Significance
Distinct molecular subtypes of pancreatic cancer have recently been identified with the squamous subtype exhibiting a particularly poor prognosis. Precision-medicine approaches are needed in pancreatic cancer due to its dismal prognosis. Accordingly, novel and specific dependencies in these aggressive subtypes need to be identified. This study uncovers a group of transcription factors which form an interdependent network driving the squamous subtype via subtype-specific super enhancers. These factors include deltaNp63 which we show specifically cooperates with BHLHE40, HIF1A, and RXRA to control transcription in the squamous subgroup. Importantly, an epigenetic signature identified in this study is capable of accurately identifying squamous subtype samples in pancreatic cancer patient-derived xenograft tumors.
Keywords: deltaNp63, pancreatic cancer, super enhancers, transcription factors, BHLHE40
Abstract
Molecular subtyping of cancer offers tremendous promise for the optimization of a precision oncology approach to anticancer therapy. Recent advances in pancreatic cancer research uncovered various molecular subtypes with tumors expressing a squamous/basal-like gene expression signature displaying a worse prognosis. Through unbiased epigenome mapping, we identified deltaNp63 as a major driver of a gene signature in pancreatic cancer cell lines, which we report to faithfully represent the highly aggressive pancreatic squamous subtype observed in vivo, and display the specific epigenetic marking of genes associated with decreased survival. Importantly, depletion of deltaNp63 in these systems significantly decreased cell proliferation and gene expression patterns associated with a squamous subtype and transcriptionally mimicked a subtype switch. Using genomic localization data of deltaNp63 in pancreatic cancer cell lines coupled with epigenome mapping data from patient-derived xenografts, we uncovered that deltaNp63 mainly exerts its effects by activating subtype-specific super enhancers. Furthermore, we identified a group of 45 subtype-specific super enhancers that are associated with poorer prognosis and are highly dependent on deltaNp63. Genes associated with these enhancers included a network of transcription factors, including HIF1A, BHLHE40, and RXRA, which form a highly intertwined transcriptional regulatory network with deltaNp63 to further activate downstream genes associated with poor survival.
Distinct molecular subtypes in cancer are defined by different deregulated pathways, mutational profiles, and aberrant transcriptional programs that may potentially be leveraged to optimize therapy and elucidate mechanisms in a disease that is characterized by a particularly high degree of heterogeneity (1). Molecular stratification of breast and colorectal cancer, for example, revolutionized therapy for these malignancies and extended our knowledge about the pathways and mechanisms involved in disease development and progression (2–4). Recently, analyses in pancreatic cancer, which has a particularly low survival rate, uncovered various molecular subtypes with different characteristics and prognoses (5–10).
Collisson et al. (6) used human and mouse samples in addition to pancreatic cancer cell lines to identify recurrent patterns of gene expression and identified three subtypes, referred to as classical, exocrine-like, and quasimesenchymal, with the latter being correlated with a particularly poor prognosis. Optimization of molecular stratification by filtering stromal profiles further grouped the molecular subtypes of pancreatic cancer into classical-like and basal-like, with the latter corresponding to the worse prognosis seen in the quasimesenchymal subtype (7). Extending these analyses to include mutational backgrounds of patients and DNA methylation states in addition to gene expression revealed four subtypes, including the highly aggressive squamous subgroup (8). Further analysis confirmed the identification of specific patterns of expression with one molecular subtype, irrespective of nomenclature, representing a small subgroup of pancreatic cancer patients with a particularly poor prognosis (9, 10).
While more advances are being made in the analytical aspect of subtyping pancreatic cancer, the molecular mechanisms underlying these gene signatures are still largely unclear. Bailey et al. (8) identified deltaNp63 expression as a hallmark of the squamous subtype, which overlaps with its basal-like counterpart and profoundly correlates with worse prognosis (9). p63 is a transcription factor (TF) of the p53 family which has two major isoforms, including the transactivation domain-containing isoform, TAp63, and the shorter isoform, deltaNp63 (11). DeltaNp63 was reported to play a crucial role in keratinocyte differentiation and its expression was shown to be a defining feature of basal cells and squamous cell carcinomas (12–14). Concordantly, deltaNp63 is expressed in many squamous-like cancers, including esophageal squamous cell carcinoma (15, 16), head and neck squamous carcinoma (17), and lung squamous cell carcinoma (18, 19). p63 was found to drive epidermal differentiation through distal regulatory elements associated with its target genes (20). However, to date a role for deltaNp63 in pancreatic cancer has remained largely unclear.
An early report described an up-regulation of deltaNp63 in a group of pancreatic cancer cells displaying a squamous-like phenotype, while normal pancreata were completely devoid of deltaNp63 expression (21). Interestingly, depletion of the histone demethylase KDM6A led to the activation of super enhancers regulating deltaNp63 and enriched for a more squamous-like phenotype in pancreatic cancer cells (22). Super enhancers are clusters of distal regulatory elements which are highly enriched for transcription factor binding and have a high potential to affect target genes (23–25). Our previous work identified tissue-specific patterns of gene expression which were particularly coupled to transcription factor and cofactor recruitment to distal enhancer regions rather than occupancy in the proximal promoter region (26–28).
In this study, we performed an unbiased analysis of epigenomic gene activation profiles and identified deltaNp63 as a major driver of gene activation in a particularly lethal subtype of pancreatic cancer. We report that L3.6pl and BxPC-3 pancreatic cancer cell lines represent appropriate cell culture models of the squamous molecular subtype described in patients. Here, we uncovered a major dependence of subtype-specific super enhancers on deltaNp63. Furthermore, we confirmed the analyses of our model cell lines and significantly expanded the relevance of the findings by comparing our results with data from patient-derived xenograft (PDX) samples. Using this approach we identified 45 super enhancers that signify the squamous subgroup and are associated with genes that are highly deltaNp63 dependent and correlate with poor prognosis in pancreatic cancer. Among these genes, we uncovered a highly interactive transcriptional regulatory hub, including deltaNp63, HIF1A, RXRA, and BHLHE40, where these factors activate one another as well as downstream genes. Altogether, our study elucidates underlying mechanisms by which deltaNp63 drives gene expression patterns associated with the squamous molecular subtype in pancreatic cancer and identify a number of super enhancers that may potentially be used to identify this subgroup to stratify patients with poorer prognosis in a simple and accessible manner.
Results
DeltaNp63 Is a Major Driver of Differential Gene Activation in Specific Pancreatic Cancer Cell Lines and Patient-Derived Xenografts.
