Significance
This work shows that the amidated terminal ends of the secreted hypocretin (HCRT) peptides (HCRTNH2) are autoantigens in type 1 narcolepsy, an autoimmune disorder targeting HCRT neurons. The autoimmune process is usually initiated by influenza A flu infections, and a particular piece of the hemagglutinin (HA) flu protein of the pandemic 2009 H1N1 strain was identified as a likely trigger. This HA epitope has homology with HCRTNH2 and T cells cross-reactive to both epitopes are involved in the autoimmune process by molecular mimicry. Genes associated with narcolepsy mark the particular HLA heterodimer (DQ0602) involved in presentation of these antigens and modulate expression of the specific T cell receptor segments (TRAJ24 and TRBV4-2) involved in T cell receptor recognition of these antigens, suggesting causality.
Keywords: narcolepsy, TCR, autoimmunity, DQ0602, tetramer
Abstract
Type 1 narcolepsy (T1N) is caused by hypocretin/orexin (HCRT) neuronal loss. Association with the HLA DQB1*06:02/DQA1*01:02 (98% vs. 25%) heterodimer (DQ0602), T cell receptors (TCR) and other immune loci suggest autoimmunity but autoantigens are unknown. Onset is seasonal and associated with influenza A, notably pandemic 2009 H1N1 (pH1N1) infection and vaccination (Pandemrix). Peptides derived from HCRT and influenza A, including pH1N1, were screened for DQ0602 binding and presence of cognate DQ0602 tetramer-peptide–specific CD4+ T cells tested in 35 T1N cases and 22 DQ0602 controls. Higher reactivity to influenza pHA273–287 (pH1N1 specific), PR8 (H1N1 pre-2009 and H2N2)-specific NP17–31 and C-amidated but not native version of HCRT54–66 and HCRT86–97 (HCRTNH2) were observed in T1N. Single-cell TCR sequencing revealed sharing of CDR3β TRBV4-2-CASSQETQGRNYGYTF in HCRTNH2 and pHA273–287-tetramers, suggesting molecular mimicry. This public CDR3β uses TRBV4-2, a segment modulated by T1N-associated SNP rs1008599, suggesting causality. TCR-α/β CDR3 motifs of HCRT54–66-NH2 and HCRT86–97-NH2 tetramers were extensively shared: notably public CDR3α, TRAV2-CAVETDSWGKLQF-TRAJ24, that uses TRAJ24, a chain modulated by T1N-associated SNPs rs1154155 and rs1483979. TCR-α/β CDR3 sequences found in pHA273–287, NP17–31, and HCRTNH2 tetramer-positive CD4+ cells were also retrieved in single INF-γ–secreting CD4+ sorted cells stimulated with Pandemrix, independently confirming these results. Our results provide evidence for autoimmunity and molecular mimicry with flu antigens modulated by genetic components in the pathophysiology of T1N.
Whereas genetic (1–6), epidemiological (7–11), and pathophysiological (12–14) studies implicate autoimmunity in response to flu infection in type 1 narcolepsy (T1N), a disease caused by hypocretin (HCRT) neuronal loss (15–17), the autoantigen involved is still unknown. Unlike other autoimmune diseases, autoantibodies to HCRT cell proteins, HCRT itself (18–21), or other targets, such as TRIB2 (22, 23) or HCRT receptor 2 (HCRTR2) (24–27) have not been consistently found. This has led to the suggestion that HCRT cell loss may be T cell-mediated, with limited or no involvement of autoantibodies.
A large recent genome-wide association study analysis of over 5,500 patients across multiple ethnicities confirmed the primordial importance of the HLA DQ0602 heterodimer in disease predisposition, with important secondary associations in T cell receptor (TCR) loci α and β, and other immune loci, such as perforin (28). Of additional interest is the observation that TCR polymorphisms associated with T1N are quantitative trait loci (QTL) for TRAJ24 (decreasing), TRAJ28, and TRVB4-2 (increasing) usage in peripheral T cells in both controls and patients (28). A significant L to F coding polymorphism (underlined in sequences throughout the paper) located within the antigen binding complementarity-determining region (CDR) 3 loop of TRAJ24-expressing TCRs is also associated with T1N. These effects suggest that T cell responses involving TRAJ24- or TRAJ28- and TRVB4-2–bearing TCRs are likely bottleneck responses in the causative autoimmune T cell responses leading to HCRT cell death. Overall, the genome-wide association study results suggest the importance of DQ0602 presentation of antigens to specific CD4+ T cells by antigen presenting cells and, considering the involvement of perforin, likely subsequent CD8+ destruction of HCRT cells (28). CD8+ mediation of HCRT cell death has also been shown to cause T1N in an animal model (29).
Recently, much has been learned regarding what may trigger T1N autoimmunity. Onset of T1N in children, which can often be timed to the week, is seasonal and peaks in spring and summer (10). Onset has also been associated with Streptococcus pyogenes infections (9, 30) and influenza A (10), suggesting the trigger may be a winter infection, followed by a delay in peaking at 5 mo once sufficient HCRT cell loss has occurred (>80%) and symptoms appear. Most strikingly, prevalence of T1N increased several-fold in mainland China and Taiwan, with a similar delay following the 2009–2010 “Swine Flu” H1N1 influenza pandemic (pH1N1) (10, 31–33), although this association is less clear in other countries (11). Similarly, cases of T1N have been triggered by the pH1N1 vaccine Pandemrix in Europe with relative risk increasing 5- to 14-fold in children and adolescents and 2- to 7-fold in adults after vaccination (33, 34). Because Pandemrix is an AS03 adjuvanted vaccine containing the artificially produced reassortant strain X-179A, a mix of PR8, an old H1N1 strain derived from pre-2009 seasonal H1N1, and A/California/07/2009 containing key H1N1 2009 surface proteins (hemagglutinin, HA and neuraminidase, NA) (35), flu proteins are likely critically involved in triggering T1N. The fact that HLA and TCR genetic associations are universal (6, 36–39) is also consistent with a flu trigger, because influenza A infections occur on a global basis (40). As has been illustrated above, both the H1N12009 pandemic and the Pandemrix vaccination also exhibited variable effects across different countries, thus demanding the consideration of additional factors to fully explain T1N occurrences.
Based on the above information, T cell reactivity to various autoantigens has also been explored, starting with HCRT itself, with various results reported as follows. In 2013, homology between DQ0602-binding sequences pHA275–287 and HCRT56–68/HCRT87–99, sequences encoding the C-terminal end of secreted hypocretin-1 and hypocretin-2 was noted and mimicry suggested, although part of the results published showing differential ELISpot reactivity in narcolepsy versus controls were later retracted (18, 41). A lack of differential ELISpot CD4+ T cell reactivity (measured by INF-γ and IL-17) to HCRT53–67 and HCRT86–97 was subsequently found by Kornum et al. (19), who tested 22 cases and 23 DQ0602 controls with 6 known HCRT sequences binding DQ0602 (detection limit of 1 in 10,000 cells). Similar results were found by Ramberger et al. (20), who tested CD4+ cells of 15 patients and 13 DQ0602 controls after an 8-d culture amplification with HCRT peptide pools in carboxyfluorescein succinimidyl ester (CFSE), followed by FACS. These authors found three potentially reactive subjects in patients and none in controls, suggesting no differential effects.
The situation changed a few months ago, thanks to work published by Latorre et al. (42). In this work, the authors used an ultrasensitive technique to detect autoantigen T-cell responses that involves polyclonal expansion and cloning of CD45RA−CD4+ T cell lines, followed by screening of these lines for proliferation as a surrogate of reactivity to autoantigen peptide pools presented by autologous B cells. Screening peripheral blood mononuclear cells (PBMCs) of 19 T1N cases [15 with documented HCRT deficiency, defined by low hypocretin-1 in the cerebrospinal fluid (CSF)] and 13 DQ0602 controls, Latorre et al. found strong line reactivity to HCRT in all patients versus no or limited responses in 13 controls, with significantly higher reactivity in T1N. Although less strikingly different, increased T cell reactivity in narcolepsy was also found with TRIB2, a previously proposed autoantigen. Further characterization of the identified autoreactive cell lines showed autoreactive CD4+ T cells to be mostly DR-restricted and very rare: <1–89.7 cells per 106 CD4+ cells. TCR sequencing, although limited, revealed Vβ sequences without any clear pattern. Latorre et al. also screened these same cell lines for proliferative responses to seasonal influenza A antigens and found comparable responses in patients and controls, concluding that flu mimicry could not be detected.
