Significance
Enterovirus D68 (EV-D68) is an emerging pathogen that primarily causes childhood respiratory infections and is linked to neurological diseases. It was unclear how the virus uncoats and delivers its genome into a host cell to establish viral replication. Using high-resolution cryoelectron microscopy, we showed that acid induces structural rearrangements of EV-D68 to initiate genome release from the virus. Structural analyses delineated a viral uncoating pathway that involves multiple distinct conformational states. Particularly, the structure of a previously unknown uncoating intermediate enabled the identification of potential molecular determinants that facilitate EV-D68 uncoating. These results advance the knowledge of cell entry of EV-D68 and open up possibilities for developing antiviral therapeutics that impede structural rearrangements of the virus.
Keywords: enterovirus D68, cryoelectron microscopy, virus uncoating, conformational changes, acidification
Abstract
Enterovirus D68 (EV-D68) belongs to a group of enteroviruses that contain a single positive-sense RNA genome surrounded by an icosahedral capsid. Like common cold viruses, EV-D68 mainly causes respiratory infections and is acid-labile. The molecular mechanism by which the acid-sensitive EV-D68 virions uncoat and deliver their genome into a host cell is unknown. Using cryoelectron microscopy (cryo-EM), we have determined the structures of the full native virion and an uncoating intermediate [the A (altered) particle] of EV-D68 at 2.2- and 2.7-Å resolution, respectively. These structures showed that acid treatment of EV-D68 leads to particle expansion, externalization of the viral protein VP1 N termini from the capsid interior, and formation of pores around the icosahedral twofold axes through which the viral RNA can exit. Moreover, because of the low stability of EV-D68, cryo-EM analyses of a mixed population of particles at neutral pH and following acid treatment demonstrated the involvement of multiple structural intermediates during virus uncoating. Among these, a previously undescribed state, the expanded 1 (“E1”) particle, shows a majority of internal regions (e.g., the VP1 N termini) to be ordered as in the full native virion. Thus, the E1 particle acts as an intermediate in the transition from full native virions to A particles. Together, the present work delineates the pathway of EV-D68 uncoating and provides the molecular basis for the acid lability of EV-D68 and of the related common cold viruses.
Enteroviruses (EVs) are a genus of single-stranded RNA viruses with a positive-sense RNA genome surrounded by an icosahedral capsid shell (1). EVs from seven species, EV-A, EV-B, EV-C, EV-D, rhinovirus (RV) A, RV-B, and RV-C, are causative agents of a variety of human diseases (2, 3). These viruses include polioviruses, coxsackieviruses, RVs, EV-A71, and EV-D68. Among these, EV-D68 is a globally emerging human pathogen that mainly causes respiratory infections in young children (4–7). It has also been closely linked to neurological diseases (7–10). The development of effective vaccines and antiviral treatments against EV-D68 has been difficult due to limited knowledge of the molecular mechanisms of virus infection. In particular, despite recent progress in studying receptor-dependent cell entry of EV-D68 (11–16), it remains unclear how the virus uncoats and delivers its genome into host cells.
Enterovirus capsids are assembled from 60 copies of viral proteins VP1, VP2, VP3, and VP4 with pseudo T = 3 icosahedral symmetry (17, 18). The VP1, VP2, and VP3 subunits, each having an eight-stranded β-barrel “jelly-roll” fold, form the icosahedral shell with an outer diameter of ∼300 Å (17, 18). The capsid inner surface is decorated by 60 copies of VP4, together with the N termini of VP1, VP2, and VP3. During EV infection, host factors, such as cellular receptors and endosomal acidification, trigger EV uncoating by altering the capsid structure (19, 20). The uncoating process has been proposed to proceed via a structural intermediate, the A (altered) particle (21–23), characterized by loss of VP4 and by externalization of the VP1 N-terminal residues (21, 24–27). These structural changes precede viral penetration of the membranes of intracellular compartments (21, 28, 29). This facilitates genome release from the A particles into the cytosol of host cells and the production of emptied particles (22, 30, 31).
EV-D68 shares features, including acid lability, with rhinoviruses that are respiratory viruses from the species RV-A, RV-B, and RV-C (4, 12). However, sequence comparisons have shown that EV-D68 is more closely related to members of the species EV-A, EV-B, and EV-C (4). These viruses, as exemplified by polioviruses, are resistant to the acidic environment of the human gastrointestinal tract (1). It has been established that acid treatment of RVs in vitro causes structural alterations of the virus and often leads to the formation of uncoating intermediates, including A particles and emptied particles (32–34). Moreover, endosomal acidification acts as an important cue for RV uncoating in host cells (19, 22, 35). By analogy with RVs, it is probable that acid initiates uncoating of EV-D68.
Here we report that an EV-D68 isolate from the 2014 outbreak in the United States is particularly sensitive to acid. Acid treatment causes dramatic conformational changes of full native virions to form A particles. Moreover, cryoelectron microscopy (cryo-EM) analyses of the virus at neutral pH and following acid treatment show that EV-D68 uncoating proceeds via multiple structural intermediates. These include a previously unknown structural state, the expanded 1 (E1) particle, which retains ordered VP1 N-terminal residues and ordered VP4. Weak interpentameric interactions and the absence of a pocket factor are among potential structural determinants that facilitate the conversion of the virus to the A-particle state via the E1-particle intermediate. These observations provide a structural basis for the uncoating process and acid sensitivity of EV-D68 and related viruses.
Results and Discussion
Current EV-D68 Strains Are Acid-Sensitive.
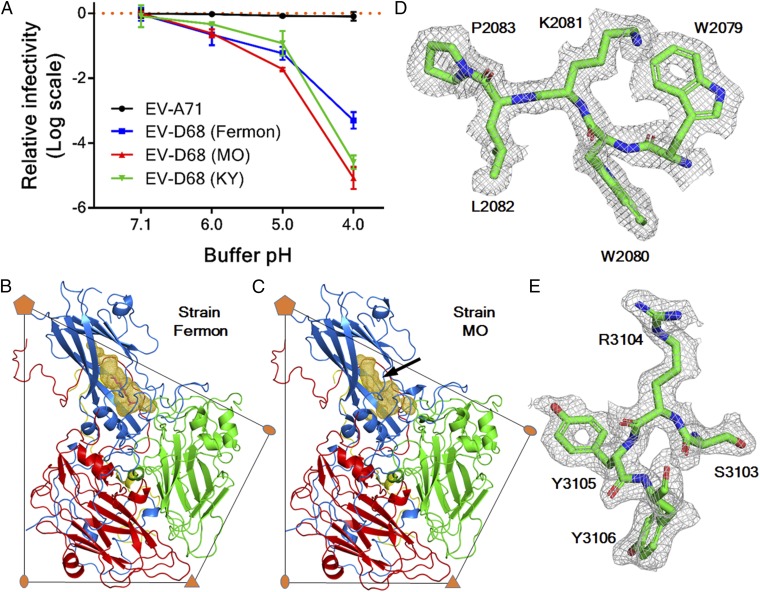
The effect of acid treatment on virus infectivity was examined using a plaque assay. It was found that EV-A71 is acid-resistant and retained infectivity at low pH (pH 4 to 6) (Fig. 1A). In contrast, the tested EV-D68 strains are acid-labile, including the prototype Fermon strain and two strains (US/MO/14-18947 and US/KY/14-18953) from the 2014 outbreak. Hereafter, US/MO/14-18947 and US/KY/14-18953 are referred to as MO and KY, respectively. Among these strains, strain MO is the most sensitive to acid (Fig. 1A).
Fig. 1.
Structure of the acid-sensitive EV-D68 strain MO. (A) Plot of changes of viral infectivity in logarithm scale as a function of buffer pH. Data are presented as mean ± SD of triplicate measurements. The viruses were treated using buffer with a series of pH values and assayed to determine viral titers. (B and C) One protomer of the EV-D68 capsid is colored blue (VP1), green (VP2), red (VP3), and yellow (VP4). The volume of the VP1 hydrophobic pocket is colored gold. A pocket factor (magenta) is present in strain Fermon (B) but absent in strain MO (C). A black arrow indicates where the collapse of the VP1 pocket of strain MO occurs. (D and E) Typical map densities of strain MO at 2.2-Å resolution with the fitted atomic model.