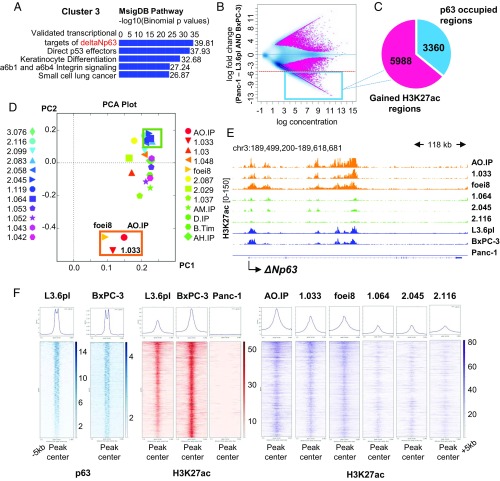
Due to previously observed different characteristics of the pancreatic cancer cell lines Panc-1, BxPC-3, and the highly metastatic L3.6pl (28–30), we performed chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) for histone 3 acetylation at lysine 27 (H3K27ac) in these commonly studied cell lines to elucidate the differences in their epigenomic landscapes. As H3K27ac marks active promoters as well as enhancers, it gives a comprehensive insight into active gene transcription and underlying mechanisms. Hierarchical clustering of H3K27ac peaks in all cell lines identified three clusters with the first and second having low and high signal, respectively, in all cell lines. Only the third cluster showed marked enrichment in BxPC-3 and L3.6pl compared with low enrichment in Panc-1 (SI Appendix, Fig. S1A). Genomic regions enrichment of annotations tool (GREAT) analysis for the 15,286 regions included in cluster 3 (out of 88,773 total regions) showed validated targets of deltaNp63 as the most significant hit for the genes associated with these regions (Fig. 1A). To further investigate regions that are specifically marked in L3.6pl and BxPC-3, we performed unbiased differential binding analysis for H3K27ac in the three cell lines and identified 9,348 regions that are significantly gained in BxPC-3 and L3.6pl and have at least a fivefold enrichment compared with Panc-1, henceforth referred to as H3K27ac-gained regions (Fig. 1B and Dataset S1). Additionally, we validated that gained H3K27ac on these regions is correlated with significantly higher gene expression levels of associated genes in L3.6pl and BxPC-3 compared with Panc-1 by RNA sequencing (RNA-seq) (SI Appendix, Fig. S1B and Dataset S2). Consistent with initial findings based on hierarchical clustering, differential occupancy analyses revealed targets of deltaNp63 to be the most highly significant pathway enriched for genes associated with H3K27ac-gained regions (SI Appendix, Fig. S1C). To determine the extent to which deltaNp63 may play a direct role in determining the differential marking of H3K27ac-gained regions, we performed ChIP-seq for p63 in L3.6pl and BxPC-3 and found that approximately one-third of the H3K27ac-gained regions are occupied by p63 (Fig. 1C). Thus, these unbiased analyses provide evidence suggesting that p63 is a major driver of genes specifically activated in L3.6pl and BxPC-3 cell lines compared with Panc-1 cells.
Fig. 1.
DeltaNp63 drives gene activation in L3.6pl and BxPC-3. (A) Associated genes with regions in cluster 3 (enriched in L3.6pl and BxPC-3) defined by GREAT analysis using the default basal plus extension association rule and showing validated targets of deltaNp63 as the top hit. (B) Binding affinity plot for H3K27ac peaks in Panc-1 compared with L3.6pl and BxPC-3 showing the regions with a significant enrichment difference in pink. The blue box highlights the regions that are significantly enriched in L3.6pl and BxPC-3 by more than a fivefold change, henceforth called H3K27ac-gained regions (9,348 regions). (C) Pie chart depicting the percentage of the 9,348 regions that are occupied by p63. (D) Principal component analysis plot for the H3K27ac profiles of 24 patient-derived xenografts named on the sides limited to the 9,348 regions in B. Orange box indicates three xenografts which clustered independently from the other samples and the green box highlights samples clustering in the opposite direction. (E) Occupancy profiles of H3K27ac at the TSS of deltaNp63 for the six xenografts highlighted in orange and green box in D, L3.6pl, BxPC-3, and Panc-1. Results show an enrichment for the three uniquely clustering xenografts, L3.6pl, and BxPC-3, while the other samples have very low occupancy of H3K27ac. (F) Average binding profiles and heatmaps depicting the p63 and H3K27ac occupancy at the H3K27ac-gained regions in L3.6pl, BxPC-3 for p63, and L3.6pl and BxPC3, Panc-1, and the six differentially clustered patient-derived xenografts for H3K27ac.
To examine if the epigenome patterns observed in L3.6pl and BxPC-3 also occur in patient tumors and not exclusively in vitro, we examined H3K27ac profiles from 24 pancreatic cancer PDXs (31). Principal component analysis using the H3K27ac-gained regions identified 3 of the 24 xenografts as forming a distinct cluster (Fig. 1D). Interestingly, these three xenografts were highly marked by H3K27ac near the transcriptional start site (TSS) of deltaNp63 compared with the rest of the samples (with three other representative samples shown as an example; Fig. 1E). Analysis of RNA-seq data confirmed the expression of p63 in these tumor samples. Isoform-specific expression was confirmed by H3K27ac occupancy at the TSS of deltaNp63, but not TAp63, with only one xenograft appearing to coexpress both p63 isoforms (SI Appendix, Fig. S1 D and E). Importantly, H3K27ac-gained regions identified in L3.6pl and BxPC3 also displayed a clear increased enrichment of H3K27ac in PDX samples expressing deltaNp63 compared with the nonexpressing ones (Fig. 1F). Thus, these data support that deltaNp63-driven epigenetic patterns observed in L3.6pl and BxPC-3 can also be found in pancreatic cancer PDXs.
L3.6pl and BxPC-3 Highly Express DeltaNp63 and Are Representative in Vitro Models for the Squamous Subtype of Pancreatic Cancer.
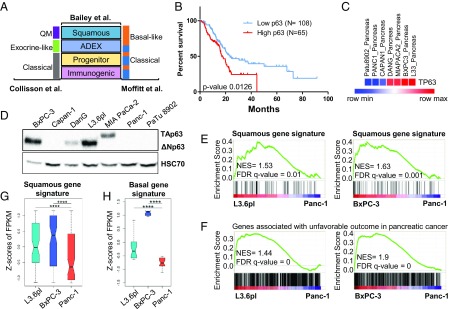
Various molecular subtypes were recently identified in pancreatic cancer based on unique gene expression profiles and distinct genomic and epigenomic signatures (Fig. 2A). One of the most aggressive subgroups of pancreatic cancer identified is the squamous subtype, which is associated with worse prognosis and high expression of deltaNp63. Indeed, patient survival data from The Cancer Genome Atlas (TCGA) confirmed that patients expressing higher p63 had a poorer prognosis than patients with lower levels (Fig. 2B). As deltaNp63 was identified in our analyses to be highly correlated with L3.6pl- and BxPC-3–specific active cis-regulatory regions, we next examined whether these cell lines may, indeed, represent the squamous subtype and serve as model systems for studying the molecular mechanisms driving this particularly aggressive tumor phenotype observed in vivo. Therefore, we evaluated the expression of p63 in different pancreatic cancer cell lines in the Morpheus database and further verified these findings at the protein level (Fig. 2 C and D and SI Appendix, Fig. S1F) (32). Notably, only BxPC-3 and L3.6pl displayed high expression of deltaNp63, with DanG showing a moderate expression, and MIA Paca-2 cells specifically expressing only the TAp63 isoform which is consistent with previous reports of predominant expression of TAp63 in the MIA PACA-2 cell line (33) (Fig. 2 C and D). To further examine whether L3.6pl and BxPC-3 faithfully represent the squamous subtype, we examined whether they expressed a squamous gene signature compared with Panc-1. Accordingly, we used the squamous gene signature defined by Bailey et al. (8) (Dataset S3) and verified that this signature is significantly enriched in both L3.6pl and BxPC-3 cell lines compared with Panc-1 cells (Fig. 2 E and G). Tendencies of enrichment for these genes were also observed in the three xenografts we identified as highly expressing deltaNp63 (SI Appendix, Fig. S1G). Notably, we also found that genes associated with an unfavorable prognosis in cancer patients were specifically enriched in the L3.6pl and BxPC-3 cell lines, providing further support that these cell lines may serve as a model for understanding the molecular mechanisms driving the aggressive characteristics of squamous-like pancreatic tumors (Fig. 2F and SI Appendix, Fig. S1H). Given that the squamous subgroup reported by Bailey et al. (8) roughly corresponds to the basal phenotype identified by Moffitt et al. (7), we also tested whether the basal gene signature was also enriched in L3.6pl and BxPC-3 and, indeed, observed a significant enrichment of the expression of these genes compared with Panc-1 (Fig. 2H). Together, these findings confirm that L3.6pl and BxPC-3 are representative in vitro model systems of squamous/basal-like pancreatic cancer.