In this work and following on our 2013 initial findings, we have continued to systematically interrogate DQ0602-restricted flu and autoantigen CD4+ responses in DQ0602 T1N patients versus matched controls. To increase sensitivity of detection, we used DQ0602 tetramers examining frequency and TCR sequences of CD4+ T cells recognizing specific HCRT and flu epitopes bound to DQ0602 tetramers. Results of our experiments now confirm our initial hypothesis of molecular mimicry between pHA273–287 and HCRT54–66/HCRT86–97, although autoreactivity is only found with the amidated, posttranslationally modified version of the antigen (HCRT54–66-NH2/HCRT86–97-NH2 denoted collectively as HCRTNH2). TCR sequences involved in these responses involves TRAJ24 and TRBV4-2, correlating with genetic effects and supporting causality. Our data suggest the importance of TCR-α/β chain-specific responses in driving autoimmunity through the hitchhiking of partner TCR-α/β cross-reactive sequences, a phenomenon that can be best visualized through TCR-α/β network analysis across epitopes and individuals.
Results
DQ0602 Restricted Epitopes of H1N1 Flu Responses in T1N Versus Controls Suggest a Role for pHA273–287 and NP17–31.
We screened overlapping 15-mer peptides for DQ0602 binding for key proteins contained in Pandemrix and pH1N1 wild-type, HA, NA, influenza RNA-dependent RNA polymerase subunit 1 (PB1), and nucleoprotein (NP) (Dataset S1), as described previously (18). These proteins were selected on the basis of their abundance in Pandemrix (enriched by vaccine design) (35, 43) and the wild-type pH1N1 (present naturally). DQ0602 peptide screening was done using peptides derived from A/reassortant/NYMC X-179A (A/California/07/2009 × NYMC X-157, reference ADE2909) and complemented with peptides of wild-type pH1N1 sequence (A/California/07/2009, reference AFM728) when polymorphism was evident, or peptides were of distinct origin (i.e., NP). Screening for these proteins identified a large number of binders (Dataset S1) using the biotin-conjugated Epstein-Barr Virus (EBV)486–500 epitope (Bio-EBV), a known binder (18, 44). Peptides that outcompeted >75% of the reference EBV peptide signal were considered binders and selected for tetramer construction and screening (18, 41).
PBMCs from six cases and four DQ0602 controls were expanded in an in vitro 10-d culture stimulated with Pandemrix (100 ng/mL) or the corresponding single peptide (6.25 μM) (see SI Appendix, Supplementary Methods and Dataset S9 for demographic information, and Dataset S11 for staining results). The control and narcoleptic subjects were all known post-Pandemrix vaccinees, except for one case who developed T1N independent of vaccination 4 mo before blood sampling (recent-onset case).
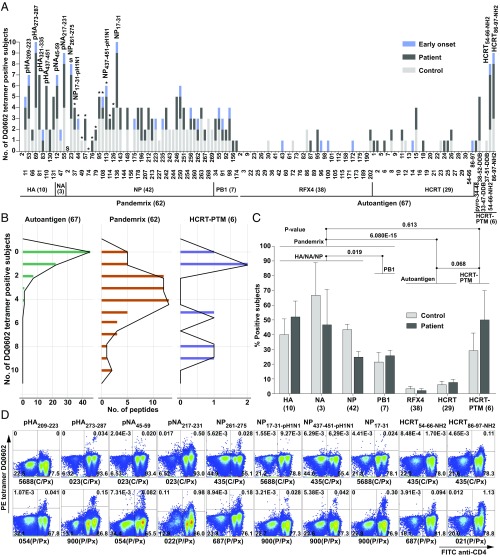
Fig. 1A shows controls and T1N patients (from 10 subjects) positive for each tetramer staining, as well as example staining (Fig. 1D) with selected tetramers. Tetramer positivity ranged from none to 10 positives for some tetramers (Fig. 1B). A global analysis of tetramer (peptides binned into respective proteins) reactivity confirmed (Fig. 1C) that post-Pandemrix cases and controls had similar flu antigen exposure, as no difference was found across the nine vaccinated cases and controls overall (all epitopes). Significantly lower reactivity with PB1 versus HA, NP, and NA was found, likely the consequence of the lower concentration of PB1 in Pandemrix (and all vaccines) (35).
Fig. 1.
Results of primary tetramer screening in 10 subjects. (A) Tetramer positivity in post-Pandemrix patients (dark gray, from five total), post-Pandemrix controls (light gray, from four total), and one early-onset patient (onset = 0.3 y before blood sampling; blue, single subject). Tetramers were built one by one using peptide exchange and DQ0602-CLIP (SI Appendix, Supplementary Methods). Binder sequences are provided in Dataset S1. S, shared by A/California/07/2009 and A/Puerto Rico/8/1934 (PR8); asterisk (*), from A/California/07/2009; others are of PR8 origin. Last six peptides are posttranslationally modified sequences of prepro-HCRT, including N-pyroglutamate (pyro) HCRT34–48, HCRT38–52, HCRT33–47, and HCRT37–51 with double disulfide bond (DDB), C-terminal amidated HCRT54–66 and HCRT86–97, denoted HCRT54–66-NH2 and HCRT86–97-NH2, as in secreted, mature hypocretin-1 and -2. (B) Distribution of positive subjects per antigen. (C) Mean percentage of positive subjects for DQ0602 binders in each protein in post-Pandemrix subjects. (D) Example of FACS staining for various selected tetramers. The entire FACS staining plots are provided as Dataset S11. CD3+ T cells are shown with frequency. C, control; C/Px, post-Pandemrix control; P, patient; P/Px, post-Pandemrix patient.
X-179A–derived tetramers with the highest frequencies of specific CD4+ T cells included pHA273–287, pNA217–231 (pH1N1 specific epitope), NP17–31 (PR8 specific epitope), and NP261–275 (shared epitope between A/California/07/2009 and PR8) (Fig. 1D), suggesting these epitopes are immunodominant (Fig. 1A, NP epitopes from pH1N12009 instead of PR8 indicated by asterisks). pHA273–287 was notable as it was previously reported to have homology with HCRT sequences HCRT54–66 and HCRT86–97 (18, 41). Interestingly, among immunodominant epitopes, responses to three epitopes—pHA273–287 (of H1N12009 origin), NP17–31 (of PR8 origin), and NP261–275 (shared with A/California/07/2009 and PR8)—were preliminarily increased in Pandemrix-vaccinated cases versus controls, with reactivity in the early-onset case (Fig. 1A). These three epitopes were thus selected for additional investigation.
Additional analysis of 24 cases and 14 DQ0602 controls confirmed significant overabundance of T cells reactive to pHA273–287 and NP17–31 but not NP261–275 tetramers (Fig. 2 A and C, SI Appendix, Fig. S1, and Dataset S11), suggesting that these two epitopes could be involved in the recruitment of differential CD4+ T cell populations related to T1N. This was also the case in post-Pandemrix subjects only (11 cases of mean age 17 ± 2 y vs. 8 controls of mean age 27 ± 6 y) (SI Appendix, Fig. S1D), an important control as these subjects had similar exposure to pH1N1 around the first wave of the pH1N1 pandemic infection. Flu reactivity in T1N is not broadly shared across all H1N1 epitopes, because other influenza epitopes, including pNA217–231, NP261–275, and NP17–31-pH1N1, the homologous sequence of PR8 NP17–31 in pH1N12009 (A/California/07/2009, 2 amino acid difference in the core) (Dataset S2), showed little to no significant differences overall and in subgroups (SI Appendix, Fig. S1 A–C).
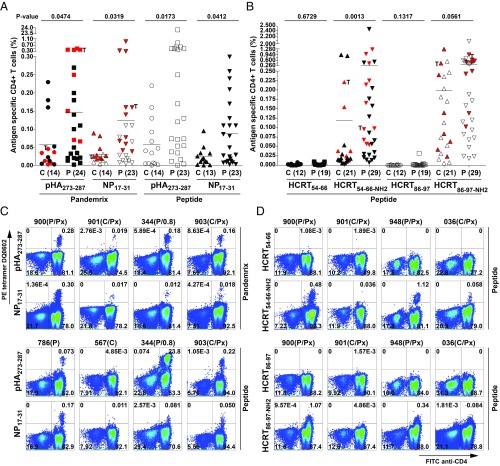
Fig. 2.