Cryoelectron micrographs showed that there was more than 95% of full particles in a purified sample of strain MO (prep A), which was prepared employing two rounds of density-gradient centrifugation (Materials and Methods). The virus structure (dataset A_Native) was determined at 2.2-Å resolution using 16,021 particles (Fig. 1 and SI Appendix, Fig. S1 and Table S1). In many EV structures, a hydrophobic pocket within the VP1 jelly roll accommodates a fatty acid-like molecule (the “pocket factor”) that regulates viral stability (36–38). In strain MO, the pocket factor was found to be absent from the VP1 hydrophobic pocket, which is partially collapsed (Fig. 1). Compared with strain Fermon, the structure of strain MO shows that Ile1217 (numbering is based on the amino acid sequence of strain Fermon throughout this paper) in the VP1 GH loop moves into the pocket with the Cα atom shifted by 2.0 Å. Such a shift would cause clashes with a pocket factor were this factor bound in the pocket (SI Appendix, Fig. S1).
Acid Induces EV-D68 Uncoating.
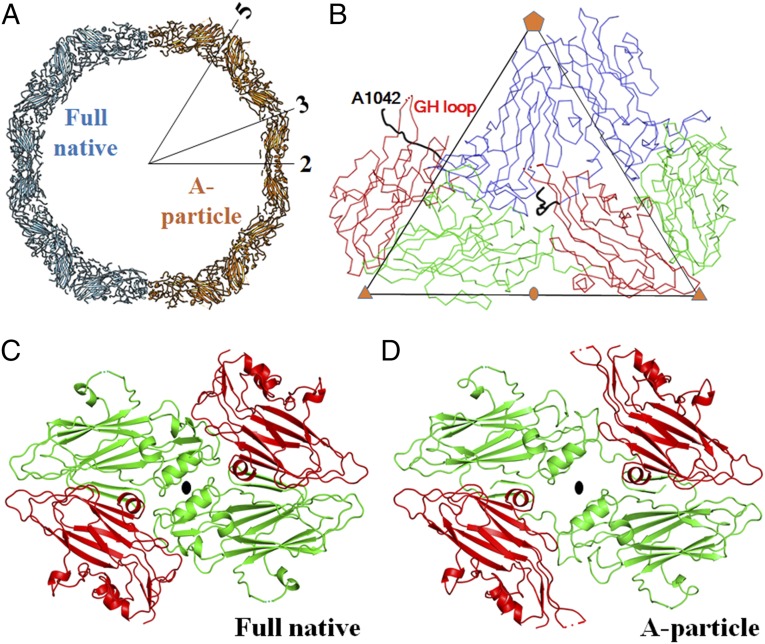
Consistent with the low stability of strain MO, a virus sample (prep B), which was not as intensively purified as prep A, yielded a mixture of full and empty particles within one fraction of about 0.5 mL after density-gradient centrifugation (SI Appendix, Fig. S2A). The full and empty particles refer to those having inner densities at their centers and those lacking inner densities at their centers, respectively. To study acid-triggered structural changes of the virus, prep B was treated with either a pH 5.5 buffer (dataset B_RT_Acid) or a neutral pH buffer (dataset B_RT_Neu) at room temperature (RT) for 20 min. Two-dimensional (2D) classification of particle images (dataset B_RT_Neu) showed the presence of full and empty particles with a ratio of about 2.0:1 (full:empty) (SI Appendix, Fig. S2B). In contrast, 2D class averages of particle images in dataset B_RT_Acid showed the presence of empty particles and a particle form that contains the genome but exhibits a thinner capsid shell than full native virions (SI Appendix, Fig. S2 C and D). The ratio between the new form of particles and empty particles was about 1.7:1, suggesting that the acid treatment had mostly induced the conversion of full native virions to the new particle form. Icosahedral reconstructions of the new form of particles (3,708 particles) and empty particles (2,150 particles) in dataset B_RT_Acid were determined to 3.3- and 3.8-Å resolution, respectively (SI Appendix, Fig. S3 and Table S1). These two forms of particles are both expanded by about 11 Å in diameter relative to full native virions. They also show significantly different capsid structures from full native virions, with a root-mean-square deviation (rmsd) of 4.6 Å (empty particles) and 5.7 Å (new form of particles) when aligning icosahedral symmetry axes (SI Appendix, Table S2). The rmsd between any two structures was calculated based on aligning equivalent Cα atoms unless otherwise specified. Moreover, the VP1 N-terminal residues 1001 to 1041 and a majority of VP4 residues, which are well-ordered in the map of full native virions, become disordered (or missing) in the map of the new particle form. Thus, the new particle form represents the A particle, a proposed uncoating intermediate known to exist in other EVs (25, 26, 34). The capsid structure of empty particles resembles A particles with an rmsd of 0.8 Å when aligning icosahedral symmetry axes (SI Appendix, Table S2), as has been previously reported (26). These empty particles include a small portion of VP0-containing native empty particles and a large portion of emptied particles that are formed after full native virions have released the viral genome (SI Appendix, Fig. S4). The VP2-containing emptied particles have an expanded capsid structure, whereas VP0-containing native empty particles often exhibit nearly the same capsid structure as full native virions do (39, 40).
To mimic the environment for virus uncoating in host cells, prep B was treated with a pH 5.5 (late endosomal pH) buffer at physiological temperature (33 °C) for 20 min (dataset B_33_Acid). Similar to the aforementioned observation in the case of room temperature incubation, A particles and emptied particles were present in dataset B_33_Acid (SI Appendix, Table S3). The cryo-EM structures of A particles (23,082 particles) and emptied particles (19,325 particles) were determined to 2.7- and 2.9-Å resolution, respectively (SI Appendix, Fig. S5 and Table S1). When full native virions were converted into A particles, a VP2 helix (residues 2091 to 2098) and its counterpart in an icosahedral twofold related VP2 molecule were shifted away from this twofold axis, opening up roughly rectangularly shaped pores around the twofold axes on A particles (with a pore size of about 9 × 18 Å) (Fig. 2). Similar pores were observed on emptied particles (with a size of about 8 × 29 Å). These pores might function as sites where the genomic RNA exits, partially because a single-stranded RNA, assuming no secondary structures, has a size of slightly less than 8 × 10 Å when looking in the direction normal to the planar aromatic bases of the RNA. More importantly, the VP1 N-terminal residues 1042 to 1051 reside in the capsid interior of full native virions. In contrast, these residues are displaced by an rmsd of 23.6 Å in A particles such that residues 1044 to 1051 traverse the capsid shell and that residue 1042 lies on the particle outer surface (Fig. 2). The VP3 GH loop, residues 3170 to 3188 excluding the disordered residues 3178 to 3183, at the particle exterior is rearranged (rmsd 9.6 Å) to adopt an extended conformation and interact with the VP1 N-terminal residues (Fig. 2). These changes result in the externalization of the VP1 amphipathic helix (disordered in the A-particle structure) through a pore around the quasi-threefold axis at the base of the “canyon” (18). The VP1 amphipathic helix (about 25 amino acids at the N terminus) was previously shown to insert into host cell membranes (21, 41), as do also VP4 molecules (29).
Fig. 2.
Acid-induced structural rearrangements of EV-D68 facilitate virus uncoating. (A) Structural comparison of the full native virion (pH 7.2) and A particle (formed at pH 5.5). For each of these two structural states, a half-capsid is represented as a slab about 20-Å thick. The numbers 5, 3, and 2 indicate an icosahedral fivefold axis, threefold axis, and twofold axis, respectively. (B) The Cα backbone representation of two neighboring protomers of the A-particle structure is colored blue (VP1), green (VP2), and red (VP3). The VP1 N-terminal residues 1042 to 1052, which are externalized through a quasi-threefold axis, are highlighted in black. (C and D) Compared with full native virions (C), pores are formed around the icosahedral twofold axes in A particles (D).
