Significance
Whether epigenetics can facilitate persistence of acquired adaptive traits remains hotly debated. Here we use a primitive multicellular animal, a reef-building coral, to address this question for gene body methylation, which is a common epigenetic modification in organisms ranging from sunflowers to humans. We show that gene body methylation is altered by the environment in an unexpectedly simple way that reflects a shifting balance between expressions of housekeeping and environmentally responsive genes, which helps acclimatization. Whether these changes contribute to longer-term adaptation remains an open question: While the majority of fixed epigenetic differences between populations do not appear to be due to fixation of plastic changes, their transgenerational inheritance cannot be completely ruled out.
Keywords: DNA methylation, coral, reciprocal transplant, gene expression, fitness
Abstract
Gene body methylation (GBM) has been hypothesized to modulate responses to environmental change, including transgenerational plasticity, but the evidence thus far has been lacking. Here we show that coral fragments reciprocally transplanted between two distant reefs respond predominantly by increase or decrease in genome-wide GBM disparity: The range of methylation levels between lowly and highly methylated genes becomes either wider or narrower. Remarkably, at a broad functional level this simple adjustment correlated very well with gene expression change, reflecting a shifting balance between expressions of environmentally responsive and housekeeping genes. In our experiment, corals in a lower-quality habitat up-regulated genes involved in environmental responses, while corals in a higher-quality habitat invested more in housekeeping genes. Transplanted fragments showing closer GBM match to local corals attained higher fitness characteristics, which supports GBM’s role in acclimatization. Fixed differences in GBM between populations did not align with plastic GBM changes and were mostly observed in genes with elevated FST, which suggests that they arose predominantly through genetic divergence. However, we cannot completely rule out transgenerational inheritance of acquired GBM states.
Gene body methylation (GBM) is a taxonomically widespread epigenetic modification the function of which remains enigmatic (1, 2). Only the detrimental effect of GBM is well understood: GBM causes hypermutability in protein-coding regions (3). Indeed, in humans, GBM is the primary driver of deleterious parent-age–related mutations (4). To merit pervasive evolutionary conservation, the fundamental biological function of GBM must be important enough to outweigh this risk (1).
One remarkable feature of GBM throughout Metazoa is its bimodal distribution among functional categories of genes: GBM is high in ubiquitously expressed housekeeping genes and low in genes that are regulated, depending on the context (2, 5). It has been proposed that GBM suppresses intragenic transcription initiation in highly expressed genes (6, 7), while other authors argued that GMB simply accumulates in actively transcribed chromatin (8). Still, the correlation of GBM level with gene expression (9, 10) is weak and cannot explain the striking bimodality of GBM among functional groups of genes. The ecological genomic literature has long expected that GBM (as well as other epigenetic marks) can respond to the environment and assist acclimatization by modulating gene expression, possibly across generations (11–13). Still, thus far the only well-documented case of GBM responding to the environment and resulting in phenotype change is determination of castes in social insects, depending on diet (14). Beyond this highly taxon-specific example, GBM response to the environment has been reported in a recent paper comparing symbiotic and symbiont-free states of a sea anemone (10) and in a paper on coral acclimatization to acidic conditions (15); notably, no clear correlation between changes in GBM and changes in gene expression has been recovered. One way to address the role of GBM in adaptation would be to examine differences between populations, as has been recently done in plants (16, 17); however, this interpretation requires disentangling the effect of environment from the effect of genetic divergence among populations (18). As for transgenerational plasticity, for GBM to be involved in this process it must be both responsive to the environment and heritable. This somewhat contradictory combination of properties has not yet been demonstrated for GBM in any study system.
Here, we used a reciprocal transplantation framework (19) to test for the roles of GBM in local adaptation and acclimatization in a reef-building coral Acropora millepora. Corals are basal metazoans of the phylum Cnidaria with many ancestral genomic features common to all multicellular animals, including humans (20, 21). Most importantly for this study, corals possess the typical metazoan gene body methylation (9) that is lacking in highly evolutionarily derived models such as Drosophila and Caenorhabditis (5). In addition, the possibility of clonal replication via fragmentation of coral colonies makes disentangling the effects of genotype and environment very straightforward, solving a major problem of ecological epigenetics (18).