Results of tetramer screening using pHA273–287, NP17–31, HCRT54–66, HCRT54–66-NH2, HCRT86–97, and HCRT86–97-NH2 in 35 patients and 22 controls. Percentage of CD4+ cells positive for pHA273–287 and NP17–31 tetramers (A) and HCRT54–66/HCRT54–66-NH2 and HCRT86–97/HCRT86–97-NH2 tetramers (B) in controls versus patients. PBMCs of subjects were grown in Pandemrix or the cognate peptide for 10 or 14 d in two independent experiments, and as results were repeatable and did not differ statistically, the mean is presented in each dot. T, twins. Subjects in red are those with single-cell sorting and TCR-sequencing data available. Number of subjects tested is in parenthesis. Example of pHA273–287 and NP17–31 tetramer staining (C) and HCRT54–66, HCRT54–66-NH2, HCRT86–97, and HCRT86–97-NH2 tetramer staining (D) in a few subjects (for all results, see Dataset S11). CD3+ T cells are shown with frequency. C, control; C/Px, post-Pandemrix control; P, patient; P/Px, post-Pandemrix patient. Interval is shown if patient is early onset (<1.0 y).
Narcolepsy Cases Have Increased CD4+ T Cell Autoreactivity to Amidated HCRT54–66-NH2 and HCRT86–97-NH2 Epitopes.
We next tested whether T1N was associated with autoreactivity involving two proteins known to be primarily colocalized with HCRT, HCRT itself and regulatory factor X4 (RFX4), a transcription factor abundant in HCRT cells (45). HCRT and RFX4 binders were tested in the 10 subjects as above, with results suggesting no or little reactivity, consistent with T cell tolerance toward autoantigen peptide sequences (Fig. 1 A and B). Interestingly, while the CD4+ T cell reactivity to native versions of self-peptides, such as native HCRT and RFX4, was significantly lower (P < 10−14) than that of influenza peptides (Fig. 1C), the reactivity to the amidated version of HCRT was comparable to influenza-derived peptides (Fig. 1B).
Because prepro-HCRT is cleaved and posttranslationally modified in the region where homologous sequences HCRT54–66 and HCRT86–97 are located (resulting in secreted hypocretin-1 and -2), most notably C-amidated, a transformation necessary to its biological activity, we also tested HCRT54–66-NH2 and HCRT86–97-NH2, modified peptides that also bind DQ0602 with higher affinity (Dataset S1) (18). To our surprise, T cell reactivity to these fragments was high in both DQ0602 controls and T1N subjects (Fig. 2B and Dataset S11). Of notable interest was the fact that HCRT54–66-NH2 staining was generally restricted to distinct populations (not unlike what was observed with flu antigens), while HCRT86–97-NH2 staining was more heterogeneous, suggestive of large amounts of low affinity tetramer cross-reactivity (Fig. 2D and Dataset S11). These data were later expanded to 29 cases and 21 DQ0602 positive controls, and a significantly higher percentage of HCRT54–66-NH2 tetramer-positive cells was found in T1N (Fig. 2B), with marginally higher values for HCRT86–97-NH2. Subanalysis of 13 post-Pandemrix cases (mean age 17 ± 1 y) versus 10 post-Pandemrix controls (mean age 27 ± 6 y) confirmed significantly higher reactivity for HCRT86–97-NH2 and marginally higher values for HCRT54–66-NH2 (SI Appendix, Fig. S1F). Even more strikingly, in 7 recent onset cases (22 ± 6 y) versus 21 controls (23 ± 3 y), higher reactivity was found to HCRT54–66-NH2, suggesting an important role in the cause of T1N (SI Appendix, Fig. S1G).
TCR Sequencing of Flu Tetramer-Positive Cells Indicate Bias in TCR-α/β Usage Consistent with in Vitro Clonal Expansion.
Tetramer-reactive CD4+ T cells to pHA273–287, NP17–31, HCRT54–66-NH2, and HCRT86–97-NH2 epitopes from 14 to 20 subjects were next FACS single-sorted and their TCR (TCR-α/β) sequenced in control and T1N patients (including the post-Pandemrix subjects and one early-onset case, mentioned above). Subjects sequenced are denoted in red in Fig. 2 A and B. A large overlap of subjects was sequenced for all tetramers (see Datasets S4 and S9 for details). For comparison and reference frequencies, ∼106 peripheral memory CD4+CD45RA− T cells of the same subjects were sequenced for TCR-α and TCR-β V and J segment baseline usage in 126 subjects (28) (Dataset S4).
As expected, we found strong significant bias in TCR-α/β usage (TRAV, TRAJ, and TRBV families) specific of each epitope, except for HCRT54–66-NH2 and HCRT86–97-NH2, where a large amount of sequence sharing was found across the two highly homologous HCRT epitopes (also called HCRTNH2 as a group for future reference) and sequence usage diversity was higher in comparison with flu epitopes (SI Appendix, Fig. S2 and Dataset S3). For example, TCR-α usage of pHA273–287 was highly enriched in TRAJ23 [Mantel Haenszel odds ratio (OR) = 16.1], TRAV13-1 (OR = 17.0), and TRAV35 (OR = 23.2), while TCR-β usage for the epitope was enriched in TRBV19 (OR = 25.6) and TRBV4-3 (OR = 21.0) in comparison with the same subject’s CD4+CD45RA− cell peripheral usage, without differences in control versus T1N. Similarly, NP17–31 TCR cells were enriched in TRAV12-2 (OR = 18.6), TRAV8-6 (OR = 11.6), TRBV4-2 (OR = 42.8), TRBV7-9 (OR = 445.1), and TRBV20-1 (OR = 4.0) (Dataset S3) without differences in control versus T1N.
TCR Sequencing of C-Amidated HCRT Tetramer-Positive Cells Indicate Broad TCR Reactivity and Has Consequently More Limited Bias in TCR-α/β Usage.
Sequencing of amidated HCRT+ tetramers gave a different picture (SI Appendix, Fig. S2 and Dataset S3) in comparison with the flu-positive tetramer. First, although increased usage of specific TCR segments was found, usage diversity was much higher (SI Appendix, Fig. S2), notably when HCRT86–97-NH2 tetramers were used, giving a typically less distinct population with likely lower TCR affinity (Fig. 2D and Dataset S11). Second, in cases where HCRT54–66-NH2 and more rarely HCRT86–97-NH2 tetramers gave a clearly separated population on FACS plots (Dataset S11), sequencing resulted in almost uniform TCR sequences (even less diverse than with flu antigens), with no usage sequence sharing (patients 022, 948, 051, and 685 for HCRT54–66-NH2 and controls 567 and 391 for HCRT86–97-NH2 in SI Appendix, Fig. S2, with corresponding FACS plots in Dataset S11).
Shared CDR3 and CDR3 k-Mer Analysis Reveals Enriched k-Mers Across Flu and HCRTNH2 Tetramers.
pHA273–287 and HCRT54–66-NH2/HCRT86–97-NH2 share weak epitope homology (18, 41), a result confirmed using tools.iedb.org/cluster/reference/ (46), with three EBV sequences as anchor (Fig. 3E). Based on this, mimicry has been proposed but not demonstrated (18, 41). To search for mimicry across pHA273–287, NP17–31, HCRT54–66-NH2, and HCRT86–97-NH2, we first analyzed sharing of CDR3α, CDR3β, and CDR3α/β pairs across epitopes. Dataset S4 shows cross tabulations of shared sequences across epitopes.
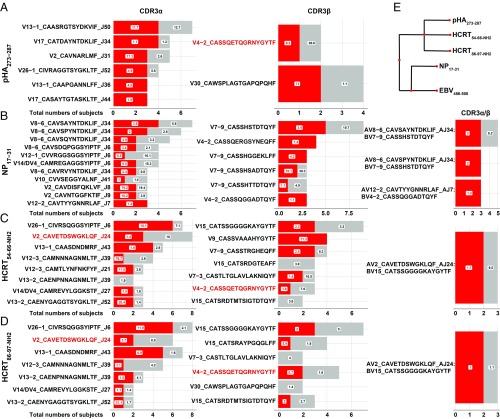
Fig. 3.
Shared public CDR3α, CDR3β, and CDR3α/β sequences for pHA273–287 (A), NP17–31 (B), HCRT54–66-NH2 (C), and HCRT86–97-NH2 (D) across subjects. Red, number of patients with corresponding CDR3; gray, number of controls with same CDR3 (per horizontal axis). Public clones in these cases are CDR3s found in at least three individuals. Box numbers indicate median percentage when the CDR3 occurred across patients and controls. (E) pHA273–287, NP17–31, HCRT54–66-NH2, and HCRT86–97-NH2 sequence homology assessed using tools.iedb.org/cluster/reference/ (46), with three EBV sequences used as anchor. Note that high similarity of HCRT54–66-NH2 and HCRT86–97-NH2 also share homology with pHA273–287 (18, 41), as previously reported, but not NP17–31.
