichment in enzymes associated with pyrimidine metabolism within cluster 1, the cluster most suppressed with aging (Fig. 4E).
Ex Vivo Activation of Aged T Cells Is Partially Rescued by Addition of Metabolites in One-Carbon Metabolism.
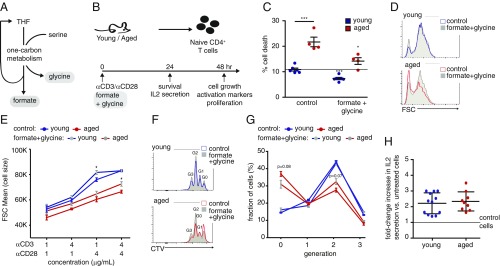
The markedly compromised induction of enzymes in one-carbon metabolism led us to hypothesize that the addition of metabolites in this pathway would improve the outcome of activation of naïve T cells. To test this idea, we examined whether addition of glycine and formate (Fig. 5A) to naïve aged CD4+ T cells during initial activation could improve their survival and function (Fig. 5B). Addition of formate and glycine to growth media reduced cell death to levels similar to those found in untreated young cells (Fig. 5C). For both young and aged CD4+ T cells, cell growth positively correlated with the strength of the activation signal. Under a weak stimulation (1 μg/mL anti-CD3 and anti-CD28), supplementation of formate and glycine slightly increased cell growth in both populations (SI Appendix, Fig. S5A). Under a strong stimulation (with 4 μg/mL anti-CD3 and anti-CD28), metabolite supplementation increased cell growth only in aged T cells, suggesting that under these conditions young T cells have maximized their growth potential (Fig. 5 D and E). CD69 expression was lower in aged T cells compared with young, and slightly increased with the treatment under a weak stimulation (SI Appendix, Fig. S5 B and C). To test the effect of metabolite supplementation on proliferation, young and aged T cells were labeled with CellTrace Violet (CTV) before stimulation. The rate of proliferation increased with the stronger stimulation conditions and was lower in aged T cells under both conditions (Fig. 5 F and G and SI Appendix, Fig. S5D). Addition of formate and glycine to aged T cells modestly enhanced proliferation (Fig. 5 F and G). IL-2 secretion was evaluated under strong stimulus and was similarly increased by metabolite supplementation to young and aged T cells (Fig. 5H). These studies demonstrate that metabolites, which restore one-carbon metabolism, were sufficient to rescue survival and function of aged T cells ex vivo.
Fig. 5.
Adding back metabolites of one-carbon metabolism improves activation and survival of aged T cells ex vivo. (A) Mitochondrial one-carbon metabolism: One carbon from serine is transferred to tetrahydrofolate (THF) to form glycine and methylene-THF that is further oxidized to formate. (B) Naïve CD4+ T cells were isolated from young and aged mice, as described in Fig. 1A, and activated with plate-bound anti-CD3/anti-CD28 (4 μg/mL each, unless otherwise indicated). Culture media were supplemented with formate + glycine, or no metabolites (control cells). Cells were harvested at 24 or 48 h postactivation and analyzed by flow cytometry. (C) Percentage of cell death (measured by 7AAD incorporation). (D) Representative flow cytometry plots showing activation-induced cell growth. (E) Cell size as a function of anti-CD3 and anti-CD28 concentration. To assess proliferation, cells were loaded with CTV and analyzed by flow cytometry at 48 h postactivation. Cell divisions were detected by dilution of the CTV. (F) Representative flow cytometry plots showing dilution of CellTrace Violet staining in young and aged T cells. (G) Quantitation of cell division, showing the percentage of cells in each generation. (H) IL-2 secretion was analyzed by measuring IL-2 concentration in growth media. A pool of two experiments, showing the ratio between IL-2 levels in the media of treated vs. untreated cells. *P < 0.05, ***P < 0.001 (Student’s t test comparing each treatment group to its untreated control, and the aged controls to young controls, when marked by a line). Data are representative of at least two independent experiments.
Discussion
In this study we performed a side-by-side comparison of mitochondrial biogenesis, intracellular metabolites, and quantitative proteomics in young versus aged T cells. We found cell-intrinsic defects in metabolism during the activation of aged naïve CD4+ T cells, including evidence of lower glycolysis and attenuated induction of one-carbon metabolism. Importantly, addition of metabolites in one-carbon metabolism partially rescued defects in activation of aged CD4+ T cells.
To investigate intrinsic deficits in aged naïve CD4+ T cells, we purified naïve CD4+ cells from aged mice and analyzed their activation ex vivo using anti-CD3/anti-CD28. This ex vivo approach eliminated the effect of other potential age-related factors, such as reduced efficiency of antigen uptake and/or presentation (21) and the increase in immune suppressor populations (e.g., regulatory T cells and myeloid derived suppressor cells) (22).