To investigate the scope of acidic conditions suitable for initiating virus uncoating, cryo-EM analysis was performed on the more homogeneous prep A using a variety of pH conditions, incubation times, and incubation temperatures (Materials and Methods, Table 1, and SI Appendix, Fig. S6 and Table S4). A particle population consisting of nearly exclusively A particles was observed after treating the virus for ∼3 min at pH 6.0 and at RT. This demonstrated a nearly complete conversion of full native virions into A particles in a fast reaction process. In contrast, the production of A particles was partially completed when increasing the pH to 6.5 or reducing the reaction time to ∼30 s. In agreement with these results and with the aforementioned porous structure of A particles, particle stability thermal release (PaSTRy) assays showed that upon acid treatments, the viral genome of EV-D68 strain MO but not EV-A71 became more exposed to RNA-binding dye molecules at low temperatures (SI Appendix, Fig. S7 A–C). Furthermore, consistent with the disordering (or absence) of VP4 in icosahedral reconstructions of A particles, acid-treated particles of strain MO had a significantly reduced level of VP4 than nontreated particles did (SI Appendix, Fig. S7D). Thus, structural and functional characterizations show that acid-induced structural changes facilitate EV-D68 uncoating.
Table 1.
Effect of acid treatments on the conformational states of EV-D68 strain MO
| Dataset | A_Acid_1 | A_Acid_2 | A_Acid_3 | A_Acid_4 |
| Temperature | 4 °C | RT* | RT* | RT* |
| Duration† | 3 min | 3 min | 30 s | 3 min |
| pH | 6.5 | 6.5 | 6.0 | 6.0 |
| No. of full particles | 32,972 | 68,112 | 100,113 | 19,798 |
| No. of empty particles | 891 | 2,389 | 4,687 | 184 |
| Percentage of full particles | 97.4 | 96.6 | 95.5 | 99.1 |
| Percentage of full native virions‡ | 76.6 ± 0.4 | 40.2 ± 0.2 | 37.4 ± 0.4 | 3.5 ± 0.1 |
| Percentage of A particles‡ | 7.3 ± 0.2 | 56.4 ± 0.2 | 58.1 ± 0.4 | 95.6 ± 0.1 |
| Percentage of E1 particles‡ | 13.5 ± 0.5 | —§ | —§ | —§ |
Room temperature.
Approximate time from the initiation of acid treatment to the state of being quenched by freezing.
Percentage among all particles. Data are presented as mean ± SD; 3D classifications were performed six times.
Not identified.
Identification of Multiple Structural Intermediates.
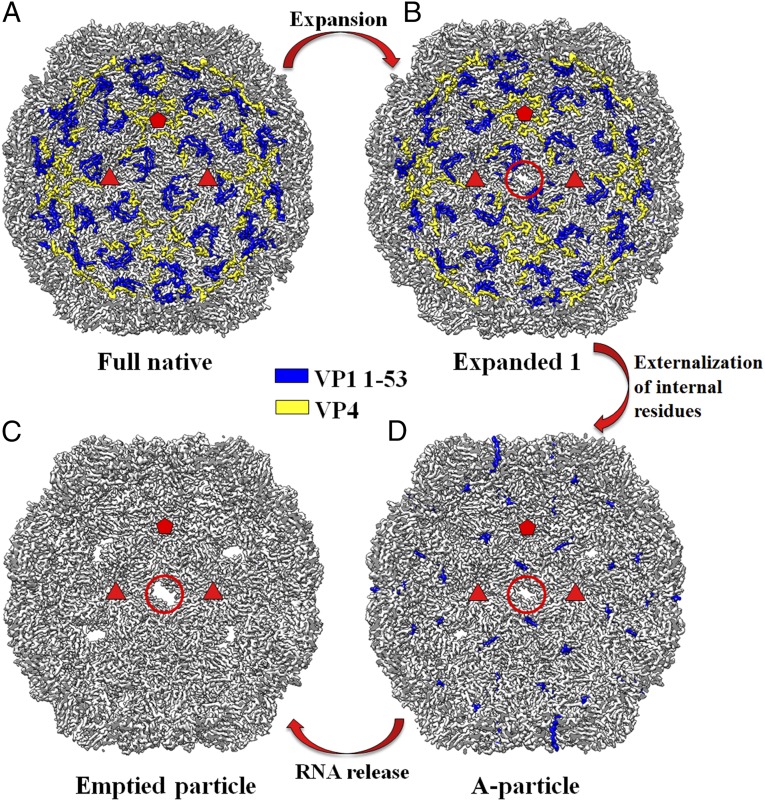
At neutral pH, prep B contains a heterogeneous particle population as mentioned above. To analyze the sample heterogeneity at neutral pH in detail, 2D classification and subsequently 3D classification of particle images (dataset B_4_Neu) resulted in six different structural states (Materials and Methods and SI Appendix, Fig. S8 and Tables S3 and S5). These structures were determined at 3.2- to 3.3-Å resolution (SI Appendix, Fig. S9 and Table S5). They differ mostly in particle size and in internal regions, including the VP1 N-terminal residues 1001 to 1053 as well as VP4. The two predominant states are full native virions (52% of all particles) and emptied particles (20% of all particles) (SI Appendix, Table S5). This observation suggests that a portion of full native virions might have uncoated to produce emptied particles during virus preparation. Consistent with this prediction, two uncoating intermediates have also been identified from the whole particle population. One intermediate was found to be A particles (about 9%), whereas the other represents a previously undescribed structural state (about 5%), named here the “expanded 1 particle.” Unlike A particles, the VP1 N-terminal residues 1001 to 1053 (excluding the disordered residues 1016 to 1019) and VP4 residues 4030 to 4057 are ordered in E1 particles, as is the case for full native virions (Fig. 3 and SI Appendix, Fig. S10). Nevertheless, E1 particles are expanded by about 8 Å in diameter with respect to full native virions. The rmsd between these two structures when aligning icosahedral symmetry axes is 4.4 Å, whereas the rmsd between the structures of A particles and E1 particles is 3.3 Å (SI Appendix, Table S6). Thus, distinct structural intermediates (E1 and A) are involved in EV-D68 uncoating (Fig. 3).
Fig. 3.
EV-D68 uncoating proceeds via multiple distinct structural intermediates. A cutaway view of each of four structural states of the EV-D68 capsid during virus uncoating. Shown are the full native virion (A), expanded one particle (B), A particle (D), and emptied particle (C) when looking into the particle along an icosahedral twofold axis. The ordered regions of VP1 N-terminal residues 1001 to 1053 are colored blue. The ordered regions of VP4 are colored yellow. Two red triangles and one red pentangle outline the limit of an icosahedral asymmetric unit. A red circle indicates the presence of a pore around the icosahedral twofold axis. A detailed structural comparison of the ordered VP1 N-terminal residues and VP4 between full native virions and the E1 particles is shown in SI Appendix, Fig. S10. Structural differences in regions near the twofold axes between the E1 particles and A particles are illustrated in SI Appendix, Fig. S12.
In addition, two other structural states, each of which accounted for about 7% of all particles, were identified (SI Appendix, Fig. S11A). The rmsd between the full native virion and each of these two states was found to be 1.0 to 1.1 Å (SI Appendix, Table S6). These two states either lacked inner densities or had rod-like densities in the particle interior (SI Appendix, Fig. S11B). These states might be abortive products during virus assembly.
Conformational Changes of EV-D68 During Uncoating.