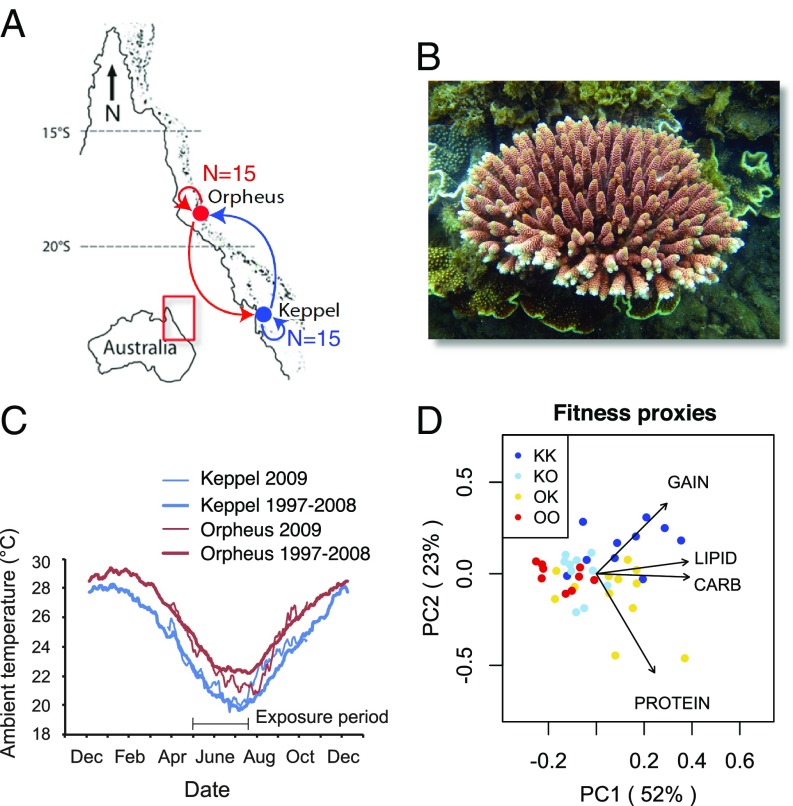
We have compared corals from two reefs, Orpheus Island in the central sector of the Great Barrier Reef (GBR) and Keppel Island in the south of the GBR (Fig. 1A). These reefs are notably different in temperature (Fig. 1B) as well as several other abiotic parameters (22) and host slightly genetically divergent A. millepora populations (23, 24). From each reef, 15 coral colonies, each representing a single genetically distinct diploid individual (a genet), were halved and reciprocally transplanted between the sites for three winter months (Fig. 1 A–C). In this way, each genet was simultaneously exposed to two distinct reef conditions.
Fig. 1.
Overview of the experiment. (A) Design of reciprocal transplantation. (B) A. millepora. (C) Temperature profiles at the transplantation sites, historical (thick lines) and during the year of experiment (thin lines). (D) Principal component analysis of the four fitness proxies: weight gain and lipid, protein, and carbohydrate content. In the sample names, the first letter is the origin location and the second letter is transplantation location.
We analyzed genome-wide gene expression by sequencing 3’-mRNA tags (TagSeq; ref. 25) and DNA methylation by sequencing DNA fragments bound to the methyl-binding domain (MBD-seq; ref. 9) in 44 coral fragments (n =11 from each experimental group) following 3 mo of transplantation. We also measured several fitness proxies and estimated genetic distances between genets based on genetic polymorphisms detected in the MBD-seq data. All experimental corals were dominated by symbionts of genus Cladocopium (ref. 26, formerly clade C), which was established by analyzing Symbiodiniaceae-matching MBD-seq reads.
For both Keppel- and Orpheus-origin corals fitness proxies were higher at Keppel (Fig. 1D), possibly due to a higher concentration of inorganic nutrients there (22, 23). For Keppel-origin corals the difference in performance was predominantly in weight gain (i.e., skeletal growth) while for Orpheus-origin corals the difference was mostly in protein content (Fig. 1D).
To determine the characteristic level of GBM for each gene [“MBD score” (9)] we used MBD-seq results for 12 samples [3 per each experimental group (K, Keppel; O, Orpheus), KK, KO, OK, and OO, Fig. 1D] for which we sequenced both MBD-captured and flow-through fractions. MBD score was calculated using the method for differential gene expression analysis based on the negative binomial distribution (DESeq2; ref. 27) as logarithm with the base 2 of each gene’s enrichment in captured relative to flow-through fractions. Thus, MBD score of zero corresponds to a gene that was equally represented in captured and flow-through fractions; highly methylated genes that were more prevalent in the captured fraction received positive MBD scores, and lowly methylated genes that were more prevalent in the flow-through fraction received negative MBD scores. We validated these results by bisulfite sequencing 13 amplicons representing low- to middle-level MBD scores and found that an MBD-score range from −3 to 1 corresponded to the range of methylation levels between 1% and 30% (SI Appendix, Fig. S1B). Genome-wide validation of our MBD-seq data was also provided by the facts that the MBD score exhibited the expected bimodal distribution across genes (SI Appendix, Fig. S1A) and showed strong correlation with the established proxy of historical methylation level, the ratio of observed to expected numbers of CpG dinucleotides (Fig. 2D) (5). Relative quantification results based only on MBD-captured read data were nearly identical to those obtained when using both captured and flow-through fractions (SI Appendix, Fig. S2), and so for the remaining 32 samples we sequenced only the MBD-captured fraction.