A large number of CDR3 motifs were shared between HCRT54–66-NH2 and HCRT86–97-NH2 tetramers across CDR3α (11.8%), CDR3β (9.3%), and both CDR3α/β (3.6%), a result that is not surprising considering that HCRT54–66-NH2 and HCRT86–97-NH2 only differ in their N-amidated C-terminal end, L-NH2 (hypocretin-1) and M-NH2 (hypocretin-2) in P9 of their binding repertoire (Dataset S2). In contrast, pHA273–287, a viral epitope that has some weak homology with HCRT54–66-NH2 and HCRT86–97-NH2 (Fig. 3E) shared very few of the same clones with HCRT54–66-NH2 and HCRT86–97-NH2: for example, only 1.9% and 1.5% for CDR3α, 0.7% and 1.8% for CDR3β, and none and 0.3% (two instances) for CDR3α/β with HCRT54–66-NH2 and HCRT86–97-NH2, respectively. Potential sharing was even lower with NP17–31, which shared a few clones with pHA273–287, and at even lower level with HCRTNH2.
To explore differential CDR3 k-mer usage per epitopes and disease status, we extracted peptide contact residues for each of the CDR3α/β motif derived from pHA273–287, NP17–31, and HCRTNH2 across individuals. Both linear and discontinuous k-mers (3- to 5-mer) of peptide contact CDR3s as in GLIPH (47) were extracted. Tetramer-derived unique CDR3 k-mers were compared with a large database of CDR3 k-mers derived from bulk sequencing of ∼106 CD4+CD45RA− T cells obtained from the same 35 narcolepsy and 22 DQ0602 control subjects (see Dataset S5 for a list of paratopes significant at the P < 0.05 false-discovery rate value for each epitope). Because CDR3β sequences are more unusual than CDR3α, more CDR3β than CDR3α paratopes were enriched across all epitopes (Dataset S5). Interestingly, paratope motifs SQG, S.GR, and SQGR were significantly enriched in narcolepsy versus controls in pHA273–287 tetramer-positive cells (Dataset S5). This motif is unique to TRBV4-2 and TRBV4-3 families and reflected a significant enrichment. SI Appendix, Fig. S3 also shows preferential usage of specific amino acids in tetramers versus control sequences at various lengths, an analysis that was not revealing beyond the significant k-mers presented in Dataset S5. We note that in comparison with k-mers generated from bulk sequencing of total CD4+ memory cells, tetramer-specifc CDR3α/β k-mers were significantly enriched and reflected the clonal response to same peptide specificity (SI Appendix, Fig. S3 and Dataset S5).
Commonly Used CDR3α and CDR3β Segments Observed with pHA273–287 and NP17–31 Tetramers.
We next analyzed sharing of CDR3α, CDR3β, and CDR3α/β pairs across patients and controls, with all “public” CDR3 clones found at least three times across subjects (Fig. 3). Not surprisingly, sharing of public CDR3α was more frequent than sharing of public CDR3β and exact CDR3α/β pairs were rarely seen across subjects. In pHA273–287-specific clones, six different CDR3αs were recruited across three individuals or more, with TRAV2-CAVNARLMF-TRAJ31, TRAV26-1-CIVRAGGTSYGKLTF-TRAJ52, TRAV13-1-CAAPGANNLFF-TRAJ36, and TRAV17-CASAYTGTASKLTF-TRAJ44 being overrepresented in narcolepsy cases compared with controls. Consistent with usage patterns in Fig. 3, CDR3αs from TRAV families, such as TRAV13-1, TRAV26-1, and TRAV17 were dominant. On the other hand, with respect to CDR3βs sharing in pHA273–287-specific clones, while only one clone (TRBV30-CAWSPLAGTGAPQPQHF) met our threshold (shared at least three times), there was another clone TRBV4-2-CASSQETQGRNYGYTF that was shared across one narcolepsy case and one control (Dataset S4). This clone was retained for analysis because of its extensive sharing (eight individuals) in both control and narcolepsy HCRTNH2 tetramers, as detailed below.
Among the NP17–31 tetramer-specific CDR3α clones, dominant TRAV8-6 usage was found with 5 of 11 clones that were shared in three or more individuals all bearing TRAV8-6 and TRAJ34/TRAJ6. Additionally TRAV12 was used in clones that were shared. Finally, among NP17–31 tetramer-specific CDR3β clones, TRBV7-9-CASSHSTDTQYF public CDR3 was preferentially used across eight individuals (including five patients).
Sharing for viral epitope pHA273–287 and NP17–31 tetramers was as expected from usage preference reported in SI Appendix, Fig. S2. The most commonly used segments in enriched clones were TRAV8-6, TRAJ34, TRBV7-9, and TRBV4-2 for NP17–31. Usage diversity for pHA273–287 was biased toward TRAV13-1, TRAV17, and TRBV4-2, with more limited sharing of TRBV for this epitope. We also noted frequent usage of TRAV26-1 CDR3α across all epitopes; DQ0602 is a strong trans-QTL of TRAV26-1 segment usage, probably because it interacts through its CDR1α with DQ0602 (48).
HCRTNH2 Tetramers Most Frequently Use TRAJ24 CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24.
As mentioned above, in narcolepsy cases, TRAJ24, TRAJ28, and TRBV4-2 are of special interest because genetic QTL locations modulating their usage are strongly associated with narcolepsy. While TRAJ28 did not show over representation, we observed extensive sharing of specific TRAJ24 and TRBV4-2 fragments in CD4+ T cells reactive to HCRTNH2 tetramers among cases and controls (Fig. 3). The most striking finding was extensive sharing of TRAJ24 CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 in patients and controls with HCRT54–66-NH2 and HCRT86–97-NH2, respectively (Fig. 3 C and D). This is of significance considering that TRAJ24L polymorphism (TRAV2-CAVETDSWGKLQF-TRAJ24) is protective (in comparison with TRAJ24F), an effect replicated across many studies (2, 5, 28). Furthermore, from our bulk sequencing data, we observed that TRAJ24 is a low-usage segment (0.87%), suggesting this is highly significant, as it is highly enriched in the HCRTNH2-reactive public CD4+ T cell response. Datasets S6 and S7 report on all TRAJ24 and TRBV4-2 clones found in all cultures, respectively. As can be seen in the datasets, the TRAV2-CAVETDSWGKLQF-TRAJ24 CDR3α generally occurs in the context of diverse CDR3βs containing TRBV15, TRBV2, TRBV29-1, and TRBV9 in controls and TRBV15, TRBV3-1, TRBV7-2, and TRBV20-1 in patients. Only TRBV15-CATSSGGGGKAYGYTF CDR3β was shared by two patients and one control following HCRTNH2 culture and tetramer isolation. Of note, TRAV2-CAVETDSWGKFQF-TRAJ24 (the equivalent CDR3α that would carry TRAJ24F, the genetically narcolepsy-associated allele) was not observed in any subject, control, or T1N.
Single-Cell Sorting and TCR Sequencing of INF-γ+, Pandemrix-Reactive CD4+ T Cells Confirms the Importance of pHA273–287 and NP17–31 as Immunodominant Peptides of the HLA Class II Response to Pandemrix, and Suggest Mimicry with HCRT CD4+ Responses.
INF-γ+ (Pandemrix-reactive) CFSE-low CD4+ cells from PBMCs in eight patients and eight controls were single-cell–sequenced after Pandemrix stimulation (overlapping with the sample above used in tetramers). This experimental design amplifies all MHC class II-mediated responses, with DQ0602-mediated responses likely in the minority, notably because DRB1 responses are typically dominant among class II responses. To limit this problem, one patient repeated twice was also cultured with Pandemrix in the presence of anti-DR and anti-DP antibodies, restricting the reactivity to a DQ response (DQ0602 and the other translocated allele). Upon completion of these experiments, CDR3 TCR-α and TCR-β sequences were matched with tetramer-derived sequences to search for commonality (Dataset S8). Strikingly, we found that sequences of pHA273–287 and NP17–31 tetramer-positive cells, in many cases sharing both TCR-α and -β chains, were present in these cultures, most notably in the cultures that had been blocked by anti-DR and anti-DP antibodies, validating our tetramer based assays. Interestingly, we also found a significant number of sequences shared with those of HCRT54–66-NH2 and HCRT86–97-NH2 tetramer-positive cells, suggesting mimicry with one or several unknown peptide variants in Pandemrix (Datasets S8 and S10).
Network Analysis of CDR3α/β Pairs Reveals That HCRTNH2 Response Is Dominated by a Central Node of TRAJ24 CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 That Is Closely Intertwined with a Major Node Using TRBV4-2 CDR3β TRBV4-2-CASSQETQGRNYGYTF Across HCRTNH2 and pHA273–287 Tetramers.