The identification of multiple structural states of the capsid in prep B suggests that EV-D68 uncoating proceeds via a series of conformational changes. Moreover, in vitro reconstitution of acid-initiated EV-D68 uncoating under different conditions offers a means to dissect structural changes of the virus in the authentic uncoating process in vivo triggered by endosomal acidification. Unlike the observed proportion of E1 particles (∼5%) under neutral pH as mentioned above, the E1 particles (∼14% of all particles) were captured following acid treatment of prep A at pH 6.5 and at 4 °C (Table 1). The structure of the E1 particles formed under this condition is similar to that of the E1 particles in prep B, with an rmsd of 0.5 Å between all of the equivalent Cα atoms (SI Appendix, Table S3). When increasing the reaction temperature, a significant increase of the proportion of A particles was observed, whereas the E1 particles were not identified (Table 1). Thus, it is reasonable to assume that virus uncoating is initiated by particle expansion of full native virions to form E1 particles, producing pores on the capsid. Subsequent loss of VP4 through these pores and externalization of the VP1 N termini result in A particles. Ultimately, the genomic RNA is released to yield emptied particles (Fig. 3).
Particle expansion of full native virions to E1 particles is achieved primarily through rigid-body movements of capsid proteins. The centers of mass of VP1, VP2, VP3, and VP4 are translated away from the particle center by 4.5, 3.4, 3.9, and 4.7 Å, respectively, while these proteins are rotated by 2.8°, 3.3°, 2.8°, and 3.3°, respectively. This yields a structure which has an rmsd of 1.1 Å (VP1), 1.3 Å (VP2), 1.4 Å (VP3), and 0.3 Å (VP4) compared with the E1 particle (SI Appendix, Table S7). During particle expansion, the buried surface areas between VPs within a protomer (defined as VP1, VP2, VP3, and VP4) stay roughly unchanged (SI Appendix, Table S8). This observation provides the basis for superimposing one protomer of the full native virion structure with a protomer in the E1-particle structure. VP2, VP3, VP4, and the VP1 regions distant from the fivefold axis are well-superimposable (rmsd 1.3 Å), whereas the VP1 β-barrel and loops near the fivefold axis are rearranged in a hinge-like manner with a shift of 0.9 Å and a rotation of 3.0°.
Pores with a size of about 6 × 15 Å are created around the twofold axes in E1 particles, as a result of the displacement of the VP2 residues 2091 to 2098 near the twofold axis with respect to full native virions (SI Appendix, Fig. S12). These changes impair the interactions between neighboring pentamers. More importantly, two β-strands at the VP2 N terminus (residues 2013 to 2026) in one pentamer, together with a β-strand at the VP1 N terminus (residues 1017 to 1020) and the VP3 β-strands C, H, E, and F in another pentamer, form a seven-stranded interpentamer β-sheet that spans from the capsid outer surface to the inner surface in full native virions (SI Appendix, Fig. S13A). This sheet and its symmetry-related equivalents help hold adjacent pentamers together and provide structural stability to the VP1 N termini in the capsid interior (37, 42). Particle expansion disrupts this interpentamer β-sheet and weakens the interpentamer contact in E1 particles (SI Appendix, Figs. S13B and S14). These effects are due primarily to rearrangements of VP2 residues 2012 to 2017 (rmsd 4.0 Å), VP2 residues 2026 to 2030 (rmsd 4.3 Å), and VP1 residues 1020 to 1026 (rmsd 8.8 Å) with respect to full native virions (SI Appendix, Figs. S10 and S15). Furthermore, the VP3 GH loop in E1 particles becomes partially disordered and moves outward (away from the virus center) with an rmsd of 5.6 Å (residues 3180 and 3186 to 3188) (SI Appendix, Figs. S15 and S16). These changes might precede pore opening at the base of the canyon, similar to what was previously proposed (43). Thus, structural alterations from full native virions to E1 particles not only lead to impaired pentamer–pentamer interactions (SI Appendix, Table S9) but also prime the exiting of VP4 and the externalization of the VP1 N termini.
Superposition of equivalent protomers in E1 particles and A particles shows that VP2, VP3, and the fivefold distant regions of VP1 are well-aligned, with an rmsd of 1.1 Å. However, the VP1 β-barrel and fivefold proximal loops undergo hinge-like motions with a rotation of 1.9°. The VP2 C-terminal tail (residue 2242 to the carboxyl end) at the capsid exterior in E1 particles becomes internalized in A particles (SI Appendix, Fig. S12). This generates enlarged pores around the twofold axes, facilitating the exiting of VP4 molecules (44). Moreover, the internalized VP2 C-terminal tail would clash with the N-terminal residues 1042 to 1044 of VP1 in a neighboring, fivefold related protomer if the VP1 N terminus were to stay stationary (SI Appendix, Fig. S12). Structural reorganization of the VP3 GH loop and VP1 GH loop in A particles with rmsd values of 11.5 Å (residues 3170 to 3178) and 2.7 Å (residues 1202 to 1207), respectively, with respect to E1 particles creates room near the quasi-threefold axes (SI Appendix, Figs. S16 and S17). Collectively, these conformational changes from E1 particles to A particles promote the externalization of VP1 N termini through holes at the base of the canyon (Fig. 3 and SI Appendix, Fig. S16).
The final stage of uncoating involves genome release from a specific pore around a twofold axis in A particles, generating emptied particles (45, 46). Given the high structural similarity between these two states as mentioned above, structural analyses make it difficult to identify the trigger that initiates RNA release. Nevertheless, this process has been reported to depend on interactions of A particles with host cell membranes (30, 41, 47), disruption of the secondary structures of the viral RNA (48), and electrostatic repulsion between the negatively charged RNA and negatively charged residues lining the inner capsid surface (26).
Molecular Basis for Acid Lability.
Unlike members of the EV-A, EV-B, and EV-C species that have a well-formed pocket factor with a long aliphatic chain, the VP1 hydrophobic pockets of RVs and EV-D68 either contain a pocket factor with a short aliphatic chain (RV-A2, RV-A16, and EV-D68 strain Fermon) or cannot accommodate a pocket factor at all due to the pocket being collapsed as in RV-B3, RV-B14, and EV-D68 strain MO (49). Furthermore, RV-A2, RV-A16, and RV-C15 have particularly small interaction areas at the pentamer–pentamer binding interface (SI Appendix, Table S9). In comparison with other EVs, the above-mentioned structural features of EV-D68 and RVs would lead to enhanced conformational fluctuations of the capsid (50) and promote irreversible conformational changes from the full native virion to an E1 particle. As mentioned above, the E1-particle state primes the externalization of the VP1 N termini and loss of VP4, which are major features of the A-particle state. The E1 particles were only identified under a mild reaction condition (pH 6.5 at 4 °C) but not under relatively harsh conditions (e.g., pH 6.0 to 6.5 at RT) (Table 1), suggesting the transient and unstable properties of this uncoating intermediate under physiological conditions. Low-pH conditions probably facilitate the conversion of E1 particles into A particles in three ways:
-
i)
Particle expansion from full native virions to E1 particles could lead to influx of protons through transient openings around the fivefold axes, causing conformational changes of fivefold proximal regions at the capsid interior, including VP4. Similar changes that precede the release of VP4 have been reported for RV-B14 under acidic conditions (51).
-
ii)
Protons could enter into the capsid through pores around the twofold axes and disrupt polar interactions that stabilize the VP1 N termini, which are already destabilized in E1 particles relative to full native virions. Acid is also known to modulate the conformational states of the GH loops in VP1 and VP3 around the quasi-threefold axes (51, 52). These acid-induced changes would probably drive the externalization of the VP1 N termini.
-
iii)
The E1-particle state shows partially disrupted seven-stranded interpentamer β-sheets because of rearranged VP1 and VP2 N termini. Structural rearrangements of these regions have been proposed to regulate the accessibility of interpentameric histidine residues to the acidic environment (53). This may further impair the interactions at the pentamer–pentamer interface, as is observed in A particles with respect to E1 particles (SI Appendix, Table S9).