Fig. 2.
L3.6pl and BxPC-3 represent the squamous subtype. (A) Graphical representation of the molecular subtypes recently defined in pancreatic cancer. (B) Kaplan–Meier plot showing the percent survival in pancreatic cancer patients (TCGA database) expressing high and low levels of p63 and showing significantly worse survival rates in patients with high levels of p63. Patients include all molecular subtypes and are not classified. (C) Heatmap depicting the general expression patterns of p63 in different pancreatic cancer cell lines (Morpheus database). (D) Western blot analysis of the levels of p63 in various pancreatic cell lines with HSC70 as a loading control. (E and F) GSEA plots comparing the enrichment of the squamous gene signature (E) and genes associated with unfavorable outcome in pancreatic cancer (F) in L3.6pl and BxPC-3 compared with Panc-1 using the FPKM values of all expressed genes with the normalized enrichment score (NES) and FDR indicated on the graph. (G) Box plot showing the Z scores of FPKM values of the squamous gene signature in L3.6pl, BXPC-3, and Panc-1. n = 3. ****P ≤ 0.0001. (H) Box plot showing the Z scores of FPKM values of the basal gene signature in L3.6pl, BXPC-3, and Panc-1. n = 3. ****P ≤ 0.0001.
Depletion of DeltaNp63 Alters the Molecular Identity of Squamous Pancreatic Cancer Cells.
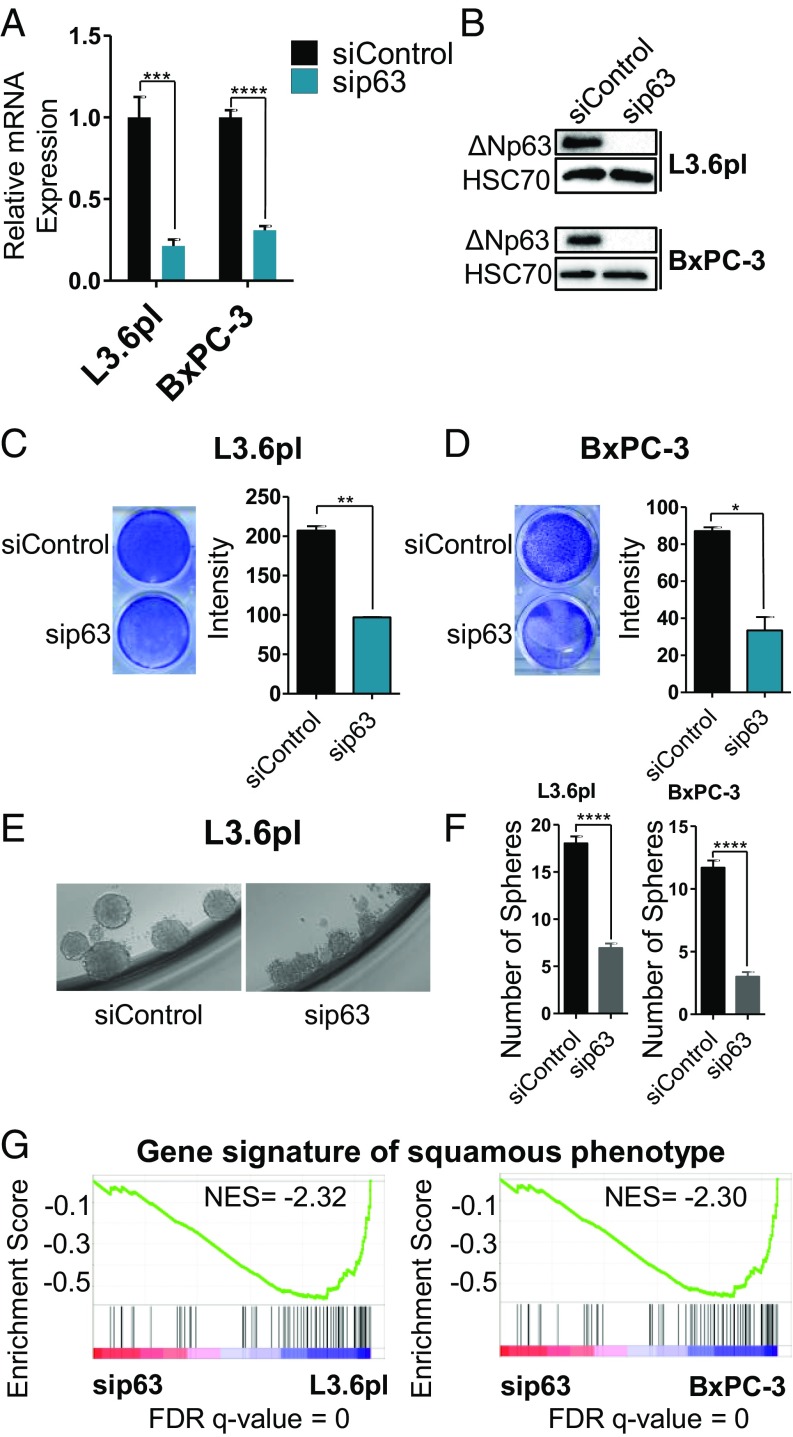
To investigate the role of deltaNp63 in gene activation in L3.6pl and BxPC-3 cells, we depleted deltaNp63 by siRNA-mediated knockdown and validated its down-regulation at the mRNA and protein levels (Fig. 3 A and B). Interestingly, knockdown of deltaNp63 led to a marked decrease in proliferation in both L3.6pl and BxPC-3 cells (Fig. 3 C and D). Consistent with the lack of p63 expression in Panc-1, knockdown of p63 in this line had no effect on proliferation (SI Appendix, Fig. S2 A and B). Interestingly, knockdown of TAp63 in MIA Paca-2 had also no effect on proliferation, supporting distinct roles of the two major isoforms of p63 in pancreatic cancer (SI Appendix, Fig. S2 C and D). Moreover, sphere formation in L3.6pl and BxPC-3 was significantly impaired upon p63 knockdown, with the few remaining spheres that were formed displaying a more diffuse and less defined structure, particularly in the case of L3.6pl (Fig. 3 E and F). This implies that deltaNp63 plays a role in driving a more aggressive phenotype in both of these cell lines. Interestingly, we observed that the TAp63-expressing MIA PACA-2 cells form very diffuse aggregates in sphere formation assays, further suggesting opposing roles of the p63 major isoforms (SI Appendix, Fig. S2E). To further understand how deltaNp63 drives this phenotype, we performed RNA-seq in both L3.6pl and BxPC-3 cell lines upon knockdown of deltaNp63. Remarkably, deltaNp63 down-regulation led to the reversal of the enrichment of the squamous gene signature, validating a clear and central role of deltaNp63 in driving the activation of these genes (Fig. 3G). Furthermore, gene set enrichment analysis (GSEA) identified MYC and HIF1A as target pathways of deltaNp63 (SI Appendix, Fig. S2F and Dataset S4). Notably, deltaNp63 depletion mimicked a switch from a mesenchymal to a luminal phenotype defined in breast cancer. Moreover, an enrichment of pathways with decreased tumorigenesis was observed in cells with less deltaNp63 (SI Appendix, Fig. S2G and Dataset S5). Top genes that are regulated in L3.6pl and BxPC-3 are provided in Dataset S6.