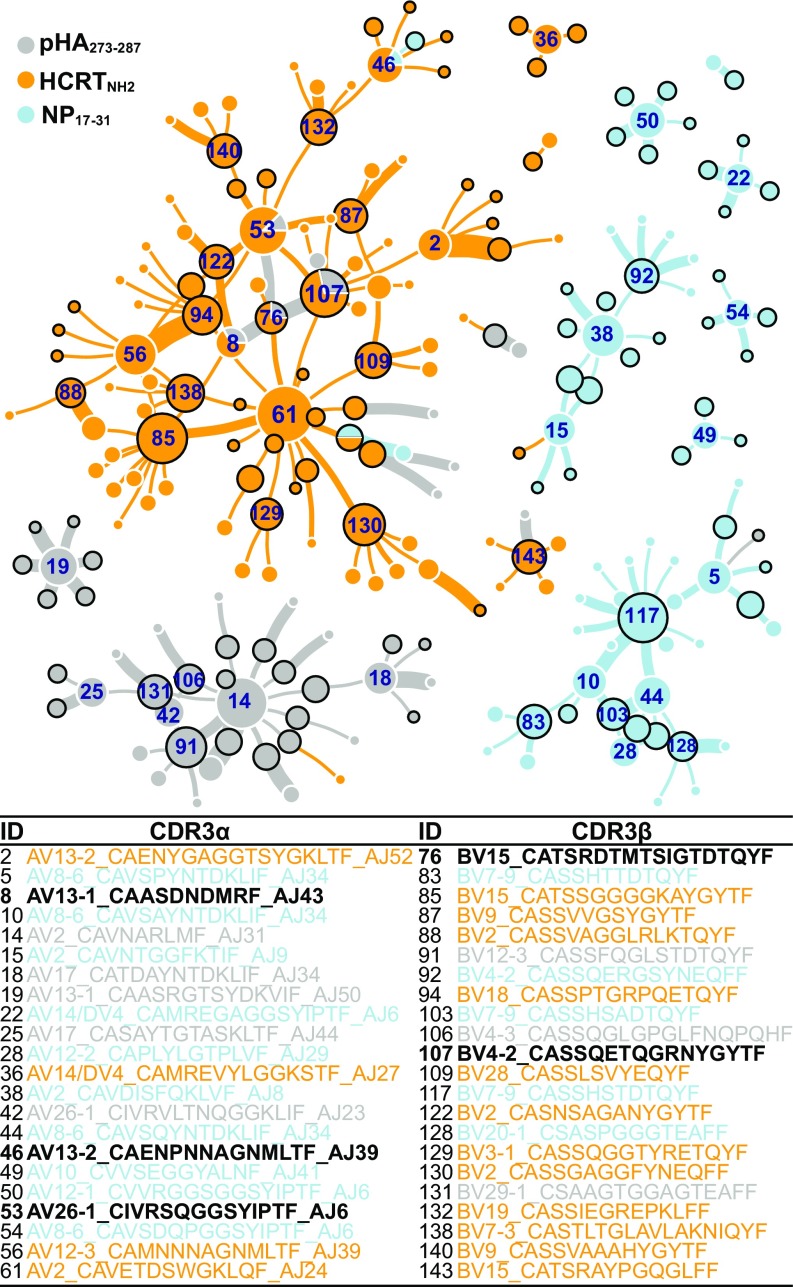
In Fig. 4, paired CRD3α and CRD3β found in various tetramers across antigens but present at least in three or more individuals (from Fig. 3, public CDR3s) were used as inputs to build a recursive tree traversal algorithm and the resulting CDR3 pairs used to construct a network plot. The recursive tree traversal algorithm was implemented to find all CDR3s that pair with each of the public CDR3s.
Fig. 4.
Network clustering of public TCR segments obtained through tetramer sequencing. Public CDR3s were used as input to a recursive tree traversal algorithm, which recursively extracts all pairs until no pairs remained. Each node represents a CDR3α (white external circle) or a CDR3β (black external circle) segment, with size proportional to occurrences. The network is seeded by nodes that contain only public CDR3s (CDR3s found in at least three different individuals) that connect to each other, then can connect once to any CDR3 partner found fewer than three times. Lines connecting these clusters represent heterodimers, with thickness proportional to clone number. Color-coding corresponds to type of tetramer (pHA273–287, HCRTNH2 or NP17–31) yielding specific clones.
These shared connections were plotted as a network plot, with the thickness of connecting lines reflecting number of clones found with each denoted heterodimer. Tables in Fig. 4 indicate key segments used in this connectome. As can be seen in the figure, the nodes in this connectome resolve into respective antigen specificities. For example, two clearly separated clusters (with central nodes labeled as 38 and 117) are NP17–31 reactive clones; these clones predominantly use TRAV2, TRBV4-2, TRBV7-9, TRAV8-6, TRAJ6, TRAJ34, and TRBV20. Note in the figure the near absence of connections to any of the HCRTNH2 reactive CDR3s. In contrast, clusters of HCRTNH2 and pHA273–287 were tightly connected and intertwined (with central node labeled 61 in Fig. 4), reflecting sequence homology between these epitopes, increased sharing of specific CDR3α and CDR3β (Dataset S4), and two groups of antigen-specific heterodimers (HCRTNH2 and pHA273–287 -specific) connected through a few discrete cross-reactive nodes.
The HCRTNH2 reactive node is centered around the identified CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 (Fig. 4, node labeled as 61). As mentioned above, this TCR-α chain connects with a large variety of diverse TCR-β sequences, suggesting that HCRTNH2 may interact more primarily with TCR-α. The CDR3βs that are paired with TRAV2-CAVETDSWGKLQF-TRAJ24 (#61 in Fig. 4) are dominated exclusively by HCRTNH2 reactive nodes such as TRBV15-CATSSGGGGKAYGYTF (#85 in Fig. 4), TRBV7-3-CASTLTGLAVLAKNIQYF (#138 in Fig. 4), TRBV3-1-CASSQGGTYRETQYF (#129 in Fig. 4), TRBV2-CASSGAGGFYNEQFF (#130 in Fig. 4), TRBV28-CASSLSVYEQYF (#109 in Fig. 4), and TRBV18-CASSPTGRPQETQYF (#94 in Fig. 4) as expected.
In addition, two tightly intertwined CDR3β (TRBV15-CATSRDTMTSIGTDTQYF #76 and TRBV4−2-CASSQETQGRNYGYTF #107 in Fig. 4) and two CDR3α (TRAV13-1-CAASDNDMRF-TRAJ43 #8 and TRAV26-1-CIVRSQGGSYIPTF-TRAJ6 #53 in Fig. 4) sequences are bireactive: that is, they are found in some instances in the context of HCRTNH2 tetramers, and in other in the context of pHA273–287 tetramers (Fig. 4 and Dataset S4). The finding is most striking for one TRBV4-2 (found in one patient and one control with pHA273–287 and in three patients and five controls for HCRTNH2) and several other TRAV26-1 sequences (Fig. 4 and Dataset S4). These cross-reactive sequences are also connected with the TRAV2-CAVETDSWGKLQF-TRAJ24 (Fig. 4, node labeled as 61) HCRTNH2 tetramer dominated network both directly and indirectly. Presumably TCR-α/β containing these chains have high affinity for pHA273–287 and HCRTNH2 in various combinations. Importantly, however, although we found heterodimers containing two presumably pHA273–287 and HCRTNH2 cross-reactive CDR3α and -β sequences (Fig. 4), these combinations were never retrieved in single cells by either pHA273–287 or HCRTNH2 tetramers.
Discussion
In this work, we screened two potential autoantigens, HCRT and RFX4, for DQ0602 binding and T cell reactivity using tetramer staining of PBMCs of T1N cases and controls. These human proteins were selected because of their high enrichment in HCRT cells (45). We found only limited TCR reactivity directed toward any segment of the native RFX4 or HCRT peptide sequence, but strong reactivity to the N-amidated, posttranslationally modified (PTM) C-terminal end of hypocretin-1 and -2 (denoted HCRT54–66-NH2 and HCRT86–97-NH2, or, as a group, as HCRTNH2) (Fig. 1). HCRTNH2 are homologous peptides differing only in their C-terminal–amidated residue (L-NH2 for HCRT1 and M-NH2 for HCRT2) that are secreted by HCRT neurons after undergoing amidation of their C-terminal end, a PTM necessary for biological activity (49). These amidated peptides likely have very short half-lives, with degradation involving C-terminal peptidases, as the main HCRT metabolite found in CSF is a truncated hypocretin-1 fragment that lacks its C-terminal–amidated end (50). The observation of higher reactivity of the C-terminal end of hypocretin-1 and -2, HCRTNH2, extends other observations made in other autoimmune diseases, such as rheumatoid arthritis, celiac disease, and type 1 diabetes (51), indicating that PTMs are frequently present in autoantigens. It is also noteworthy that HCRTNH2 have higher affinities for DQ0602 in comparison with nonamidated HCRT (Dataset S1) (18), a property that could also play a role. The last amidated amino acid is projected to be positioned in P9, an important residue that contacts both DQ0602 and the TCR. The current explanation is that PTMs do not occur sufficiently in the thymus during central tolerance establishment (52) and thus increase the risk of autoimmunity.