Sequence analyses show that acid-labile enteroviruses have, on average, six more histidine residues in capsid proteins than acid-resistant enteroviruses do (SI Appendix, Table S10). In particular, within the species EV-D, the differences between EV-D68 and its acid-resistant relatives, EV-D70 (12) and EV-D94 (54), involve amino acid substitutions at 10 positions where a His is found in D68 but not in D70 or D94 (SI Appendix, Table S11). Seven out of these 10 His residues are more than 93% conserved among 469 EV-D68 strains. It is therefore probable that some of these histidine residues act as a pH sensor to regulate the acid sensitivity of EV-D68, as has previously been reported for other picornaviruses, including rhinovirus (51) and foot-and-mouth disease virus (55).
Implications for Cell Entry of EV-D68.
Previous reports have identified sialic acid (a carbohydrate moiety) and intercellular adhesion molecule 5 (ICAM-5), a sialic acid-containing glycoprotein, as cellular receptors for EV-D68 (11, 14–16). The present work indicates that endosomal acidification might serve as a trigger for EV-D68 uncoating in host cells. Sialic acid receptor binding to the Fermon strain has previously been shown to cause ejection of the pocket factor that destabilizes the virus (14), resulting in a virus that is much like strain MO because both structures show the absence of a pocket factor. The infectivity of the Fermon strain is slighted impaired at pH 6.0 (Fig. 1), suggesting that endosomal acidification alone is probably insufficient for uncoating of this strain. This observation raises the possibility that sialic acid receptor binding increases the pH threshold for inducing EV-D68 uncoating. Likewise, strain KY uncoats after incubation with soluble ICAM-5 at pH 6.0, whereas the uncoating process is less efficient when the virus is incubated without receptor molecules at pH 6.0 (16). Hence, cellular receptors (e.g., sialic acid or ICAM-5) and endosomal acidification probably assist EV-D68 entry into host cells in a synergistic manner, as has previously been proposed for other EVs (56, 57). In this way, receptor binding can prime EV-D68 uncoating, which then occurs in intracellular compartments that have a suitable environment (e.g., acidic pH) for genome release. Nevertheless, the synergistic action of cellular receptors and low pH might depend on virus strains. It was observed that unlike strain Fermon, strain MO has low stability even at physiological temperatures (SI Appendix, Fig. S7). Moreover, conversion of full native virions to A particles can take place at early endosomal pH in vitro (Table 1). Thus, endosomal acidification alone is probably sufficient for triggering uncoating of strain MO during cell entry.
In summary, cryo-EM analyses of the acid-sensitive EV-D68 have shown the involvement of multiple structural intermediates in the viral uncoating pathway. A set of high-resolution structures presented here provides the basis for developing antiviral therapeutics that would interfere with structural rearrangements of EV-D68 during cell entry by stabilizing the virus. Moreover, the identification of the expanded, E1, particles, a missing link between the full native virions and the A particles, suggests that the conformational fluctuations of enterovirus capsids may account for the differences in acid lability among enteroviruses.
Data Deposition.
The atomic coordinates of A_Native-full, B_RT_Acid-Aparticle, B_RT_Acid-Emptied, B_33_Acid-Aparticle, B_33_Acid-Emptied, B_4_Neu-Full-Native, B_4_Neu-E1, B_4_Neu-Aparticle, B_4_Neu-Emptied, B_4_Neu-Abortive1, B_4_Neu-Abortive2, and Acid_A_1-E1 have been deposited in the Protein Data Bank (ID codes 6CSG, 6CS6, 6CSA, 6CS4, 6CSH, 6CRR, 6CS3, 6CRS, 6CRU, 6CRP, 6CS5, and 6MZI). The cryo-EM maps of A_Native-full, B_RT_Acid-Aparticle, B_RT_Acid-Emptied, B_33_Acid-Aparticle, B_33_Acid-Emptied, B_4_Neu-Full-Native, B_4_Neu-E1, B_4_Neu-Aparticle, B_4_Neu-Emptied, B_4_Neu-Abortive1, and B_4_Neu-Abortive2 have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-7599, EMD-7593, EMD-7598, EMD-7589, EMD-7600, EMD-7569, EMD-7583, EMD-7571, EMD-7572, EMD-7567, and EMD-7592). The cryo-EM maps of A_Acid_1-Full-Native, A_Acid_1-Aparticle, A_Acid_1-E1, A_Acid_2-Full-Native, A_Acid_2-Aparticle, A_Acid_3-Full-Native, A_Acid_3-Aparticle, and A_Acid_4-Aparticle have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-9053, EMD-9054, EMD-9055, EMD-9056, EMD-9057, EMD-9058, EMD-9059, and EMD-9060).
Materials and Methods
Viruses.
EV-A71 (strain MY104-9-SAR-97, GenBank accession no. ABC69262.1) was provided by Jane Cardosa (Universiti Malaysia Sarawak, Sarawak, Malaysia). EV-D68 strain Fermon CA62-1 (strain Fermon, GenBank accession no. AY426531.1) was provided by M. Steven Oberste (Centers for Disease Control and Prevention, Atlanta, GA). Two EV-D68 strains, US/MO/14-18947 (strain MO, GenBank accession no. AIS73051.1) and US/KY/14-18953 (strain KY, GenBank accession no. AIS73057.1), from the 2014 outbreak in the United States, were obtained from BEI Resources, National Institute of Allergy and Infectious Diseases, National Institutes of Health. All viruses were propagated in human rhabdomyosarcoma (RD) cells (ATCC CCL-136) and stored at −80 °C.
Virus Growth and Purification.
A sample (prep A) for structure determination of the full EV-D68 virion (strain MO) was prepared in the following way (49). RD cells were infected with EV-D68 at a multiplicity of infection of about 0.01. Cells and supernatant were harvested at 3 d post infection and then separated by centrifugation (10,000 × g for 30 min at 4 oC). Cell pellets were subjected to multiple cycles of freezing and thawing followed by centrifugation to remove cell debris. The resultant supernatant was combined with the original supernatant and used for ultracentrifugation at 278,000 × g (Ti 50.2 rotor) for 2 h at 4 °C. The resultant pellets were resuspended in buffer A (250 mM Hepes, 250 mM NaCl, pH 7.5) and treated sequentially with 5 mM (final concentration throughout the treatments) MgCl2, 10 µg/mL DNase, 7.5 mg/mL RNase, 0.8 mg/mL trypsin, 15 mM EDTA, and 1% (wt/vol) sodium N-lauryl-sarcosinate. The crude virus sample was sedimented through a potassium tartrate density gradient [10 to 40% (wt/vol)]. A band in the middle of the tube was extracted and subjected to buffer exchange. The resultant sample was further purified using an iodixanol (OptiPrep; Sigma) density gradient [10 to 51% (vol/vol)] at 175,000 × g (SW 41 Ti rotor) for 2 h at 4 °C. Electron micrographs of the final sample verified the presence of more than 95% of full particles (SI Appendix, Fig. S1A).
A virus preparation (prep B) that contained a heterogeneous population of EV-D68 particles was used for structural studies at both neutral pH and acidic pH. Procedures for virus infection, sample collection, initial centrifugation, and treatment of cell pellets followed the same procedures as described above. Subsequently, polyethylene glycol 8000 (PEG8000) [40% (wt/vol) stock solution] and NaCl (powder) were added into the original supernatants (after infection) to reach a final concentration of 8% PEG8000 and about 500 mM NaCl. After low-speed agitation at 4 °C for about 6 h, the mixture was spun down. The resultant pellets were resuspended in buffer A, which was combined with the previous supernatant from the step that dealt with the cell pellets. The remaining steps were the same as mentioned above, except that the crude virus sample was purified through only one round of density-gradient centrifugation using the iodixanol gradient.
Acid Sensitivity Assay.
Four different viruses were used, including the acid-resistant EV-A71, acid-labile EV-D68 strain Fermon, and two EV-D68 isolates (strains MO and KY) from the 2014 outbreak in the United States. Purified viruses were treated with phosphate-citrate buffer (100 mM Na2HPO4, 50 mM citric acid) at pH 4.0, 5.0, 6.0, or 7.1 at 33 °C for about 45 min. The resultant samples were neutralized back to pH 7.1 to 7.2 using 400 mM Na2HPO4 and 200 mM citric acid (pH 7.4) before being assayed for determination of viral titers.