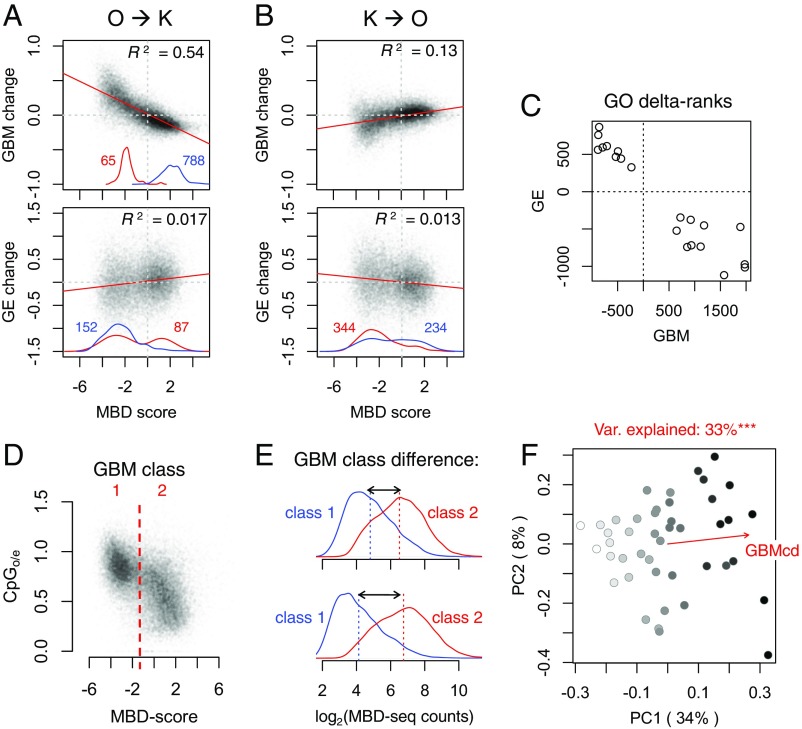
Fig. 2.
GBM response to transplantation is negatively correlated with GE and mainly consists of changes in genome-wide GMB disparity among genes. (A and B) Log2 fold changes in GBM (Top) and GE (Bottom) in response to transplantation (away relative to home), plotted against MBD score for each gene. All correlations are significant at the P < 0.0001 level. Red and blue curves near the x axis are density plots of significantly up (red)- and down (blue)-regulated genes, numbers of which are given in corresponding colors (there were none for the K → O GBM comparison). (C) Plot of delta ranks for 20 Gene Ontology categories that were significantly enriched with either up-regulated genes (positive delta rank) or down-regulated genes (negative delta rank) in both GE and GBM analyses. (D) Scatter plot of the measure of depletion of CpG dinucleotides (CpGO/E, known to correlate negatively with the historical methylation level) against MBD score for each gene. The MBD-score threshold defining the two GBM classes is indicated. (E) Calculation of GBMcd (gray double arrows), a proxy of genome-wide GBM disparity among genes. Each panel shows two density plots of log2-MBD-seq counts for the two GBM gene classes (D); the mean for each class is indicated by a dashed line. Top shows the sample with the lowest GBMcd and Bottom shows the sample with the highest GBMcd. (F) Principal coordinate biplot of GBM colored according to GBMcd. The red vector shows the direction of correlation of GBMcd with the ordination. The title of the plot identifies the proportion of variance (Var) in GBM explained by GBMcd. Significant difference ***P < 0.001.
For GBM, we analyzed 27,084 genes (of 32,045 total annotated genes in the Acropora digitifera genome, which was used as a reference) with mean counts across samples equal to or greater than 20. Gene expression (GE) analysis used the dataset published previously (28), which included 19,706 genes. Multivariate analysis of GE and GBM (SI Appendix, Fig. S3) revealed that GBM is more consistent across fragments of the same original colony than GE, resulting in the estimate of broad-sense heritability of 0.79 compared with 0.64 for GE (SI Appendix, Fig. S3C). In addition, significant effects of origin and transplantation site were observed for GE and GBM, both more pronounced for GE (SI Appendix, Fig. S3C).