Fig. 3.
Depletion of deltaNp63 hampers growth and reverses enrichment of gene expression profiles associated with the squamous subtype. (A) Gene expression analysis of deltaNp63 upon depletion of p63 after 48 h shown as relative mRNA expression and normalized to the unregulated housekeeping gene (GAPDH). n = 3. (B) Western blot analysis for p63 in L3.6pl and BxPC-3 48 h after depletion of p63 to validate its down-regulation. HSC70 is shown as loading control. (C and D) Crystal violet staining showing the proliferation of cells after 48 h of depletion of p63 compared with control for L3.6pl (C) and BxPC-3 (D) with relative area fraction shown in the bar graph. Data are represented as mean ± SEM. n = 2. (E and F) Sphere formation assay analysis with E showing a representative change in the sphere structure upon p63 depletion and F showing the numbers of the spheres formed in L3.6pl and BxPC-3 after 500 cells were seeded in 96-well plate for 7 d. Data are represented as mean ± SEM. n = 24. (G) GSEA plots comparing the enrichment of the squamous gene signature in sip63 compared to sicontrol of the respective cell lines, L3.6pl and BxPC-3. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
DeltaNp63 Exerts Its Effects Through Activation of Super Enhancers.
To elucidate the mechanism by which deltaNp63 exerts its marked effect on cell proliferation, gene activation, and pancreatic cancer cell fate specification, we examined the occupancy of deltaNp63 throughout the genome and identified numerous deltaNp63-occupied regions (20,679 peaks). Many of these regions intersected with H3K27ac and open chromatin regions identified by assay for transposase-accessible chromatin (ATAC) sequencing (SI Appendix, Fig. S3 A and B). Interestingly, very few of these regions were associated with transcriptional start sites (TSSs) and GREAT analysis revealed that the majority of deltaNp63 peaks were distal (Fig. 4A and SI Appendix, Fig. S3 C and D). This distal pattern of occupancy implied that deltaNp63 mainly exerts its effects via enhancer activation. As depletion of deltaNp63 severely affects the transcriptional program of the cells and dramatically alters their molecular identity, we hypothesized that deltaNp63 may occupy and potentially nucleate super enhancers (SEs), as these have been reported to be major drivers of cell identity (34). In concordance with the different gene activation profiles of Panc-1 compared with L3.6pl and BxPC-3, distinct super enhancers were identified in Panc-1 compared with the other two cell lines, which generally showed the same patterns and tendencies (Fig. 4B and SI Appendix, Fig. S3 E and F). Interestingly, the majority of super enhancers in L3.6pl contained at least one peak of deltaNp63, with approximately a quarter of them having three or more peaks (Fig. 4C).
Fig. 4.
DeltaNp63 exerts its effects through activation of super enhancers. (A) Bar graph from GREAT analysis showing the distribution of p63 peaks of L3.6pl in relation to the TSS. (B) Enhancers in L3.6pl ranked based on BRD4 signal intensity using the ROSE algorithm defining 624 super enhancers. (C) Bar graph showing the percentages of SEs that are occupied by any number of peaks of deltaNp63 (≥1), by at least more than two peaks, or three peaks, with gray depicting regions that are common between SEs and COREs. (D) Venn diagram showing the overlap between genes associated with SEs and COREs and genes down-regulated upon depletion of p63 (q ≤ 0.05, fold change ≤ −2). Associated genes were identified by GREAT analysis using the basal plus extension association rule using default settings. (E) Box plot showing the log fold change upon depletion of p63 in the 45 subtype-specific super enhancer regions with the gene name on the Left for L3.6pl and Right for BxPC-3. Genes in red are significantly down-regulated (q < 0.05) by >1.5-fold, orange by 1.2- to 1.5-fold, genes in green are significantly up-regulated, and gray indicates insignificant changes.
Taking into consideration the potential bias in identifying super enhancers, which is dependent on the stitching of regions and the intensity of the factor used to rank the enhancers (35), we compared these results using a new algorithm to identify clusters of regulatory elements (COREs). COREs are determined using a machine learning approach to consider different windows between enhancers for stitching and does not require intensity of factors for ranking enhancers (https://www.biorxiv.org/content/early/2018/03/20/222562). Interestingly, we observed a high overlap between COREs and SEs, with COREs also showing the same high degree of occupancy by deltaNp63 (SI Appendix, Fig. S3 G and H). Notably, genes associated with COREs and/or SEs containing more than two peaks of deltaNp63 displayed a particularly high dependence on deltaNp63 (SI Appendix, Fig. S4 A and B). Notably, ChEA and enrichR consensus predicted p63 to be an upstream activator of super enhancers, and highly significant ontology terms associated with super enhancers included squamous cell carcinoma, confirming the role of SEs in defining the squamous subtype (SI Appendix, Fig. S4 C and D). Concordantly, deltaNp63-dependent genes associated with SEs and/or COREs included genes that are associated with epidermal differentiation like keratins and integrins (Fig. 4D). To identify the super enhancer regions that are driven by deltaNp63, we intersected the super enhancer regions in L3.6pl with the H3K27ac-marked regions gained in the both squamous cell lines compared with Panc-1, as well as the super enhancers identified in the patient-derived xenograft samples, since these more accurately represent in vivo squamous-like pancreatic tumors (SI Appendix, Fig. S4E). Consequently, we identified 93 SEs that were common for all these regions. We further filtered the SEs that were specifically enriched compared with the other patient-derived xenografts which clustered separately from the squamous samples and did not express deltaNp63 (SI Appendix, Fig. S4F). In this way we identified 45 super enhancer regions that were associated with the squamous subtype with high confidence. Interestingly, most genes associated with these regions showed a significant dependence on deltaNp63 (Fig. 4E). These genes included most notably FAT atypical cadherin 2 (FAT2), nectin cell adhesion molecule 1 (NECTIN1), and hypoxia inducible factor alpha subunit (HIF1A). These findings are in concordance with a squamous phenotype where hypoxic pathways are enriched and adhesion factors play a role in the development of the aggressive phenotype (8).