We found that mitochondrial mass and activation are reduced in stimulated aged compared with young T cells. Reduced mitochondrial activation may impair functionality by dysregulating critical early signaling events. For example, calcium buffering by the mitochondria at the immune synapse extends Ca+2-dependent signaling of key T cell activators NF-κB and NFAT (23). In addition, mitochondrial reactive oxygen species induce cytokine production through activation of NF-κB and AP-1 (24).
Our mass spectrometry-based analysis of intracellular metabolites was performed in an enriched metabolic environment (culture media), containing ample glucose (11 mM). However, aged T cells failed to properly activate intracellular metabolism. The cause of the decreased glycolytic flux in the aged T cells remains unclear. One possible mechanism for impaired glucose metabolism is reduced glucose uptake. Weakened TCR and costimulatory signaling in aged T cells may compromise the relocation of glucose transporters to the plasma membrane (25). Smaller pools of glycolytic intermediates could also reflect lower activity of glycolytic enzymes in aged T cells. However, our proteomic quantitation suggests that the enzymes involved in glycolysis, the TCA cycle, and the PPP are induced in activated aged T cells similarly to young cells. Possible alternate models include altered allosteric regulation of glycolysis, changes in ATP demand, or differences in protein localization. Importantly, not all intracellular metabolites were reduced in aged T cells. In fact, we identified an induction in metabolites of FAO. Stimulation of young T cells induces a metabolic switch in which these T cells stop burning fats and start synthesizing them (9). The increase in FAO metabolites in aged T cells further illustrates how metabolic adaptation is impaired in aged T cells.
Our previous studies defined one-carbon metabolism as the most induced metabolic pathway in T cell mitochondria during early activation (8). Moreover, genetic interference with one-carbon metabolism and/or limited substrate supplies reduced T cell expansion and survival (8, 14). Here, our analysis of the mitochondrial proteome highlighted one-carbon metabolism as particularly blunted in aged T cells. We demonstrate that providing metabolites in one-carbon metabolism to aged T cells resulted in improved activation and survival ex vivo. In agreement, in aged human fibroblasts, epigenetic changes suppress SHMT2 expression, and the related respiratory defect is reversed by supplementation of glycine (26). Further studies are needed to determine whether aged cells treated ex vivo by metabolite supplementation will perform better when introduced back into the aged mice.
Our studies define how stimulation-induced metabolic adaptation, a critical factor for successful T cell activation, is defective in aged T cells. It is not clear whether frail humans have a reduced naïve T cell pool. However, our findings suggest that in addition to studying the naïve T cell repertoire and pool size in the frail versus healthy aged, quantifying the metabolic fitness of the cells has the potential to identify new strategies for improving T cell function in the elderly.
Materials and Methods
Mice.
Young (7–10 wk old) and aged (20–22 mo old) C57BL/6 mice were purchased from the National Institute on Aging. Female mice were used in all experiments unless otherwise stated. Experimental mice were housed in specific pathogen-free conditions at Harvard Medical School and used in accordance with animal care guidelines from the Harvard Medical School Standing Committee on Animals and the National Institutes of Health.
Culture and Stimulation of Naïve CD4+ T Cells.
Naïve T cells were isolated and activated ex vivo, as previously described (8). All details are given in SI Appendix.
Flow Cytometry.
All details for cell processing and staining are given in SI Appendix. Data were analyzed on an LSR II (BD Biosciences) using standard filter sets and FlowJo software (TreeStar).
Electron Microscopy: Sample Preparation, Data Collection, and Analysis.
T cells were processed as previously described (8). All details are given in SI Appendix. Imaging was done using a JEM-1400 transmission electron microscope (JEOL), and analyzed using Volocity 3D (PerkinElmer).
Metabolomics.
Cellular metabolites in activated CD4+ T cells were quantified by liquid chromatography/MS. All details are given in SI Appendix. Data were analyzed using the MAVEN software suite (27). Metabolite abundance was normalized to cell count.
Proteomics Sample Preparation and Data Analysis.
All details on sample preparation and data extraction are given in SI Appendix. Fold-change data were Lowess normalized and collapsed into unique protein representation to average fold change of all peptides. Proteins were clustered based on the expression levels in young T cells using the Agglomerative Clustering function from the SciKit-Learn machine learning library in Python (https://scikit-learn.org/stable/). Clustering analysis compared a protein’s abundance relative to naïve T cells at 4 and 24 h following activation, in each of two experimental replicates (so that four data points per protein were used for clustering). Values from the two replicates were averaged for drawing the curves in Figs. 3 and 4. All proteins were included in this analysis.