Relevant to T1N, we also found that patients have higher numbers of CD4+ T cells binding HCRT54–66-NH2 when presented by DQ0602, the major HLA allele associated with T1N (Fig. 2). Recent onset cases also showed significantly higher numbers of T cells recognizing HCRT86–97-NH2, a peptide differing only in its C-terminal amino acid from HCRT54–66-NH2. These results indicate that autoreactivity to amidated HCRTNH2 fragments is common in both patients and controls but occurs at higher levels in patients. These findings could reflect causality, cross-reactivity secondary to HCRT cell loss via another mechanism, or an epiphenomenon. Strikingly however, not only was sharing of “public” CDR3 sequences found across patients [as recently reported for gluten-reactive T cells in celiac disease (53)], but the top CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 involved in the recognition of HCRTNH2 in controls and patients uses TRAJ24 (Fig. 3), a segment marked by SNPs rs1154155G and rs1483979 strongly associated with T1N risk. This genetic association is the strongest association after HLA across all ethnicity (1–6) and, as TRAJ24 is normally only used rarely (0.87% of repertoire) (28), the result is striking and suggests causality.
These results extend on recent findings by Latorre et al. (42), who found increased polyclonal CD4+ T cell reactivity to HCRT in 19 patients but not 13 DQ0602 controls. In this work, however, reactivity was primarily DR-restricted and was to diverse HCRT fragments, not just the C-terminal–amidated end. Cellular phenotypes were also Th1, as expected for cellular-mediated autoimmune responses. Because DR is not genetically associated with T1N, this broader reactivity is unlikely primary to narcolepsy pathophysiology, and may rather result from epitope spreading following a DQ response. Interestingly, unlike our finding with DQ0602 tetramers, extended reactivity to HCRT was found only in patients and not in controls, suggesting that perhaps the autoimmune reaction, which we hypothesize is initiated by DQ0602-flu and DQ0602-HCRT reactivity (see below), becomes broader and more intense as the disease develops.
We also note that the Latorre et al. (42) finding will need confirmation, notably because many of the autoreactive T1N cases included were atypical. Indeed, 16 reported cases with typical cataplexy and low CSF hypocretin-1 (T1N) contained 2 HLA-DQ0602− subjects. HLA negativity in HCRT deficiency is extremely rare, with less than 15 cases documented worldwide (54). Similarly, three HLA-DQ0602− cases without cataplexy (so called T2N, non-HCRT–deficient) were also autoreactive. Nonetheless, these experiments suggest that polyclonal HCRT epitopes involving HLA-DR play a role in the disease process as it progresses to full disease. Rare CD8+ clones reactive to HCRT were also found, suggesting a possible involvement of this antigen in CD8+-mediated cell killing of HCRT cells.
Besides showing that HCRTNH2 is a major autoantigen in narcolepsy, our study found that T1N is associated with differential reactivity to at least two flu peptides, pHA273–287 and NP17–31, derived from X-179A, the reassortant strain used in Pandemrix (Fig. 2). Pandemrix has been linked with T1N onset in northern Europe (34); however, the latter correlation varies across countries, suggesting interactions with other factors (33). Of particular interest was the fact that one peptide, pHA273–287, is specific of 2009H1N1, while another peptide NP17–31, was derived from the vaccine backbone sequence strain PR8, an old seasonal H1N1 strain originally derived from 1918H1N1. The NP17–31 sequence is representative of seasonal H1N1 before the 2009 pandemic flu (descendant of 1918H1N1, which disappeared around 1957 and reappeared in 1976) and seasonal H2N2 “Asian flu” strains that circulated from 1956 to 1958. Whether or not involved in mimicry, our data also suggest that flu epitopes derived from multiple flu strains (pH1N1, PR8-like, H3N2) may be needed to favor immune response toward T1N autoimmunity (10, 32, 33, 35, 55). In this context, Pandemrix vaccination coincidental with pH1N1 pandemic or other strains may also have been a perfect storm in some countries. The confluence of exposure to multiple epitopes could also explain why the administration of Arepanrix, an adjuvanted vaccine similar to Pandemrix that was used in Canada, has not exhibited a strong link to T1N (33, 56).
As noted in Fig. 3E, pHA273–287 has significant homology with HCRT54–66 and HCRT86–97 sequences and their binding repertoire (18), making it the most suitable mimicry candidate with HCRTNH2 (Fig. 3). This homology is reflected by increased sharing of CDR3α and CDR3β sequences in the corresponding tetramers, which was very high within HCRTNH2 peptides (only different in one amino acid, projected to be in P9), substantial between HCRTNH2 and pHA273–287 but almost absent between HCRTNH2 and NP17–31 or between pHA273–287 and NP17–31. This is particularly visible in sequences of tetramer-positive cells that are found in at least three independent individuals (Fig. 3) because almost identical sequences are retrieved for HCRT54–66-NH2 and HCRT86–97-NH2 (HCRTNH2) (Fig. 3), validating the use of this stringent filter to reduce noise and eliminate any risk of contamination. This almost linear relationship suggests that the more homology the mimic has with the autoantigen sequence, the most likely similar TCR sequences are recruited by cross-reactivity. Surprisingly, however, the number of epitope-specific k-mer paratopes shared between sequences was similar (Dataset S5). We also tried to examine whether TCR paratope motifs common to HCRTNH2 and pHA273–287 recognition were more abundant in HCRTNH2+ tetramers from patients versus controls; however, no compelling observations were made.
Most revealing was a network analysis examining clustering of public CDR3α and CDR3β pairs across pHA273–287, NP17–31, and HCRTNH2 tetramers (Fig. 4). Using this analysis, clearly separate clusters of connected CDR3α−CDR3β heterodimers emerged for NP17–31 tetramers and a subset of pHA273–287 heterodimers. Most striking was the presence of four pHA273–287 cross-reactive sequences in the midst of the main HCRT cluster, suggesting significant pHA273–287–HCRTNH2 cross-reactivity. In contrast, cross-reactivity with NP17–31, although present in two instances within the HCRT cluster, was very peripheral (Fig. 4). The pHA273–287–HCRTNH2 cross-reactive nodes (#53, #8, #107, and #76 in Fig. 4) include two CDR3α chains and two CDR3β chains. These TCRs are present as TCR-α/β heterodimers in pHA273–287-binding TCRs (denoted as gray connections in Fig. 4), and as single chains (with other partners) in HCRTNH2 binding. We were unable to find any TCR-α/β heterodimers containing the same two of four cross-reactive TCR-α and -β segments reactive to both HCRTNH2 and pHA273–287 tetramers.
One likely hypothesis is that these cross-reactive combinations, even if not yet retrieved in both HCRTNH2 and pHA273–287 tetramer-sequenced pools, are truly cross-reactive to both HCRTNH2 and pHA273–287 in vivo, and that this will be detected with increased amounts of single-cell tetramer sequencing. We, however, prefer the hypothesis that TCR-α with one cross-reactive TCR-α or -β chain (not both) will be sufficient for cross-reactivity to occur. This hypothesis is supported by our CDR3α/β network analysis, which reveals that many CDR3α or CDR3β can accommodate a large diversity of partnering chains in the recognition of any specific peptide (Fig. 4). This is, for example, the case for CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 (#61 in Fig. 4), which recognizes HCRTNH2 in association with distinct CDR3βs and for CDR3β TRBV15-CATSSGGGGKAYGYTF, which recognizes HCRTNH2 in association with a large number of CDR3αs. This is also observed in NP17–31 (#38 and #117 in Fig. 4) or pHA273–287 (#14 and #91 in Fig. 4) specific-clusters, where an occasional α- or β-chain is a strong node connecting with multiple partners.
Similarly, a recent analysis of ∼80 available TCR-pMHC structures (26 MHC class II) has shown that, whereas there is always at least one CDR3β contact with pMHC, there are cases where no CDR3α contact occurs (47, 57). It may thus be that promiscuity of CDR3β could involve a CDR3α cross-reactive with HCRTNH2. Reverse peptide docking has also been reported in some cases (57), with CDR3β but not CDR3α contacting peptide residues. Similarly, an autoreactive TCR–MHC class II complex found extreme amino-terminal positioning of the TCR over the antigen-binding platform of the MHC molecule, suggesting a link between atypical TCR docking modes and autoreactivity (58), although this has not been found in other autoreactive structures. TCR interactions with HLA-peptide complexes are thus diverse (59) and do not always need tight contacts through both TCR-α and -β chains. In this configuration, TCR-α/β containing either a cross-reactive TCR-α, a TCR-β, or both could be the seed of pH1N1 and HCRT cross-reactivity, resulting in molecular mimicry and autoimmunity. Studies involving transfection of these heterodimers in TCR reporter-containing TCR-deficient Jurkat cell lines, with activation with cognate pHA273–287 and HCRTNH2 ligands displayed by DQ0602, will be needed to confirm whether a single cross-reactive TCR-α/β or both are needed for cross-reactivity.