Super Enhancers in the Squamous Subtype Are Dependent on DeltaNp63.
To validate that the super enhancers which we identified are dependent on deltaNp63, we performed chromatin immunoprecipitation followed by quantitative real-time PCR on selected regions in those enhancers after depletion of deltaNp63. Specifically, we examined enhancers associated with FAT2, NECTIN1, and HIF1A due to their high dependence on deltaNp63 and their high relevance to the squamous phenotype. We observed two p63-occupied regions upstream of the FAT2 which were occupied by a peak of p63 gene in two separate SEs and corresponding to ATAC peaks in L3.6pl and H3K27ac peaks in L3.6pl, BxPC-3, and the three squamous patient-derived xenografts (Fig. 5A). We validated FAT2 down-regulation in both L3.6pl and BxPC-3 by qRT-PCR and the occupancy of these regions by deltaNp63, which was lost upon its depletion (Fig. 5 B and C). Consistent with a dependence of these enhancers on deltaNp63, H3K27ac occupancy at these enhancer regions was significantly decreased upon down-regulation of p63 (Fig. 5D). This was also seen for the other investigated enhancers, including the NECTIN1 super enhancer with two deltaNp63 peaks and an enhancer region upstream of HIF1A (SI Appendix, Fig. S5).
Fig. 5.
Super enhancers in the squamous subtype are highly dependent on deltaNp63. (A) Occupancy profiles at the FAT2 gene, which was identified to be highly dependent on deltaNp63 and associated with subtype-specific SEs. Profiles shown are for p63 in L3.6pl and BxPC-3, ATAC-seq in L3.6pl, H3K27ac in L3.6pl and BxPC-3, and six xenografts (three in orange, which express deltaNp63-expressing tumors, and three in green as a representative sample of the opposing clustering xenografts), in addition to the region files for SEs in L3.6pl and BxPC-3. Enlarged snapshots of the regions highlighted by the red dotted lines are shown on the Right with black arrows showing the regions examined for p63 enrichment and the red arrows for H3K27ac. (B) Gene expression analysis for FAT2 following depletion of p63 for 48 h shown as relative mRNA expression and normalized to an unregulated housekeeping gene (GAPDH). n = 3. (C and D) Validation of p63 (C) and H3K27ac (D) enrichment by ChIP-qPCR at the two highlighted regions in A (−3.7 kb from TSS of FAT2 and −46.5 kb from TSS of FAT2) in control and p63-depleted cells after 48 h. Red dotted line represents average IgG signal in control and p63-depleted cells. Data are represented as mean ± SEM. n = 2–3. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; ns, not significant.
DeltaNp63 Cooperates with Other Transcription Factors to Activate Target Genes Associated with Worse Prognosis.
Given the crucial role of deltaNp63 in defining a tumor subtype characterized by poorer prognosis, we evaluated the association of the deltaNp63-dependent SE-associated genes with prognosis using data from the TCGA research network (https://cancergenome.nih.gov/). Interestingly, increased expression of many deltaNp63-dependent genes displayed a significant correlation with poorer prognosis in pancreatic cancer patients (SI Appendix, Figs. S6 and S7A). As transcription factors frequently function cooperatively in lineage specification (36), we examined the list of genes associated with our identified enhancers and evaluated the expression of transcription factors contained within that list using the Morpheus database. Remarkably, the super enhancer-driven transcription factors HIF1A, basic helix–loop–helix family member E40 (BHLHE40), and retinoid X receptor alpha (RXRA) were more highly expressed in L3.6pl and BxPC-3 cells compared with Panc-1 (SI Appendix, Fig. S7B). Consequently, we asked if this specific expression pattern may help to form a transcriptional network underlying the marked effects of deltaNp63 in our system. Accordingly, we utilized genome occupancy data for HIF1A, RXRA, and BHLHE40 from the ReMAP database and filtered out all regions that did not overlap with the gained H3K27ac regions in L3.6pl and BxPC-3 compared with Panc-1 [fold change >4, false discovery rate (FDR) <0.5]. Then, using this information, together with our RNA-seq results, we constructed a regulatory network containing deltaNp63-dependent associated genes, along with the deltaNp63-dependent SE-associated genes, and extended the network by transcription factor-target query function using the Cytoscape iRegulon app (Fig. 6A). Notably, many of the target genes were affected by a combination of these transcription factors, which also showed reciprocal regulation patterns with many of the transcription factors binding and activating one another. To validate the role of the members of this regulatory network in our system, we depleted BHLHE40, HIF1A, and RXRA in L3.6pl cells and observed a significant dependence of deltaNp63 target genes FAT2 and NECTIN1, on each member of this network, albeit to varying degrees (Fig. 6B). We also observed an interconnected tendency of dependence of deltaNp63 on the other members of the network, particularly BHLHE40 (Fig. 6B and SI Appendix, Fig. S7C). Consistently, proliferation of L3.6pl was similarly affected by knockdown of BHLHE40 as deltaNp63, with the cells showing similar morphological changes upon knockdown of deltaNp63 or BHLHE40 (SI Appendix, Fig. S7 D–F). Moreover, we validated the cooccupation by the members of the transcriptional factor network at the enhancers of FAT2 and NECTIN1, which we identified to be enriched with and dependent on deltaNp63 (Fig. 6C). Accordingly, we conclude that deltaNp63 drives the expression of central target genes via the activation of super enhancers associated with downstream transcription factors such as HIF1A, BHLHE40, and RXRA. The activation of these deltaNp63-dependent genes enables the further indirect or cooperative activation of additional downstream target genes.
Fig. 6.
(A) Network depicting the interactions of HIF1A, BHLHE40, RXRA, and p63, where common genes are activated by all those transcription factors, which also activate each other. TF-target interactions were extracted from the iRegulon app of Cytoscape in addition to the genes associated with peaks from these transcription factors that intersect with H3K27ac-gained regions (with a lower fold change threshold of 4). (B) Gene expression analysis for deltaNp63, BHLHE40, HIF1A, RXRA, FAT2, and NECTIN1 following depletion of p63, BHLHE40, HIF1A, and RXRA for 48 h shown as relative mRNA expression and normalized to an unregulated housekeeping gene (GAPDH). n = 3. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; ns, not significant. (C) Validation of BHLEHE40, HIF1A, and RXRA enrichment at two enhancer regions for FAT2 (−3.7 kb from TSS of FAT2 and −46.5 kb from TSS of FAT2), another two enhancer regions at NECTIN1 (+31.8 kb and +36.2 kb from TSS of NECTIN1), and at OLIG2 as a negative control region in L3.6pl cells. Red dotted line represents average IgG signal in control and p63-depleted cells. n = 3.