Surprisingly, the change in GBM in response to transplantation from Orpheus to Keppel consisted mainly of genome-wide reduction of disparity between highly and lowly methylated genes: Highly methylated genes became less methylated and lowly methylated genes became more methylated (Fig. 2A, Top). This change was mirrored by less pronounced but clearly reciprocal GBM adjustment in Keppel corals transplanted to Orpheus (Fig. 2 B and D and SI Appendix, Fig. S4A). Both exons and introns underwent this change (SI Appendix, Fig. S5). Absolute changes in methylation included both positive and negative shifts (SI Appendix, Fig. S6 A and B), indicating that the change in relative GBM levels among genes was not due to biased genome-wide methylation increase or decrease. Methylation levels of intergenic regions and repeated elements remained relatively constant compared with those of genic regions (SI Appendix, Fig. S6 C and D).
Remarkably, GE changes correlated with lowly and highly methylated gene classes (environmentally responsive and housekeeping, respectively) and were in the opposite direction relative to GBM changes (Fig. 2 A and B, Bottom). Just like GBM changes, GE changes were reciprocal between transplantation directions but were of higher magnitude (SI Appendix, Fig. S4B). Although the correlation between GE and GBM was weak at the individual gene level (SI Appendix, Fig. S4 C and D), it was very strong when comparing broader functional groups of genes (Gene Ontology categories, Fig. 2C and SI Appendix, Fig. S7), reflecting the partitioning of gene functions among GBM classes (5, 28).
To better characterize genome-wide GBM disparity across samples, we computed the sample-specific GBM class difference (“GBMcd”) as difference in mean log2(MBD-seq counts) between highly and lowly methylated gene classes (Fig. 2 D and E). GBMcd was significantly different among genets (linear model R2 = 0.68, P = 0.00013) and aligned nearly perfectly with the first principal coordinate of GBM variation for the whole experiment, explaining 33% of total variation (Fig. 2F). For comparison, the next principal coordinate explains only 8% of variation.
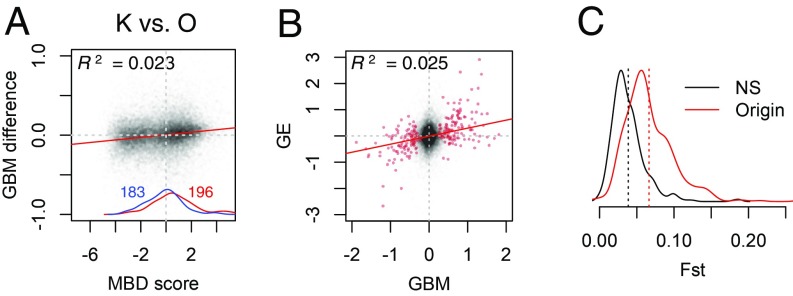
If the specific plastic changes in GBM that we have observed were contributing to fixed GBM differences between populations, perhaps via transgenerational inheritance, stable origin-specific differences (i.e., differences between all Keppel-origin and all Orpheus-origin fragments, irrespective of the site of transplantation) would be similar to plastic changes observed when transplanting a coral from Orpheus to Keppel (Fig. 2A). Stable GBM differences did follow a weak “seesaw” pattern resembling a plastic response but were in the opposite direction (Fig. 3A), with several gene ontology categories (notably, transcription factors) significantly regulated in the opposite way (SI Appendix, Fig. S7). Perhaps a better match would be observed if plastic changes were integrated over a longer period of time or sampled at some other, more critical, time point. Still, it is notable that the majority of genes that showed a significant GBM difference between populations were in the middle of the MBD-score range (Fig. 3A, red and blue density curves), displayed positive correlation between GBM and GE differences (Fig. 3B), and were associated with elevated FST (Fig. 3C), all of which is unlike genes involved in plastic response (Fig. 2 A and C and SI Appendix, Figs. S4 C and D and S8). While these results do not rule out the possibility of transgenerational inheritance of plastic GBM changes, they do indicate that the mechanism that gave rise to the main bulk of fixed GBM differences between our populations was not related to the mechanism of plasticity and was most likely genetic divergence. We checked whether genes with significant GBM differences between populations demonstrated correlated differences in CpG content but found no association (SI Appendix, Fig. S9), which suggests between-population variation in intensity of methylation rather than CpG polymorphism. Stronger GBM might be accumulating in more highly expressed alleles, perhaps over evolutionary timescales, which would explain the positive association between GBM and GE divergence (Fig. 3C).