Cryoelectron Microscopy.
About 2.8 µL of sample was applied onto a 400-mesh continuous carbon grid (Ted Pella). Immediately after blotting for about 8 s, the grid was vitrified in liquid ethane that was precooled by liquid nitrogen. Frozen, hydrated particles that were embedded in a thin layer of vitreous ice were imaged with a K2 Summit direct electron detector (Gatan) using a Titan Krios transmission electron microscope (FEI) operating at 300 kV. Cryo-EM data on strain MO were automatically collected using the program Leginon (58). The dose rate was kept at ∼8 e−⋅pixel−1⋅s−1 for data collection. For structure determination of full native virions using prep A, movies of frozen, hydrated virus particles (dataset A_Native) were collected at a nominal magnification of 22,500× in superresolution mode with defocus values ranging from 0.3 to 3.0 µm. A total electron dose of about 36 e−/Å2 was fractionated into 38 frames in every movie with a frame rate of 200 ms per frame. For initial low-pH studies using prep B, viruses were treated with phosphate-citrate buffer (100 mM Na2HPO4, 50 mM citric acid) to reach a final pH of 5.5 (dataset B_RT_Acid) or a pH of 7.2 (dataset B_RT_Neu), followed by incubation at room temperature for 20 min and, subsequently, neutralization with 400 mM Na2HPO4 and 200 mM citric acid (pH 7.8). Data were collected at a nominal magnification of 22,500× in superresolution mode. The defocus range for datasets B_RT_Acid (144 movies) and B_RT_Neu (87 movies) were 0.9 to 4.8 µm and 1.3 to 3.6 µm, respectively. For dataset B_RT_Acid, a total electron dose of about 28 e−/Å2 was fractionated into 30 frames (200 ms per frame). For dataset B_RT_Neu, a total electron dose of about 25 e−/Å2 was fractionated into 27 frames (200 ms per frame). For low-pH studies using prep B to mimic conditions during virus infection, viruses were treated at pH 5.5 similar to the aforementioned procedure except that the incubation temperature was changed to 33 °C. Data (dataset B_33_Acid) were collected at a nominal magnification of 22,500× with defocus values ranging from 0.5 to 3.5 µm. A total electron dose of about 38 e−/Å2 was fractionated into 40 frames (200 ms per frame). For analyzing the heterogeneous particle population of prep B (stored at 4 °C) at neutral pH, data (dataset B_4_Neu) were collected at a nominal magnification of 18,000× in electron-counting mode with defocus values ranging from 1.7 to 5.3 µm. A total electron dose of about 45 e−/Å2 was fractionated into 60 frames (250 ms per frame). For datasets A_Acid_1, A_Acid_2, A_Acid_3, and A_Acid_4, prep A was treated using conditions shown in Table 1 followed by plunge freezing. This process did not include the aforementioned step of backneutralization. These datasets were collected at a nominal magnification of 81,000× in electron-counting mode, resulting in a pixel size of 1.73 Å per pixel at the specimen level. In these cases, the Titan Krios transmission electron microscope was equipped with a Gatan Quantum energy filter (zero-loss energy filtering with a slit width of 20 eV) and a post GIF K2 summit direct electron detector. A total electron dose of about 28 e−/Å2 was fractionated into 42 frames (250 ms per frame). Statistics for data collection and processing have been summarized in SI Appendix, Tables S1, S4, and S5.
Image Processing.
Movie frames were subjected to motion correction using a modified version (Wen Jiang, Purdue University) of MotionCorr (59) unless otherwise stated. The aligned frames were summed to produce individual micrographs, which were used to estimate contrast transfer function (CTF) parameters using CTFFIND3 (60). For the datasets A_Native, B_RT_Acid, and B_RT_Neu, virus particles were picked from the micrographs manually using e2boxer.py in the EMAN2 program package (61). For all other datasets, particle selection was performed first manually using e2boxer.py and subsequently automatically using the program DoG Picker (62) based on templates derived from manually selected particles. Particles were subsequently boxed and extracted from the micrographs. The process was integrated into the Appion data-processing pipeline (63). The resultant particle images were subjected to 2D classification using the program RELION (64), which identified and removed some low-quality particles and separated images of full particles from those of empty particles. For instance, 2D classification of 264,850 automatically picked particles (dataset B_4_Neu) led to selection of 156,555 full particles and 80,885 empty particles.
The following reconstruction procedures were employed for all datasets except dataset B_4_Neu using the program jspr (65). In brief, particle images (8× binned, with a pixel size of 5.20 Å per pixel) were divided into two halves. For each half, random initial orientations were assigned to individual particles, allowing for reconstruction of multiple icosahedral 3D models, from which a suitable initial model was selected. The best particle orientation and center of each particle image were searched with respect to projections of the initial reference model. The reference model for the next iteration was reconstructed from particle images employing parameters for orientation and center determined in the current iteration. The refinement procedure was then extended to 4× binned and then 2× binned data. Specifically, for dataset A_Native, the procedure was extended to unbinned data. Subsequently, multiple parameters were included in the refinement process, which were parameters for particle orientation, particle center, beam tilt, defocus, scale, astigmatism, and anisotropic magnification distortion (66, 67). To achieve 3D reconstructions with the highest possible resolution, particle images (dataset B_RT_Acid only) were reextracted from micrographs that were generated by summing aligned frames 3 to 16. In this way, frames that underwent large motions and that contained limited high-resolution information due to radiation damage were discarded. Frames 3 to 16 were selected using a trial-and-error approach in which different combinations were tested, including frames 3 to 9, 3 to 16, 3 to 23, and 3 to 30. For datasets A_Native and B_33_Acid, movie frames were aligned using the program MotionCor2 (an improved version of MotionCorr) (68), in which the first frame of each movie was discarded due to large motions, and high-resolution information in late frames was down-weighted using a reported dose-weighting scheme (66). Micrographs were generated by summing aligned frames. Particle images were reextracted from individual micrographs without changing the coordinates of individual particles on every micrograph and used for reconstructing the structures of full native virions, A particles, and emptied particles at 2.17-, 2.73-, and 2.90-Å resolution, respectively. Fourier shell correlation (FSC) of two interdependently calculated half-maps (masked with a soft mask) was used to estimate the resolution of the final EM maps using an FSC cutoff of 0.143 (69, 70). The maps were sharpened by applying a negative B factor and filtered with an FSC curve-based low-pass filter (69).
The following procedures were applied to dataset B_4_Neu. After 2D classification of all particle images in the dataset, the resultant full particle images (4× binned, 6.48 Å per pixel) were used to generate initial 3D models, from which a suitable initial model was selected. The refinement process was performed using a projection-matching approach as described above. After multiple iterative cycles when the process converged, the resultant 3D model was essentially an average of all possible structural states present in the collection of full particle images. The model was low pass-filtered to 60-Å resolution and then utilized as a reference model for 3D classification of full particle images (4× binned) using the program RELION (64), where the number of classes was four and where icosahedral symmetry was imposed. Other parameters for 3D classification included an angular sampling interval of 3.7°, a translational search range of 5 pixels with a step size of 1 pixel, a regularization parameter of 2, and a total of 25 iterations. Particle images from two of the resultant four classes yielded 3D reconstructions that were nearly identical to each other by visual inspection. Thus, particle images from the two classes were combined into one class. The same process was also applied to images of empty particles. Hence, all particle images in the dataset were classified into a total of six classes (three for full particles and three for empty particles), which represented six different structural states. Procedures for cryo-EM structure determinations were the same when using each class of particle images as detailed above.