Discussion
Gene expression and epigenetic profiles in cancer cells can be affected by many factors that are intrinsic or extrinsic to the tumor. This renders the investigation of molecular subtypes in malignancies quite challenging, as systems to study the molecular mechanisms behind these subtypes are scarce. In this study, we were able to discern the same patterns of molecular subclasses observed in patients in both cell lines and patient-derived xenografts (7, 8). This confirms the high reproducibility of these stratifications and implies that these molecular characteristics are highly conserved and robust, being able to withstand extreme changes of conditions. Most importantly, these systems provide an ideal opportunity to identify and target certain dependencies specific for the more aggressive subtypes. L3.6pl is unique as it exhibits highly metastatic characteristics due to the repeated cycles of spontaneous liver metastasis undergone during its establishment (37). Thus, it is not surprising that this cell line was found to be one of the systems that is representative of the squamous subtype, as it is one of the most highly metastatic and aggressive pancreatic cell lines (38). Interestingly, the BxPC-3 pancreatic cancer cell line is devoid of KRAS mutations, which is highly uncommon in pancreatic cancer (39). Despite this disparity, both BxPC-3 and L3.6pl recapitulated the same molecular subtype and shared highly common features concerning their gene expression and epigenetic profiles. Such a phenomenon further supports a paradigm of molecular classification of diverse malignancies that may be heterogeneous in their mutational backgrounds but share common features due to shared active regulatory transcription factor circuitry.
Notably, utilization of differentially active regions identified in the cell lines representative of the squamous/basal-like subtype helped to successfully identify a subgroup of patient-derived xenografts which were, consistently, previously classified as squamous/basal by Lomberk et al. (31) using other criteria. Molecular subtyping of pancreatic cancer currently requires the use of bioinformatically complicated algorithms and are usually not particularly robust, as demonstrated by only partially overlapping results seen in major recent studies (6–9). One reason for the apparent discrepancies may be due to tumor subgroups within the larger subgroups. Indeed, our analysis implicates deltaNp63 as a major driver of gene activation defining a squamous subgroup contained within the larger basal subgroup. Thus, deltaNp63 expression may be a defining feature of a further unappreciated subgroup of basal-like pancreatic tumors expressing a more squamous gene expression signature. In this study, we report a directed approach which involves principal component analysis of a single epigenetic marker (H3K27ac) on a select set of enhancer regions that are differentially active in the squamous/basal subgroup and which successfully clustered PDX samples based on molecular subtypes. This serves as an example for an accessible method to identify regions and gene signature patterns in various samples. Future studies in the scope of molecular subtypes of pancreatic cancer will play an important role in introducing conformity and clarity to the currently diverse subtyping approaches based largely on gene expression patterns.
In this study, we were able to define subtype-specific super enhancers (Dataset S7) associated with the aggressive squamous subtype in a manner akin to lineage-specific enhancers defining cell fate in pluripotent cells (40). Consistently, our findings uncover a tightly intertwined transcriptional network downstream of deltaNp63 which resembles what has been reported for transcription factors controlling pluripotency (41–43). Accordingly, it is evident that programming of cell fates, molecular subtypes, and phenotypes is efficiently achieved using a collection of transcription factors, whereby the tight regulation of entire gene expression programs is controlled by a distinct set of master transcription factors. The identification of transcription factors that are both dependent on and activate deltaNp63 in the squamous subtype can help in optimizing therapy and shed light on the molecular mechanisms which define the squamous/basal-like subtype.
Consistent with our findings, hypoxic pathways were previously reported to be enriched in the squamous subtype (8), although a direct connection to deltaNp63 was not known. Given the major role of HIF1A in the response to hypoxia, it appears likely that it may also function in promoting the increased aggressiveness of the squamous subtype and promoting cellular plasticity under hypoxic conditions (44–46). Less is known about the role of RXRA and BLHLE40 in pancreatic cancer. A connection of BHLHE40 to hypoxia was reported in breast cancer (47). BHLHE40 was found to play a crucial role in promoting a molecular switch to proinflammation in T-helper cells (48). In the brain, BHLHE40 plays a role in promoting synaptic plasticity (49). These roles in other systems imply that BHLHE40 may also play a role in promoting cellular plasticity and leading to a poorer outcome. RXRA forms a heterodimer with peroxisome proliferator-activated receptors (PPARs), which can be targeted by PPAR inhibitors (50). RXRA also dimerizes with the vitamin D receptor and its mutation is associated with bladder cancer and melanoma (50, 51). It should be noted that the targeting of all of the members of a circuitry can lead to unexpected adverse effects. Accordingly, the role of these factors in the scope of other circuits and regions should be taken into consideration. For example, RXRA, which is a member in the circuit that we identified, correlated with favorable prognosis in pancreatic cancer patients. Thus, targeting RXRA should be approached with caution. Further studies will uncover if optimal treatment of the more aggressive subtype may include the pathways that are regulated through these transcription factors.
Based on our work, the primary mode of activation of the deltaNp63-associated transcription factor network appears to be at distal regulatory elements whereby the factors not only promote the expression of common target genes but also control the expression of one another. This enhancer-specific effect is supported by our observation that differentially active regions in L3.6pl and BxPC-3 compared with Panc-1 were mainly found at putative enhancer regions and not at transcriptional start sites. Moreover, deltaNp63 occupancy at enhancers, but not at TSS regions, was more closely correlated with the effects of p63 depletion on target gene expression. Additionally, p63 was found to be up-regulated upon loss of KDM6A in pancreatic cancer via activation of associated super enhancers (22). Altogether, our findings underscore the importance of distal regulatory elements in driving important transcriptional programs in tumorigenesis and tumor progression, thereby providing a further rationale for targeting these regions and their dependencies.
In this study, we reported deltaNp63 as an activator of gene transcription. At first glance this is surprising due to the previously assumed dominant negative role of this transactivation domain-lacking isoform (52). Interestingly, our report is joined by other studies demonstrating a role of deltaNp63 as a transcriptional activator (53–58). For example, the viral oncogene protein BamHI-A rightward frame 1 (BARF1) was shown to be exclusively transactivated by deltaNp63 and not p53 or TAp63 in epithelial tumors (54). Moreover, NECTIN1 which we reported as a highly dependent gene in squamous pancreatic cancer, was also identified to be activated via two deltaNp63-dependent enhancers in skin (55). Our findings suggest a model where deltaNp63 is able to activate target genes supported by a highly interactive transcriptional factor network. This hypothesis is consistent with the fact that we see a large number of deltaNp63-bound regions, which are not marked by H3K27ac. In contrast, those that are marked by H3K27ac in the investigated cellular systems are frequently cooccupied by RXRA, BHLHE40, and/or HIF1A. Notably, p63 peaks marked by H3K27ac, indicative of active enhancer elements, more frequently intersect with BHLHE40-bound peaks (21.3%), while deltaNp63-bound regions not cooccupied by H3K27ac only show a much lower overlap with BHLHE40 (6.8%). In particular, the high dependence of deltaNp63 on BHLHE40 and the similar effects of the knockdown of both of these factors on proliferation implies a cooperative paradigm of gene activation. Different transcriptional circuitries can also explain the differential expression of p63 isoforms in various systems. In contrast to deltaNp63, the lack of effects of TAp63 knockdown on proliferation and the inability of the TAp63 expressing MIA Paca-2 to form spheres imply that deltaNp63 and TAp63 may have opposing roles in pancreatic cancer. The differential mechanisms and roles of these two isoforms in pancreatic cancer will need to be more thoroughly investigated in future studies.