Of special interest within the cluster of four cross-reactive sequences is CDR3β TRBV4-2-CASSQETQGRNYGYTF (#107 in Fig. 4), one of the most frequently shared CDR3β segments found in HCRTNH2 tetramers, a chain also used by pHA273–287 (Fig. 3). This is functionally significant because TRVB4-2, unlike others, is rarely used overall (0.74% of repertoire) (28), and expression of this segment is linked with T1N-associated SNP rs1008599 (6, 28, 48). Examination of QTL effects for TRAV26-1 (#53 in Fig. 4) (6.0% of repertoire), TRAV13-1 (#8 in Fig. 4) (2.0%), and TRBV15 (#76 in Fig. 4) (3.5%) did not reveal any genomic effect (28), unlike the well-established TRBV4-2 (#107 in Fig. 4) effect (28, 48); thus, functional connection through genetic association with these other segments could not be tested. Of particular interest is the size of the TRAV26-1-CIVRSQGGSYIPTF-TRAJ6 CDR3α node, likely explained by the fact that TRAV26-1 is a trans-QTL for DQ0602, reflecting direct CDR1/2 interactions of TRAV26-1 with the DQ0602 molecule, notably DQβ, the most specific portion of the DQ molecule (48). Other TRAV26-1 CDR3α cross-reactive sequences were retrieved (Dataset S4), but as the corresponding CDR3β was not amplified, position in the network of Fig. 4 could not be established.
In addition to the above, a large HCRTNH2 tetramer cluster centered around CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 (#61 in Fig. 4), the most frequently used segment by HCRTNH2. This segment carries the TRAJ L allele, known to be genetically protective for T1N. The TRAJ24 CDR3 segment (#61 in Fig. 4) connects to a diverse array of HCRTNH2-specific TCR-βs. These connections only occur as heterodimers in the context of HCRTNH2-reactive heterodimers (yellow lines in Fig. 4).
Heterodimers containing CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 (#61 in Fig. 4) and cross-reactive CDR3β TRBV4-2 (#107 in Fig. 4) and TRBV15 (#76 in Fig. 4) could also be involved in molecular mimicry, but we believe this hypothesis to be unlikely as it is the narcolepsy-protective J24L allele that is associated with this cluster. This is also supported by observation that CDR3α TRAV2-CAVETDSWGKLQF-TRAJ24 is observed more frequently only in controls but not in patients (SI Appendix, Fig. S2 and Dataset S3).
A number of molecular mechanisms have been suggested to explain the strong HLA-DQ0602 and the unusual TCR loci association in T1N autoimmunity (51). This work suggests that major pathophysiological steps involve tolerance escape due to PTMs of the HCRT autoantigen and cross-reactivity with specific flu sequences, which may be favored by differential binding affinity of specific CDR3α and CDR3β to binding flu mimics and the homologous HCRT fragment when presented by DQ0602. This mechanism would be a variation of the hotspot binding molecular mimicry hypothesis (51) that we call TCR heterodimer “hitchhiking.” Although this study links specific TCR heterodimers to an autoimmune disease through TCR tetramer sequencing and genetic effects, many questions remain unanswered, and will require more extensive TCR sequencing. As found by Latorre et al. (42), it is also likely that the disease mechanism subsequently involves antigen spreading with DR presentation of multiple HCRT epitopes, and possibly CD8+ T cell cytotoxic killing of HCRT neurons. Nonetheless, this study molecularly links genetic effects to specific TCRs and suggests that analysis of public TCRs of autoimmune responses through network analysis may have value in identifying molecular mimicry.
Methods
This study was reviewed and approved by the Stanford University Institutional Review Board (Protocol #14325, Registration #5136). Informed consent was obtained from each participant. Detailed information on subjects is described in SI Appendix. Details of vaccine, peptides, purification of DQ0602, peptide exchanged, tetramer staining, TCR sequencing, and network analysis are provided in SI Appendix and Datasets S1–S11.
Supplementary Material
Acknowledgments
We thank Dr. K. Christopher Garcia for providing plasmids and insect cell lines, the Emory NIH Tetramer Core Facility (NIAID-HHSN272201300006C) for sharing DQA1*01:02/DQB1*06:02 expression constructs, Wenchao Sun from the Advanced Drug Delivery Lab (BioADD) for fast protein liquid chromatography isolation of biotin-labeled DQ0602, and Selina Yogeshwar for editing. The study was primarily funded by gifts from Wake Up Narcolepsy, Jazz Pharmaceutical, and individual patients to Stanford University. Some samples have been gathered by defunct NIH Grant P50-NS-23724 (to E.M.; ended in 2016). Bulk Sequencing, past DQ0602 screening, and some sample collection has been funded by a prior grant from Glaxo Smith Kline, supervised by the European Medical Agency (ended in 2014), with corresponding data reported in Ollila et al. (28). Note that none of the authors have been involved in the generation of the ELISpot results reported in the retracted De la Herran-Arita et al. publication (18, 41), as determined by a review of Stanford’s internal investigation report by the NIH Office of Research Integrity. This work used the Genome Sequencing Service Center by Stanford Center for Genomics and Personalized Medicine Sequencing Center, supported by the Grant Award NIH S10OD020141.
Footnotes
Conflict of interest statement: E.M. occasionally consults and has received contracts from Jazz Pharmaceuticals, is and has been a Principal Investigator on clinical trials using sodium oxybate and Solriamfetol, Jazz Pharmaceutical products, for the treatment of T1N; none of these have any scientific relationships to the study.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818150116/-/DCSupplemental.