Fig. 3.
Fixed differences in GBM and GE between populations, measured as differences between all Keppel-origin and all Orpheus-origin fragments irrespective of the site of transplantation. (A) Log2-fold differences in GBM plotted against MBD score for each gene. Red and blue curves by the x axis are density plots of significantly up- and down-methylated genes, numbers of which are given in corresponding color. (B) Correlation between fixed GBM and fixed GE log2-fold differences. Red circles are genes significantly different in GBM at the 10% false discovery rate level. (C) Density plot of between-population FST for genes showing significant fixed difference in GBM (Origin, red line) compared with 500 randomly chosen nonsignificant genes (NS, black line). Dashed vertical lines mark the mean of each dataset. All correlations with reported R2 are significant at the P < 0.0001 level.
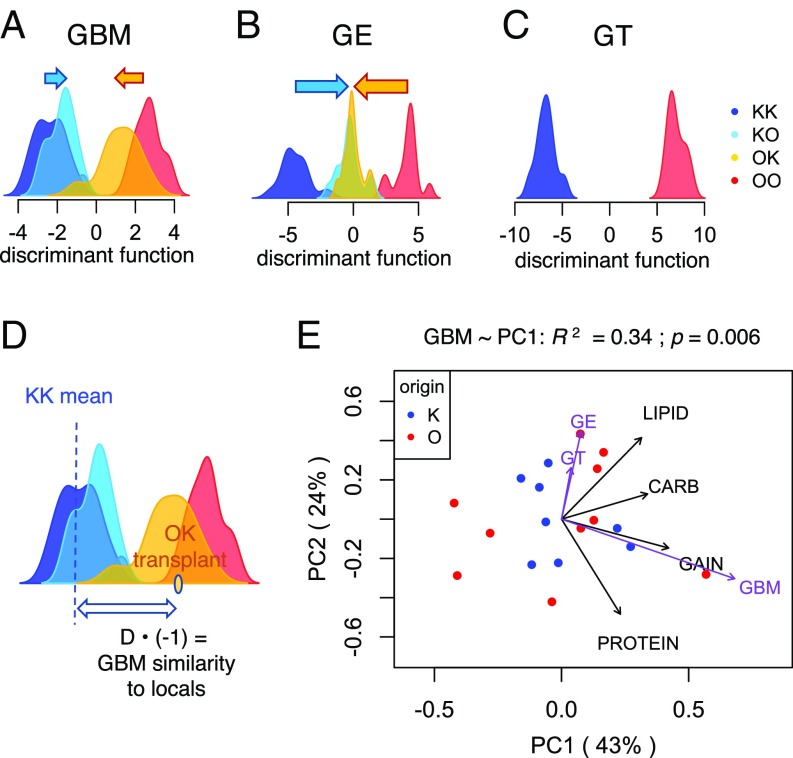
To see whether plastic GBM changes were related to acclimatization (rather than, for example, overall stress in response to transplantation) we used discriminant analysis of principal components (DAPC) (Fig. 4 A–C) to test whether GBM similarity to the local population (Fig. 4D) predicted the transplanted fragment’s fitness in the new environment. The resulting “GBM similarity to locals” was highly correlated with the first principal component of measured fitness proxies, the most strongly with weight gain (Fig. 4E). This correlation held both before and after (as shown on Fig. 4E) controlling for overall higher fitness of Orpheus corals transplanted to Keppel. This result indicates that a coral’s GBM profile reflects adaptation and acclimatization to local environmental conditions. Interestingly, the same analysis performed for GE and genotypes (GTs) failed to reveal their relationship with fitness (Fig. 4E). One explanation of this could be the difference in timescales on which the effects of GE and GT on fitness would be detectable: Compared with the 3-mo length of our experiment, GE might be linked to fitness on shorter timescales, and GTs on longer timescales.
Fig. 4.
Relationship between GBM, GE, and GT with fitness proxies in corals transplanted into a novel environment. (A–C) Density plots of discriminant function values for each variable. The functions were set to distinguish between coral fragments in their native environment (KK vs. OO) and applied to transplanted fragments (KO and OK) to quantify their adaptive plasticity (ability to shift toward local values). Arrows indicate mean plasticity. (D) Calculation of “similarity to locals” for each transplanted fragment. (E) Principal components biplot of fitness proxies in fragments transplanted to another reef, corrected for the overall higher fitness of Orpheus corals transplanted to Keppel. Fit of similarity to locals for GBM, GE, and GT onto this ordination is shown as purple vectors scaled by correlation coefficient. Only GBM fit is statistically significant.