We have identified a marked dependence of a subset of super enhancers on deltaNp63 which may open the door for specific targeting of the squamous subtype of pancreatic cancer. However, our findings also confirm the tremendous plasticity of pancreatic cancer, where a single factor is required for the activation of a whole gene signature associated with a poorer outcome. This has been further confirmed with recent findings by Somerville et al. (59) published during the revision of this work showing that deltaNp63 can reprogram the enhancer landscape in pancreatic cancer and lead to a more aggressive phenotype. Despite the fact that cancer is characterized by inter- and intratumor heterogeneity, distinct patterns of gene activation still emerge and may imply a natural selection process where certain attributes, such as overexpression of deltaNp63, lead to the selective growth or survival of these more aggressive and highly pliable tumor cells. It is likely that similar selective pressures will occur as we target the dependencies of the subtype-specific enhancers since the activation of other factors will likely lead to the activation of other compensatory gene expression programs. Future studies will be necessary to determine which factors specifically determine the gene expression patterns and cellular phenotypes of other pancreatic cancer subtypes. It will then be possible to examine the biological and therapeutic effects of subtype switching and determine whether such approaches may be useful in a therapeutic setting.
Materials and Methods
Cell Culture.
L3.6pl cells (37) were cultured in phenol-free minimum essential medium (MEM) (Thermo Fisher Scientific) supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% glutamine. Capan-1, BxPC-3, and MIA PaCa-2 were maintained in Roswell Park Memorial Institute medium (RPMI 1640; Thermo Fisher Scientific) supplemented with MEM. Panc-1, PaTu 8902, and DanG cells were maintained in high glucose GlutaMAX Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin/streptomycin. Protocols for siRNA knockdowns, proliferation assays, sphere formation assays, and harvesting of protein and RNA from cells are in SI Appendix.
ChIP, ATAC, and Library Preparations.
Chromatin immunoprecipitation was performed as described previously (26, 60). ATAC-seq was performed following the protocol of Buenrostro et al. (61). Libraries for RNA were prepared using the TruSeq RNA Library Prep kit V2 (Illumina) according to the manufacturer’s instructions. Libraries for DNA from ChIP were made using the Microplex Library Preparation kit v2 (Diagenode) according to the manufacturer’s instructions. ATAC libraries were made using the Nextera DNA Library Prep kit. Detailed protocols including number of replicates and detailed steps are in SI Appendix.
Bioinformatic Analysis.
Reads from ChIP- and ATAC-seq experiments were mapped to the reference genome assembly (hg19) using BOWTIE2/2.2.5 (62) and converted to bam files and sorted and indexed using SAMTOOLS/1.4 (63). Subsequently, reads were normalized to 1× sequencing depth using the bamCoverage tool in DEEPTOOLS/2.4.0 (64), ignoring duplicates and extending to 200 bp (500 bp for ATAC-seq) to generate occupancy profiles that were viewed with the Integrative Genomics Viewer (IGV 2.4) (65, 66). Peaks were called using MACS2/2.1.1.20160309 without building the shifting model and with cutoff of less than 0.05 (broad cutoff of 0.05 for BRD4 and H3K27ac) and input files as background (67). Reads from RNA-seq experiments were mapped using TOPHAT/2.1.0 and annotation file for hg19 was downloaded from the University of California Santa Cruz table browser (68, 69). Fragments per kilobase per million (FPKM) values were calculated and differential gene expression analysis was performed using CUFFLINKS/2.2.1 (70). Detailed protocols for the bioinformatic analyses performed in this study are available in SI Appendix.
Statistical Analysis.
For patient survival curves, the Mantel–Cox test was used to evaluate significance. For sphere formation assays and FPKM values, the Mann–Whitney test was applied. For analysis of qPCR, a nonparametric t test was used. P values are as follows: ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank G. Salinas, F. Ludewig, and S. Lutz (Transcriptome and Genome Analysis Laboratory, University Medical Center Göttingen) for performing the next generation sequencing for ChIP-seq and RNA-seq data; Z. Najafova, X. Wang, A. Kutschat, J. Henck, and all the members of the Johnsen group for their support and helpful discussions; and the Fischer group at the German Center for Neurodegenerative Diseases for performing the next generation sequencing for ATAC-seq. This work was supported by the German Academic Exchange Service (F.H.H.), Deutsche Krebshilfe (PiPAC Consortium, Grant 70112505), and the Deutsche Forschungsgemeinschaft (Grant JO 815/3-2 to S.A.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq, ChIP-seq, and ATAC-seq profiles have been deposited at ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) (accession nos. E-MTAB-7033–E-MTAB-7035).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812915116/-/DCSupplemental.