References
- 1.Han F, et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 2013;9:e1003880. doi: 10.1371/journal.pgen.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faraco J, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 2013;9:e1003270. doi: 10.1371/journal.pgen.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornum BR, et al. Common variants in P2RY11 are associated with narcolepsy. Nat Genet. 2011;43:66–71. doi: 10.1038/ng.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hor H, et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat Genet. 2010;42:786–789, and erratum (2011) 43:388. doi: 10.1038/ng.647. [DOI] [PubMed] [Google Scholar]
- 5.Hallmayer J, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41:708–711, and erratum (2009) 41:859. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ollila HM, et al. HLA-DPB1 and HLA class I confer risk of and protection from narcolepsy. Am J Hum Genet. 2015;96:136–146, and erratum (2015) 96:852. doi: 10.1016/j.ajhg.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Orozco FJ, Vicario JL, De Andres C, Fernandez-Arquero M, Peraita-Adrados R. Comorbidity of narcolepsy type 1 with autoimmune diseases and other immunopathological disorders: A case-control study. J Clin Med Res. 2016;8:495–505. doi: 10.14740/jocmr2569w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh AK, Mahlios J, Mignot E. Genetic association, seasonal infections and autoimmune basis of narcolepsy. J Autoimmun. 2013;43:26–31. doi: 10.1016/j.jaut.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aran A, et al. Elevated anti-streptococcal antibodies in patients with recent narcolepsy onset. Sleep. 2009;32:979–983. doi: 10.1093/sleep/32.8.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han F, et al. Narcolepsy onset is seasonal and increased following the 2009 H1N1 pandemic in China. Ann Neurol. 2011;70:410–417. doi: 10.1002/ana.22587. [DOI] [PubMed] [Google Scholar]
- 11.Dye TJ, Gurbani N, Simakajornboon N. Epidemiology and pathophysiology of childhood narcolepsy. Paediatr Respir Rev. 2018;25:14–18. doi: 10.1016/j.prrv.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Feltelius N, et al. A coordinated cross-disciplinary research initiative to address an increased incidence of narcolepsy following the 2009-2010 Pandemrix vaccination programme in Sweden. J Intern Med. 2015;278:335–353. doi: 10.1111/joim.12391. [DOI] [PubMed] [Google Scholar]
- 13.Ambati A, et al. Increased β-haemolytic group A streptococcal M6 serotype and streptodornase B-specific cellular immune responses in Swedish narcolepsy cases. J Intern Med. 2015;278:264–276. doi: 10.1111/joim.12355. [DOI] [PubMed] [Google Scholar]
- 14.Vaarala O, et al. Antigenic differences between AS03 adjuvanted influenza A (H1N1) pandemic vaccines: Implications for Pandemrix-associated narcolepsy risk. PLoS One. 2014;9:e114361. doi: 10.1371/journal.pone.0114361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishino S, Ripley B, Overeem S, Lammers GJ, Mignot E. Hypocretin (orexin) deficiency in human narcolepsy. Lancet. 2000;355:39–40. doi: 10.1016/S0140-6736(99)05582-8. [DOI] [PubMed] [Google Scholar]
- 16.Peyron C, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–997. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 17.Thannickal TC, et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27:469–474. doi: 10.1016/s0896-6273(00)00058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De la Herrán-Arita AK, et al. CD4+ T cell autoimmunity to hypocretin/orexin and cross-reactivity to a 2009 H1N1 influenza A epitope in narcolepsy. Sci Transl Med. 2013;5:216ra176. doi: 10.1126/scitranslmed.3007762. [DOI] [PubMed] [Google Scholar]
- 19.Kornum BR, et al. Absence of autoreactive CD4+ T-cells targeting HLA-DQA1*01:02/DQB1*06:02 restricted hypocretin/orexin epitopes in narcolepsy type 1 when detected by EliSpot. J Neuroimmunol. 2017;309:7–11. doi: 10.1016/j.jneuroim.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Ramberger M, et al. CD4+ T-cell reactivity to orexin/hypocretin in patients with narcolepsy type 1. Sleep (Basel) 2017;40:zsw070. doi: 10.1093/sleep/zsw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka S, Honda Y, Inoue Y, Honda M. Detection of autoantibodies against hypocretin, hcrtrl, and hcrtr2 in narcolepsy: Anti-Hcrt system antibody in narcolepsy. Sleep. 2006;29:633–638. doi: 10.1093/sleep/29.5.633. [DOI] [PubMed] [Google Scholar]
- 22.Cvetkovic-Lopes V, et al. Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J Clin Invest. 2010;120:713–719. doi: 10.1172/JCI41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lind A, et al. A/H1N1 antibodies and TRIB2 autoantibodies in narcolepsy patients diagnosed in conjunction with the Pandemrix vaccination campaign in Sweden 2009-2010. J Autoimmun. 2014;50:99–106. doi: 10.1016/j.jaut.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed SS, et al. Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2. Sci Transl Med. 2015;7:294ra105. doi: 10.1126/scitranslmed.aab2354. [DOI] [PubMed] [Google Scholar]
- 25.Giannoccaro MP, et al. Antibodies against hypocretin receptor 2 are rare in narcolepsy. Sleep (Basel) 2017;40 doi: 10.1093/sleep/zsw056. [DOI] [PubMed] [Google Scholar]
- 26.Luo G, et al. Absence of anti-hypocretin receptor 2 autoantibodies in post pandemrix narcolepsy cases. PLoS One. 2017;12:e0187305. doi: 10.1371/journal.pone.0187305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Heide A, Hegeman-Kleinn IM, Peeters E, Lammers GJ, Fronczek R. Immunohistochemical screening for antibodies in recent onset type 1 narcolepsy and after H1N1 vaccination. J Neuroimmunol. 2015;283:58–62. doi: 10.1016/j.jneuroim.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Ollila H, et al. 2018. Narcolepsy risk loci are enriched in immune cells and suggest autoimmune modulation of the T cell receptor repertoire. bioRxiv, 10.1101/373555. Preprint, posted July 22, 2018.
- 29.Bernard-Valnet R, et al. CD8 T cell-mediated killing of orexinergic neurons induces a narcolepsy-like phenotype in mice. Proc Natl Acad Sci USA. 2016;113:10956–10961. doi: 10.1073/pnas.1603325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longstreth WT, Jr, Ton TG, Koepsell TD. Narcolepsy and streptococcal infections. Sleep. 2009;32:1548. doi: 10.1093/sleep/32.12.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han F, Lin L, Li J, Dong XS, Mignot E. Decreased incidence of childhood narcolepsy 2 years after the 2009 H1N1 winter flu pandemic. Ann Neurol. 2013;73:560. doi: 10.1002/ana.23799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, et al. Symptoms and occurrences of narcolepsy: A retrospective study of 162 patients during a 10-year period in eastern China. Sleep Med. 2014;15:607–613. doi: 10.1016/j.sleep.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Weibel D, et al. Narcolepsy and adjuvanted pandemic influenza A (H1N1) 2009 vaccines—Multi-country assessment. Vaccine. 2018;36:6202–6211. doi: 10.1016/j.vaccine.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkanen T, Alakuijala A, Julkunen I, Partinen M. Narcolepsy associated with Pandemrix vaccine. Curr Neurol Neurosci Rep. 2018;18:43. doi: 10.1007/s11910-018-0851-5. [DOI] [PubMed] [Google Scholar]
- 35.Jacob L, et al. Comparison of Pandemrix and Arepanrix, two pH1N1 AS03-adjuvanted vaccines differentially associated with narcolepsy development. Brain Behav Immun. 2015;47:44–57. doi: 10.1016/j.bbi.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Han F, et al. HLA-DQ association and allele competition in Chinese narcolepsy. Tissue Antigens. 2012;80:328–335. doi: 10.1111/j.1399-0039.2012.01948.x. [DOI] [PubMed] [Google Scholar]
- 37.Miyagawa T, et al. New susceptibility variants to narcolepsy identified in HLA class II region. Hum Mol Genet. 2015;24:891–898. doi: 10.1093/hmg/ddu480. [DOI] [PubMed] [Google Scholar]
- 38.Mignot E, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–699. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tafti M, et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep (Basel) 2014;37:19–25. doi: 10.5665/sleep.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedford T, Cobey S, Beerli P, Pascual M. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2) PLoS Pathog. 2010;6:e1000918. doi: 10.1371/journal.ppat.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De la Herran-Arita AK, et al. Retraction of the research article: “CD4(+) T cell autoimmunity to hypocretin/orexin and cross-reactivity to a 2009 H1N1 influenza A epitope in narcolepsy”. Sci Transl Med. 2014;6:247rt241. doi: 10.1126/scitranslmed.3009995. [DOI] [PubMed] [Google Scholar]
- 42.Latorre D, et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature. 2018;562:63–68. doi: 10.1038/s41586-018-0540-1. [DOI] [PubMed] [Google Scholar]
- 43.Nicolson C, et al. An additional oligosaccharide moiety in the HA of a pandemic influenza H1N1 candidate vaccine virus confers increased antigen yield in eggs. Vaccine. 2012;30:745–751. doi: 10.1016/j.vaccine.2011.11.081. [DOI] [PubMed] [Google Scholar]
- 44.Demachi-Okamura A, et al. Epstein-Barr virus nuclear antigen 1-specific CD4+ T cells directly kill Epstein-Barr virus-carrying natural killer and T cells. Cancer Sci. 2008;99:1633–1642. doi: 10.1111/j.1349-7006.2008.00852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalal J, et al. Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 2013;27:565–578. doi: 10.1101/gad.207654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glanville J, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharon E, et al. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat Genet. 2016;48:995–1002. doi: 10.1038/ng.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakurai T, et al. Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 50.Zougman A, et al. Integrated analysis of the cerebrospinal fluid peptidome and proteome. J Proteome Res. 2008;7:386–399. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]
- 51.Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol. 2018;18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 52.Raposo B, et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat Commun. 2018;9:353. doi: 10.1038/s41467-017-02763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Risnes LF, et al. Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease. J Clin Invest. 2018;128:2642–2650. doi: 10.1172/JCI98819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han F, et al. HLA DQB1*06:02 negative narcolepsy with hypocretin/orexin deficiency. Sleep (Basel) 2014;37:1601–1608. doi: 10.5665/sleep.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cushing A, et al. Emergence of hemagglutinin mutations during the course of influenza infection. Sci Rep. 2015;5:16178. doi: 10.1038/srep16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montplaisir J, et al. Risk of narcolepsy associated with inactivated adjuvanted (AS03) A/H1N1 (2009) pandemic influenza vaccine in Quebec. PLoS One. 2014;9:e108489. doi: 10.1371/journal.pone.0108489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.La Gruta NL, Gras S, Daley SR, Thomas PG, Rossjohn J. Understanding the drivers of MHC restriction of T cell receptors. Nat Rev Immunol. 2018;18:467–478. doi: 10.1038/s41577-018-0007-5. [DOI] [PubMed] [Google Scholar]
- 58.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riley TP, et al. T cell receptor cross-reactivity expanded by dramatic peptide-MHC adaptability. Nat Chem Biol. 2018;14:934–942. doi: 10.1038/s41589-018-0130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.