Association between plastic GBM changes and GE changes (Fig. 2) prompts the question, Which one is likely to be the leading cause? Unfortunately, it cannot be conclusively resolved with our data. The magnitude of GBM changes is markedly lower than that of changes in GE (Fig. 2 A and B and SI Appendix, Fig. S4 A and B), which suggests that GE might be the primary driver. However, since GBM changes consist predominantly of adjustment of genome-wide GBM disparity (Fig. 2 E and F), it seems more plausible that they are due to modulation of some central DNA methylation mechanism rather than to collective feedbacks from transcription of individual genes. That said, one interesting possibility is that GBM and transcription influence each other in a negative feedback loop: Elevated transcription leads to higher GBM, but GBM is mildly inhibitory to transcription. This could explain the opposing relationships with transcription observed for plastic (Fig. 2 A–C) and fixed (Fig. 3B) GBM differences.
If GBM disparity change is indeed the primary response to the environment, its functional role could be the control of genome-wide balance between expressions of two broad gene classes: lowly methylated environmentally responsive (“problem-solving”) genes and highly methylated housekeeping genes. Notably, in the higher-quality environment of Keppel Island, where corals were able to attain higher fitness (Fig. 1D), GBM change was associated with suppression of the environmentally responsive genes and up-regulation of housekeeping genes (Fig. 2A and SI Appendix, Fig. S7). Conversely, at the low-quality Orpheus Island the balance shifted the other way, from housekeeping to environmental responsiveness (Fig. 2B). We propose that mediating this coarse adjustment of GE in response to the overall quality of the environment is the ecological function of GBM. Given the similarity of coral GBM to that of other animals (5, 9), this function might be generalizable to other animal phyla.
Materials and Methods
Sample Sizes.
The experiment started with 15 colonies from each of the two sites, which were halved and transplanted, resulting in a total of 60 fragments distributed across four study groups of 15 samples each: two “natives” (KK and OO), and two “transplants” (OK and KO, Fig. 1C). Although not all original samples were successfully analyzed using all approaches (SI Appendix, Table S1), in the end each of these four groups included 11 fragments representing unique coral GTs and paired by GT between “home” and “away” groups that were analyzed for GBM, GE, GT, and fitness proxies.
MBD-seq Library Preparation.
DNA was isolated from adult holobiont tissue using dispersion buffer (4 M guanadine thiocyanate, 30 mM sodium citrate, 30 mM β-mercaptoethanol) followed by phenol chloroform purification and a final cleanup with a Zymo Genomic DNA Clean and Concentrator-10 kit (no. D4011). Genomic DNA was sheared using a Misonix Sonicator 3000 to a size range of ∼200–800 bp checked by gel electrophoresis. Enrichment reactions were performed using the MethylCap kit (Diagenode no. C02020010) with an initial input of 2 μg sheared DNA per reaction. The methylated fraction was eluted from the capture beads in a single step, using High Elution Buffer. Library preparation using a NEBNext DNA library Prep Master Mix Set (E6040L) and sequencing on an Illumina HiSeq4000 were performed at the University of Texas Genome Sequencing and Analysis Facility. Our total sample size was n = 44 (22 colonies divided in half, giving 11 samples per treatment group). For the majority of these we sequenced only the enriched library eluted from the capture beads. Fold coverages from these captured libraries were used to estimate relative differences in GBM between samples. For a subset of 12 of the 44 samples, we sequenced both the captured and flow-through fractions. Fold differences between these captured and flow-through libraries were used to estimate absolute levels of methylation across genes. We did this for only a subset of samples because we were primarily interested in relative differences in GBM between groups. As relative differences could be reasonably well assessed without sequencing the flow-through (SI Appendix, Fig. S2), we chose to focus our sequencing resources on increasing sample size rather than more thorough estimates of absolute methylation levels.
TagSeq Data Processing.
Transcription was quantified using TagSeq (25, 29). The TagSeq reads were downloaded from the Short Read Archive database (accession no. SRP049522; ref. 28) and mapped against the A. digitifera genome using Short-Read Mapping Package (SHRiMP; ref. 30). Mapped reads overlapping annotated coding sequences were counted using the intersection-nonempty method in the Python framework to analyze high-throughput sequencing data (HTSeq, version 0.6.1p1; ref. 31). Normalization of raw counts and statistical analyses were performed using DESeq2 (27).