References
- 1.Zhao L, Lee VHF, Ng MK, Yan H, Bijlsma MF. Molecular subtyping of cancer: Current status and moving toward clinical applications. Brief Bioinform. April 12, 2018 doi: 10.1093/bib/bby026. [DOI] [PubMed] [Google Scholar]
- 2.Dai X, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 3.Andre F, Pusztai L. Molecular classification of breast cancer: Implications for selection of adjuvant chemotherapy. Nat Clin Pract Oncol. 2006;3:621–632. doi: 10.1038/ncponc0636. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malvezzi M, et al. European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol. 2017;28:1117–1123. doi: 10.1093/annonc/mdx033. [DOI] [PubMed] [Google Scholar]
- 6.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey P, et al. Australian Pancreatic Cancer Genome Initiative Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu; Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaferia GR, et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J. 2016;35:595–617. doi: 10.15252/embj.201592404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soares E, Zhou H. Master regulatory role of p63 in epidermal development and disease. Cell Mol Life Sci. 2018;75:1179–1190. doi: 10.1007/s00018-017-2701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 13.Nylander K, et al. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol. 2002;198:417–427. doi: 10.1002/path.1231. [DOI] [PubMed] [Google Scholar]
- 14.Koike M, et al. Molecular detection of circulating esophageal squamous cell cancer cells in the peripheral blood. Clin Cancer Res. 2002;8:2879–2882. [PubMed] [Google Scholar]
- 15.Kumakura Y, et al. Elevated expression of ΔNp63 in advanced esophageal squamous cell carcinoma. Cancer Sci. 2017;108:2149–2155. doi: 10.1111/cas.13394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye S, Lee KB, Park MH, Lee JS, Kim SM. p63 regulates growth of esophageal squamous carcinoma cells via the Akt signaling pathway. Int J Oncol. 2014;44:2153–2159. doi: 10.3892/ijo.2014.2374. [DOI] [PubMed] [Google Scholar]
- 17.Kakuki T, et al. Dysregulation of junctional adhesion molecule-A via p63/GATA-3 in head and neck squamous cell carcinoma. Oncotarget. 2016;7:33887–33900. doi: 10.18632/oncotarget.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bir F, et al. Potential utility of p63 expression in differential diagnosis of non-small-cell lung carcinoma and its effect on prognosis of the disease. Med Sci Monit. 2014;20:219–226. doi: 10.12659/MSM.890394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo Iacono M, et al. p63 and p73 isoform expression in non-small cell lung cancer and corresponding morphological normal lung tissue. J Thorac Oncol. 2011;6:473–481. doi: 10.1097/JTO.0b013e31820b86b0. [DOI] [PubMed] [Google Scholar]
- 20.Kouwenhoven EN, et al. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015;16:863–878. doi: 10.15252/embr.201439941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basturk O, et al. DeltaNp63 expression in pancreas and pancreatic neoplasia. Mod Pathol. 2005;18:1193–1198. doi: 10.1038/modpathol.3800401. [DOI] [PubMed] [Google Scholar]
- 22.Andricovich J, et al. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512–526.e8. doi: 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 24.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Najafova Z, et al. BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic Acids Res. 2017;45:127–141. doi: 10.1093/nar/gkw826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie W, et al. RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol. 2017;18:32. doi: 10.1186/s13059-017-1159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra VK, et al. Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4- and MYC-dependent manner. Nucleic Acids Res. 2017;45:6334–6349. doi: 10.1093/nar/gkx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra VK, et al. Krüppel-like transcription factor KLF10 suppresses TGFβ-induced epithelial-to-mesenchymal transition via a negative feedback mechanism. Cancer Res. 2017;77:2387–2400. doi: 10.1158/0008-5472.CAN-16-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herreros-Villanueva M, et al. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2013;2:e61. doi: 10.1038/oncsis.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomberk G, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun. 2018;9:1978. doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starruß J, de Back W, Brusch L, Deutsch A. Morpheus: A user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics. 2014;30:1331–1332. doi: 10.1093/bioinformatics/btt772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan W, Zhang Y, Chen X. TAp63γ and ΔNp63γ are regulated by RBM38 via mRNA stability and have an opposing function in growth suppression. Oncotarget. 2017;8:78327–78339. doi: 10.18632/oncotarget.18463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamdan FH, Johnsen SA. Super enhancers–New analyses and perspectives on the low hanging fruit. Transcription. 2018;9:123–130. doi: 10.1080/21541264.2017.1372044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamizu K, et al. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Reports. 2013;1:545–559. doi: 10.1016/j.stemcr.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruns CJ, Harbison MT, Kuniyasu H, Eue I, Fidler IJ. In vivo selection and characterization of metastatic variants from human pancreatic adenocarcinoma by using orthotopic implantation in nude mice. Neoplasia. 1999;1:50–62. doi: 10.1038/sj.neo.7900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura T, Fidler IJ, Coombes KR. Gene expression profile of metastatic human pancreatic cancer cells depends on the organ microenvironment. Cancer Res. 2007;67:139–148. doi: 10.1158/0008-5472.CAN-06-2563. [DOI] [PubMed] [Google Scholar]
- 39.Deer EL, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soucie EL, et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 2016;351:aad5510. doi: 10.1126/science.aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Simandi Z, et al. OCT4 acts as an integrator of pluripotency and signal-induced differentiation. Mol Cell. 2016;63:647–661. doi: 10.1016/j.molcel.2016.06.039. [DOI] [PubMed] [Google Scholar]
- 43.Xu J, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye LY, et al. Hypoxia-inducible factor 1α expression and its clinical significance in pancreatic cancer: A meta-analysis. Pancreatology. 2014;14:391–397. doi: 10.1016/j.pan.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Matsuo Y, et al. Hypoxia inducible factor-1 alpha plays a pivotal role in hepatic metastasis of pancreatic cancer: An immunohistochemical study. J Hepatobiliary Pancreat Sci. 2014;21:105–112. doi: 10.1002/jhbp.6. [DOI] [PubMed] [Google Scholar]
- 46.Shi CY, Fan Y, Liu B, Lou WH. HIF1 contributes to hypoxia-induced pancreatic cancer cells invasion via promoting QSOX1 expression. Cell Physiol Biochem. 2013;32:561–568. doi: 10.1159/000354460. [DOI] [PubMed] [Google Scholar]
- 47.Chakrabarti J, et al. The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br J Cancer. 2004;91:954–958. doi: 10.1038/sj.bjc.6602059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu F, et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med. 2018;215:1813–1821. doi: 10.1084/jem.20170155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton KA, et al. Mice lacking the transcriptional regulator Bhlhe40 have enhanced neuronal excitability and impaired synaptic plasticity in the hippocampus. PLoS One. 2018;13:e0196223. doi: 10.1371/journal.pone.0196223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halstead AM, et al. Bladder-cancer-associated mutations in RXRA activate peroxisome proliferator-activated receptors to drive urothelial proliferation. eLife. 2017;6:30862. doi: 10.7554/eLife.30862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin J, et al. Genetic variants in the vitamin D pathway genes VDBP and RXRA modulate cutaneous melanoma disease-specific survival. Pigment Cell Melanoma Res. 2016;29:176–185. doi: 10.1111/pcmr.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 53.Kajiwara C, et al. p63-dependent Dickkopf3 expression promotes esophageal cancer cell proliferation via CKAP4. Cancer Res. 2018;78:6107–6120. doi: 10.1158/0008-5472.CAN-18-1749. [DOI] [PubMed] [Google Scholar]
- 54.Hoebe E, et al. Epstein-Barr virus gene BARF1 expression is regulated by the epithelial differentiation factor ΔNp63α in undifferentiated nasopharyngeal carcinoma. Cancers (Basel) 2018;10:E76. doi: 10.3390/cancers10030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mollo MR, et al. p63-dependent and independent mechanisms of nectin-1 and nectin-4 regulation in the epidermis. Exp Dermatol. 2015;24:114–119. doi: 10.1111/exd.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang K, Wu WM, Chen YC, Lo SH, Liao YC. ΔNp63α transcriptionally regulates the expression of CTEN that is associated with prostate cell adhesion. PLoS One. 2016;11:e0147542. doi: 10.1371/journal.pone.0147542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holcakova J, et al. ΔNp63 activates EGFR signaling to induce loss of adhesion in triple-negative basal-like breast cancer cells. Breast Cancer Res Treat. 2017;163:475–484. doi: 10.1007/s10549-017-4216-6. [DOI] [PubMed] [Google Scholar]
- 58.Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S. An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PLoS One. 2009;4:e5623. doi: 10.1371/journal.pone.0005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somerville TDD, et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 2018;25:1741–1755.e7. doi: 10.1016/j.celrep.2018.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagarajan S, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karolchik D, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.