Statistical Analysis.
All analyses were performed with Prism (GraphPad).
Supplementary Material
Acknowledgments
We thank Dr. Michal Sheffer and Naftali Horwitz for discussions of proteomic data analysis. This study was supported by grants from F. Hoffmann-La Roche, Ltd. (to A.H.S. and M.C.H.), the Glenn Foundation for Medical Research (M.C.H.), the Ludwig Center at Harvard Medical School (M.C.H. and A.H.S.), and NIH Grant R01CA213062 (to M.C.H.). N.R.-H. was supported by a postdoctoral fellowship from the European Molecular Biology Organization and by the Israeli National Postdoctoral Award Program for Advancing Women In Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804149115/-/DCSupplemental.
References
- 1.Goronzy JJ, Weyand CM. Successful and maladaptive T cell aging. Immunity. 2017;46:364–378. doi: 10.1016/j.immuni.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berent-Maoz B, Montecino-Rodriguez E, Dorshkind K. Genetic regulation of thymocyte progenitor aging. Semin Immunol. 2012;24:303–308. doi: 10.1016/j.smim.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixit VD. Impact of immune-metabolic interactions on age-related thymic demise and T cell senescence. Semin Immunol. 2012;24:321–330. doi: 10.1016/j.smim.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Miller RA, et al. T cells in aging mice: Genetic, developmental, and biochemical analyses. Immunol Rev. 2005;205:94–103. doi: 10.1111/j.0105-2896.2005.00254.x. [DOI] [PubMed] [Google Scholar]
- 5.Clise-Dwyer K, Huston GE, Buck AL, Duso DK, Swain SL. Environmental and intrinsic factors lead to antigen unresponsiveness in CD4(+) recent thymic emigrants from aged mice. J Immunol. 2007;178:1321–1331. doi: 10.4049/jimmunol.178.3.1321. [DOI] [PubMed] [Google Scholar]
- 6.Bantug GR, Galluzzi L, Kroemer G, Hess C. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol. 2018;18:19–34. doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- 7.D’Souza AD, Parikh N, Kaech SM, Shadel GS. Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation. Mitochondrion. 2007;7:374–385. doi: 10.1016/j.mito.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ron-Harel N, et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan H, et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity. 2017;46:488–503. doi: 10.1016/j.immuni.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morscher RJ, et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554:128–132. doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minton DR, et al. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol Cell. 2018;69:610–621.e5. doi: 10.1016/j.molcel.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma EH, et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 2017;25:482. doi: 10.1016/j.cmet.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira DW, et al. Enhanced hepatotoxicity by acetaminophen in Vanin-1 knockout mice is associated with deficient proliferative and immune responses. Biochim Biophys Acta. 2016;1862:662–669. doi: 10.1016/j.bbadis.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Touma M, et al. Functional role for I kappa BNS in T cell cytokine regulation as revealed by targeted gene disruption. J Immunol. 2007;179:1681–1692. doi: 10.4049/jimmunol.179.3.1681. [DOI] [PubMed] [Google Scholar]
- 17.Wiehagen KR, et al. Foxp4 is dispensable for T cell development, but required for robust recall responses. PLoS One. 2012;7:e42273. doi: 10.1371/journal.pone.0042273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goud B, Gleeson PA. TGN golgins, Rabs and cytoskeleton: Regulating the Golgi trafficking highways. Trends Cell Biol. 2010;20:329–336. doi: 10.1016/j.tcb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barron M, Li J. Identifying and removing the cell-cycle effect from single-cell RNA-Sequencing data. Sci Rep. 2016;6:33892. doi: 10.1038/srep33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong C, Goldstein DR. Impact of aging on antigen presentation cell function of dendritic cells. Curr Opin Immunol. 2013;25:535–541. doi: 10.1016/j.coi.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagger A, Shimojima Y, Goronzy JJ, Weyand CM. Regulatory T cells and the immune aging process: A mini-review. Gerontology. 2014;60:130–137. doi: 10.1159/000355303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintana A, et al. Calcium microdomains at the immunological synapse: How ORAI channels, mitochondria and calcium pumps generate local calcium signals for efficient T-cell activation. EMBO J. 2011;30:3895–3912. doi: 10.1038/emboj.2011.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashizume O, et al. Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers human age-associated mitochondrial respiration defects. Sci Rep. 2015;5:10434, and erratum (2015) 5:14591. doi: 10.1038/srep10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Curr Protoc Bioinformatics. 2012;Chapter 14:Unit14 11. doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.