MBD Score and GBM Gene Classes.
For 12 samples (3 per each experimental group) we quantified absolute levels of GBM as the log2 fold difference in coverage between captured and flow-through DNA fractions while controlling for genotype, as described in ref. 9. These values were used as gene-specific MBD scores throughout the study. As expected based on previous studies (5, 9), MBD scores were bimodally distributed (SI Appendix, Fig. S1A) and negatively correlated with CpGo/e (Fig. 1D), providing genome-wide validation for our MBD-seq technique. The MBD-score threshold separating the two clouds of high point density in the scatter plot in Fig. 2D was used to delineate the two gene classes for computation of the GBMcd for each sample (Fig. 2 E and F).
Assessing Variation in GBM and Transcription.
Normalization and statistical analyses of both TagSeq and MBD-seq reads were performed with DESeq2 (27). For MBD-seq, the data for this analysis were in the form of per-gene sums of MBD-captured read counts (note that this is different from MBD score, which is a gene-specific characteristic reflecting mean methylation level, derived through comparison of MBD-captured and flow-through fractions). To assess effects of transplantation, we compared corals that were moved away vs. their clonal counterparts that were left at home (i.e., OK vs. OO for effect of transplantation from Orpheus to Keppel, and KO vs. KK for effect of transplantation from Keppel to Orpheus). For these tests, we used only fragments that were paired by genet across sites and included genet as a covariate within the model. To test for fixed population-specific effects that did not depend on the site of transplantation, we compared all Keppel-origin fragments (KK and KO) to all Orpheus-origin fragments (OO and OK), using all available data. For TagSeq, only genes with mean read count ≥3 were considered for analysis (19,706 genes), and for MBD-seq we chose genes with mean read count ≥20 (27,084 genes).
Variation in GE and GBM was explored using principal coordinate analysis (function capscale, R package vegan; ref. 32) based on Manhattan distances between individuals, which correspond to the sum of all log-fold-differences across all genes. Significance of GT, origin, and transplant effects was tested using multivariate ANOVA with 999 permutations (function adonis, R package vegan).
DAPC.
GBM data were further analyzed using DAPC implemented in the R package adegenet (33), following the procedures outlined in ref. 34. DAPC is a multivariate analysis method designed to identify between-group variation while neglecting within-group variation. We used this method to distill our multivariate MBD-seq dataset into single axes that maximized discrimination between natives from the two experimental sites (KK and OO samples). The result was a discriminant axis contrasting the GBM variation of the two native populations—with one pole designating native Keppel-like GBM patterns and the opposite pole designating native Orpheus-like patterns. The function was then applied to data from the transplanted samples, so that their positions along the discriminant axis described their similarity in GBM patterns to those of the contrasted native populations. It seems plausible that, due to either natural selection or plasticity, native corals would possess patterns optimal for their particular site. If this is true, then transplants with DAPC values closer to those of local corals would be expected to show improved fitness proxies (e.g., samples transplanted to Keppel with DAPC values closer to the “Keppel-like” pole of the discriminant axis would be expected to show higher fitness proxies and vice versa for samples transplanted to Orpheus). “DAPC similarity” values describing each transplant’s proximity along the discriminate axis to natives of its transplantation site (Fig. 4D) were computed by taking the absolute value of the difference between the transplants’ DAPC values and the mean value for natives of their respective transplant sites, converting these distances into z scores, and multiplying them by −1. The DAPC similarity value for sample X was
where, is its distance along the discriminant axis between sample X and the mean DAPC value for natives of its transplantation site, is the mean absolute distance for all transplants, and σ is the SD of absolute distance for all transplants. Correlations of these values with fitness characteristics of transplanted fragments (Fig. 4E) were computed using the function envfit [R package vegan (32)].
Data and Scripts Availability.
TagSeq, MBD-seq, and amplicon-bisulfite sequencing datasets are available through the NCBI Short Read Archive (project accession no. SRP049522). Scripts and traits data are available in a GitHub repository (https://github.com/z0on/coral_GBM_RecipTrans).
Other Methods.
SI Appendix, Supplementary Methods includes details of the transplantation experiment, fitness proxy measurements, annotation of repeated elements in the genome, MBD-seq read processing, validation of MBD-seq data with bisulfite sequencing of selected genes, analysis of genetic differentiation between populations, and determination of dominant Symbiodinium clades based on MBD-seq data.