The procedures applied to datasets A_Acid_1, A_Acid_2, A_Acid_3, and A_Acid_4 were essentially the same as mentioned above with some exceptions. Movie frames were aligned using the program MotionCor2 (68). Particles were automatically picked from dose-weighted micrographs using the program Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/). This was based on templates that were projections derived from a 3D reconstruction of full native virions of strain MO (low pass-filtered to 40-Å resolution). After 2D classification of all particle images in the dataset, full particle images (3× binned, 5.19 Å per pixel) were extracted from the micrographs and subjected to 3D classification as detailed above. To test the reproducibility of classification results (Table 1), this process was repeated six times using a regularization parameter of 2 or 4 and with the number of classes being four or six. Based on visual inspection of reconstructions derived from each class, particle images that belonged to the same structural state (full native virions, A particles, or E1 particles) were combined and used for high-resolution cryo-EM structure determination as described above. All reconstructions were determined to about 3.5-Å resolution, which enabled further verification of the identity of each structural state present in a given dataset. For each final cryo-EM map (sharpened), each of the known atomic models (full native virions, A particles, and E1 particles in dataset B_4_Neu) was fitted as a rigid body into one of 60 identical portions of the map using Coot (71). This procedure resulted in a large portion of the atomic model being well-fitted into the map density. To determine the identity of structural states, visual inspection was focused on whether the density map differed from the atomic model in specific local regions. These regions acted as markers to differentiate three structural states from each other. They included the VP3 GH loop, VP4, the N and C termini of VP1, VP2, and VP3, as well as regions surrounding the quasi-threefold and twofold axes (SI Appendix, Figs. S15 and S17). Model statistics for Acid_A_1-E1 are the following: correlation coefficient (defined in SI Appendix, Table S1), 0.832; number of atoms, 6,601; average B factor, 55.3 Å2; bond-length rmsd, 0.011 Å; bond-angle rmsd, 1.250°; and Ramachandran plot: favored, 95.2%; allowed, 4.8%; outliers, 0.0%.
Model Building and Refinement.
The same procedures were employed for all atomic structures presented in this work. The coordinates of the EV-D68 Fermon strain excluding the pocket factor and water molecules (Protein Data Bank ID code 4WM8) were used as a starting atomic model. It was manually fitted into the EM map using Chimera (72). Then, multiple cycles of model rebuilding in Coot (71) and real-space refinement against the EM map using PHENIX (73, 74) yielded an atomic model that fitted well into the map density as judged by visual inspection. A mask that contained all grid points within a radius of 5 Å around each atom of the atomic model was used to cut out a map segment from the EM map. This map segment was placed into a pseudocrystallographic unit cell with space group P1 and backtransformed into structure factors. The atomic model was subjected to refinement of atomic coordinates, B factors, and occupancy against these pseudocrystallographic structure factors in reciprocal space using REFMAC5 (75). The resultant atomic model was used for real-space refinement with 60-fold noncrystallographic symmetry constraints using PHENIX (73, 74). Water molecules were added in Coot (71). The final atomic models were validated according to the criteria of MolProbity (76). Model statistics have been summarized in SI Appendix, Tables S1 and S5. All figures were generated using PyMOL (https://pymol.org/2/) or Chimera (72). Structural comparisons were done using HOMOLOGY (77). Oligomers of capsid protomers were produced using VIPERdb (78). Buried surface areas at protein–protein interacting interfaces were calculated using PISA (79).
Particle Stability Thermal Release Assays.
PaSTRy assays were performed as previously described (49, 80). About 1 to 1.5 µg of purified EV-D68 (strain MO) or EV-A71 was incubated at a given pH (7.1, 6.5, 6.0, or 5.5) for about 3 min at RT. As a positive control, EV-D68 strain Fermon in a low-salt buffer (50 mM Tris, 2 mM CaCl2, pH 8.0) was heated to 50 °C for 3 min to produce A particles as previously reported (25). For every pH condition, buffer without virus served as a negative control. To quench the reaction, 10× NTE buffer (1× NTE: 20 mM Tris, 120 mM NaCl, 1 mM EDTA, pH 8.0), SYBR Green II dye (Life Technologies), and RNaseOUT (Life Technologies) were added to reach final concentrations of 1×, 3×, and 1 U/μL, respectively. Each reaction mixture had a volume of 50 μL. A real-time quantitative PCR system (Applied Biosystems 7300) was then used to record fluorescence intensity in triplicate at 1-°C intervals from 27 to 96 °C. The temperature at which the genomic RNA is exposed, Tr, refers to the temperature that gives the minimum of the negative first derivative of fluorescence intensity with respect to temperature.
Supplementary Material
Acknowledgments
We thank M. Steven Oberste of the Centers for Disease Control and Prevention for providing the Fermon strain of EV-D68, and Jane Cardosa of Universiti Malaysia Sarawak for supplying the EV-A71 strain MY104-9-SAR-97. We are grateful to Thomas Klose, Zhenguo Chen, Yingyuan Sun, Valorie Bowman, and Wen Jiang for help with cryo-EM analysis, and Sheryl Kelly for help preparing this manuscript. We thank Jim Baggen and Shishir Poudyal for helpful discussions. The following reagents were obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health: enterovirus D68, US/MO/14-18947 (NR-49129), and US/KY/14-18953 (NR-49132). This work was supported by NIAID Grant AI011219 (to M.G.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates of A_Native-full, B_RT_Acid-Aparticle, B_RT_Acid-Emptied, B_33_Acid-Aparticle, B_33_Acid-Emptied, B_4_Neu-Full-Native, B_4_Neu-E1, B_4_Neu-Aparticle, B_4_Neu-Emptied, B_4_Neu-Abortive1, B_4_Neu-Abortive2, and Acid_A_1-E1 reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6CSG, 6CS6, 6CSA, 6CS4, 6CSH, 6CRR, 6CS3, 6CRS, 6CRU, 6CRP, 6CS5, and 6MZI). The cryo-EM maps of A_Native-full, B_RT_Acid-Aparticle, B_RT_Acid-Emptied, B_33_Acid-Aparticle, B_33_Acid-Emptied, B_4_Neu-Full-Native, B_4_Neu-E1, B_4_Neu-Aparticle, B_4_Neu-Emptied, B_4_Neu-Abortive1, and B_4_Neu-Abortive2 reported in this paper have been deposited in the Electron Microscopy Data Bank https://www.ebi.ac.uk/pdbe/emdb (accession nos. EMD-7599, EMD-7593, EMD-7598, EMD-7589, EMD-7600, EMD-7569, EMD-7583, EMD-7571, EMD-7572, EMD-7567, and EMD-7592). The cryo-EM maps of A_Acid_1-Full-Native, A_Acid_1-Aparticle, A_Acid_1-E1, A_Acid_2-Full-Native, A_Acid_2-Aparticle, A_Acid_3-Full-Native, A_Acid_3-Aparticle, and A_Acid_4-Aparticle reported in this paper have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-9053, EMD-9054, EMD-9055, EMD-9056, EMD-9057, EMD-9058, EMD-9059, and EMD-9060).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803347115/-/DCSupplemental.
References
- 1.Racaniello VR. Picornaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 1. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 796–839. [Google Scholar]
- 2.Knowles NJ, et al. In: Picornaviridae. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Elsevier; San Diego: 2012. pp. 855–880. [Google Scholar]
- 3.Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 4.Oberste MS, et al. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol. 2004;85:2577–2584. doi: 10.1099/vir.0.79925-0. [DOI] [PubMed] [Google Scholar]
- 5.Tokarz R, et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Midgley CM, et al. EV-D68 Working Group Severe respiratory illness associated with a nationwide outbreak of enterovirus D68 in the USA (2014): A descriptive epidemiological investigation. Lancet Respir Med. 2015;3:879–887. doi: 10.1016/S2213-2600(15)00335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm-Hansen CC, Midgley SE, Fischer TK. Global emergence of enterovirus D68: A systematic review. Lancet Infect Dis. 2016;16:e64–e75. doi: 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 8.Kreuter JD, et al. A fatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135:793–796. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 9.Messacar K, et al. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet. 2015;385:1662–1671. doi: 10.1016/S0140-6736(14)62457-0. [DOI] [PubMed] [Google Scholar]
- 10.Hixon AM, et al. A mouse model of paralytic myelitis caused by enterovirus D68. PLoS Pathog. 2017;13:e1006199. doi: 10.1371/journal.ppat.1006199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uncapher CR, DeWitt CM, Colonno RJ. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 12.Blomqvist S, Savolainen C, Råman L, Roivainen M, Hovi T. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J Clin Microbiol. 2002;40:4218–4223. doi: 10.1128/JCM.40.11.4218-4223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamura T, et al. Antigenic and receptor binding properties of enterovirus 68. J Virol. 2014;88:2374–2384. doi: 10.1128/JVI.03070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Sialic acid-dependent cell entry of human enterovirus D68. Nat Commun. 2015;6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baggen J, et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc Natl Acad Sci USA. 2016;113:1399–1404, and correction (2016) 113:E1588. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei W, et al. ICAM-5/telencephalin is a functional entry receptor for enterovirus D68. Cell Host Microbe. 2016;20:631–641. doi: 10.1016/j.chom.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 18.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs R, Blaas D. Uncoating of human rhinoviruses. Rev Med Virol. 2010;20:281–297. doi: 10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 20.Bergelson JM, Coyne CB. Picornavirus entry. Adv Exp Med Biol. 2013;790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 21.Fricks CE, Hogle JM. Cell-induced conformational change in poliovirus: Externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prchla E, Kuechler E, Blaas D, Fuchs R. Uncoating of human rhinovirus serotype 2 from late endosomes. J Virol. 1994;68:3713–3723. doi: 10.1128/jvi.68.6.3713-3723.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Hogle JM, Chow M. Is the 135S poliovirus particle an intermediate during cell entry? J Virol. 2000;74:8757–8761. doi: 10.1128/jvi.74.18.8757-8761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowell RL, Philipson L. Specific alterations of coxsackievirus B3 eluted from HeLa cells. J Virol. 1971;8:509–515. doi: 10.1128/jvi.8.4.509-515.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bubeck D, et al. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol. 2005;79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren J, et al. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun. 2013;4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, et al. The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci Adv. 2016;2:e1501929. doi: 10.1126/sciadv.1501929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brabec M, et al. Opening of size-selective pores in endosomes during human rhinovirus serotype 2 in vivo uncoating monitored by single-organelle flow analysis. J Virol. 2005;79:1008–1016. doi: 10.1128/JVI.79.2.1008-1016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panjwani A, et al. Capsid protein VP4 of human rhinovirus induces membrane permeability by the formation of a size-selective multimeric pore. PLoS Pathog. 2014;10:e1004294. doi: 10.1371/journal.ppat.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garriga D, et al. Insights into minor group rhinovirus uncoating: The X-ray structure of the HRV2 empty capsid. PLoS Pathog. 2012;8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korant BD, Lonberg-Holm K, Noble J, Stasny JT. Naturally occurring and artificially produced components of three rhinoviruses. Virology. 1972;48:71–86. doi: 10.1016/0042-6822(72)90115-8. [DOI] [PubMed] [Google Scholar]
- 33.Hoover-Litty H, Greve JM. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickl-Herk A, et al. Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle. Proc Natl Acad Sci USA. 2013;110:20063–20068. doi: 10.1073/pnas.1312128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayer N, Prchla E, Schwab M, Blaas D, Fuchs R. Human rhinovirus HRV14 uncoats from early endosomes in the presence of bafilomycin. FEBS Lett. 1999;463:175–178. doi: 10.1016/s0014-5793(99)01610-5. [DOI] [PubMed] [Google Scholar]
- 36.Smith TJ, et al. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 37.Filman DJ, et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth M, Pettitt T, Symonds A, Martin J. Identification of the pocket factors in a picornavirus. Arch Virol. 2003;148:1225–1233. doi: 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 39.Basavappa R, et al. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: Structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 1994;3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc Natl Acad Sci USA. 2016;113:8997–9002. doi: 10.1073/pnas.1606595113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuthill TJ, Bubeck D, Rowlands DJ, Hogle JM. Characterization of early steps in the poliovirus infection process: Receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J Virol. 2006;80:172–180. doi: 10.1128/JVI.80.1.172-180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold E, Rossmann MG. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 Å. J Mol Biol. 1990;211:763–801. doi: 10.1016/0022-2836(90)90076-x. [DOI] [PubMed] [Google Scholar]
- 43.Strauss M, et al. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J Virol. 2015;89:4143–4157. doi: 10.1128/JVI.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katpally U, Fu TM, Freed DC, Casimiro DR, Smith TJ. Antibodies to the buried N terminus of rhinovirus VP4 exhibit cross-serotypic neutralization. J Virol. 2009;83:7040–7048. doi: 10.1128/JVI.00557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bostina M, Levy H, Filman DJ, Hogle JM. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol. 2011;85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harutyunyan S, et al. Viral uncoating is directional: Exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3′-end. PLoS Pathog. 2013;9:e1003270. doi: 10.1371/journal.ppat.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar M, Blaas D. Human rhinovirus subviral A particle binds to lipid membranes over a twofold axis of icosahedral symmetry. J Virol. 2013;87:11309–11312. doi: 10.1128/JVI.02055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandenburg B, et al. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, et al. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science. 2015;347:71–74. doi: 10.1126/science.1261962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy A, Post CB. Long-distance correlations of rhinovirus capsid dynamics contribute to uncoating and antiviral activity. Proc Natl Acad Sci USA. 2012;109:5271–5276. doi: 10.1073/pnas.1119174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giranda VL, et al. Acid-induced structural changes in human rhinovirus 14: Possible role in uncoating. Proc Natl Acad Sci USA. 1992;89:10213–10217. doi: 10.1073/pnas.89.21.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S, et al. Conformational variability of a picornavirus capsid: pH-dependent structural changes of Mengo virus related to its host receptor attachment site and disassembly. Virology. 1990;175:176–190. doi: 10.1016/0042-6822(90)90198-z. [DOI] [PubMed] [Google Scholar]
- 53.Warwicker J. Model for the differential stabilities of rhinovirus and poliovirus to mild acidic pH, based on electrostatics calculations. J Mol Biol. 1992;223:247–257. doi: 10.1016/0022-2836(92)90729-4. [DOI] [PubMed] [Google Scholar]
- 54.Smura TP, et al. Enterovirus 94, a proposed new serotype in human enterovirus species D. J Gen Virol. 2007;88:849–858. doi: 10.1099/vir.0.82510-0. [DOI] [PubMed] [Google Scholar]
- 55.Ellard FM, Drew J, Blakemore WE, Stuart DI, King AM. Evidence for the role of His-142 of protein 1C in the acid-induced disassembly of foot-and-mouth disease virus capsids. J Gen Virol. 1999;80:1911–1918. doi: 10.1099/0022-1317-80-8-1911. [DOI] [PubMed] [Google Scholar]
- 56.Nurani G, Lindqvist B, Casasnovas JM. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J Virol. 2003;77:11985–11991. doi: 10.1128/JVI.77.22.11985-11991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamayoshi S, Ohka S, Fujii K, Koike S. Functional comparison of SCARB2 and PSGL1 as receptors for enterovirus 71. J Virol. 2013;87:3335–3347. doi: 10.1128/JVI.02070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 59.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 61.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G, et al. An algorithm for estimation and correction of anisotropic magnification distortion of cryo-EM images without need of pre-calibration. J Struct Biol. 2016;195:207–215. doi: 10.1016/j.jsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 70.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 73.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossmann MG, Argos P. A comparison of the heme binding pocket in globins and cytochrome b5. J Biol Chem. 1975;250:7525–7532. [PubMed] [Google Scholar]
- 78.Carrillo-Tripp M, et al. VIPERdb2: An enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 80.Walter TS, et al. A plate-based high-throughput assay for virus stability and vaccine formulation. J Virol Methods. 2012;185:166–170. doi: 10.1016/j.jviromet.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.