Supplementary Material
Acknowledgments
This project relied on computational resources of the Texas Advanced Computing Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: TagSeq, MBD-seq, and amplicon-bisulfite sequencing datasets have been deposited in the NCBI Short Read Archive (project accession no. SRP049522) on February 28, 2018. Scripts and traits data have been deposited in a GitHub repository (https://github.com/z0on/coral_GBM_RecipTrans) on November 1, 2018.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813749115/-/DCSupplemental.
References
- 1.Zilberman D. An evolutionary case for functional gene body methylation in plants and animals. Genome Biol. 2017;18:87. doi: 10.1186/s13059-017-1230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bewick AJ, Schmitz RJ. Gene body DNA methylation in plants. Curr Opin Plant Biol. 2017;36:103–110. doi: 10.1016/j.pbi.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francioli LC, et al. Genome of the Netherlands Consortium Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarda S, Zeng J, Hunt BG, Yi SV. The evolution of invertebrate gene body methylation. Mol Biol Evol. 2012;29:1907–1916. doi: 10.1093/molbev/mss062. [DOI] [PubMed] [Google Scholar]
- 6.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neri F, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 8.Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon GB, Bay LK, Matz MV. Evolutionary consequences of DNA methylation in a basal metazoan. Mol Biol Evol. 2016;33:2285–2293. doi: 10.1093/molbev/msw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model Aiptasia. Sci Adv. 2018;4:eaat2142. doi: 10.1126/sciadv.aat2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts SB, Gavery MR. Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front Physiol. 2012;2:116. doi: 10.3389/fphys.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann GE. Ecological epigenetics in marine metazoans. Front Mar Sci. 2017;4:4. [Google Scholar]
- 13.Feil R, Fraga MF. Epigenetics and the environment: Emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 14.Yan H, et al. DNA methylation in social insects: How epigenetics can control behavior and longevity. Annu Rev Entomol. 2015;60:435–452. doi: 10.1146/annurev-ento-010814-020803. [DOI] [PubMed] [Google Scholar]
- 15.Liew YJ, et al. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci Adv. 2018;4:eaar8028. doi: 10.1126/sciadv.aar8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gugger PF, Fitz-Gibbon S, Pellegrini M, Sork VL. Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol Ecol. 2016;25:1665–1680. doi: 10.1111/mec.13563. [DOI] [PubMed] [Google Scholar]
- 17.Keller TE, Lasky JR, Yi SV. The multivariate association between genomewide DNA methylation and climate across the range of Arabidopsis thaliana. Mol Ecol. 2016;25:1823–1837. doi: 10.1111/mec.13573. [DOI] [PubMed] [Google Scholar]
- 18.Hu J, Barrett RDH. Epigenetics in natural animal populations. J Evol Biol. 2017;30:1612–1632. doi: 10.1111/jeb.13130. [DOI] [PubMed] [Google Scholar]
- 19.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–1241. [Google Scholar]
- 20.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 21.Shinzato C, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- 22.Lønborg C, et al. 2016. Marine Monitoring Program: Annual Report for Inshore Water Quality Monitoring 2014 to 2015 (Australian Institute of Marine Science and JCU TropWATER, Townsville, Australia)
- 23.Matz MV, Treml EA, Aglyamova GV, Bay LK. Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral. PLoS Genet. 2018;14:e1007220. doi: 10.1371/journal.pgen.1007220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Oppen MJH, Peplow LM, Kininmonth S, Berkelmans R. Historical and contemporary factors shape the population genetic structure of the broadcast spawning coral, Acropora millepora, on the Great Barrier Reef. Mol Ecol. 2011;20:4899–4914. doi: 10.1111/j.1365-294X.2011.05328.x. [DOI] [PubMed] [Google Scholar]
- 25.Meyer E, Aglyamova GV, Matz MV. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol Ecol. 2011;20:3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x. [DOI] [PubMed] [Google Scholar]
- 26.LaJeunesse TC, et al. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 2018;28:2570–2580.e6. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon GB, Bay LK, Matz MV. Bimodal signatures of germline methylation are linked with gene expression plasticity in the coral Acropora millepora. BMC Genomics. 2014;15:1109. doi: 10.1186/1471-2164-15-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohman BK, Weber JN, Bolnick DI. Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol Ecol Resour. 2016;16:1315–1321. doi: 10.1111/1755-0998.12529. [DOI] [PubMed] [Google Scholar]
- 30.Rumble SM, et al. SHRiMP: Accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
- 33.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenkel C, Matz MV. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat Ecol Evol. 2016;1:0014. doi: 10.1038/s41559-016-0014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.