Abstract
Background
Abundant evidence shows that triple-negative breast cancer (TNBC) is heterogeneous, and many efforts have been devoted to identifying TNBC subtypes on the basis of genomic profiling. However, few studies have explored the classification of TNBC specifically based on immune signatures that may facilitate the optimal stratification of TNBC patients responsive to immunotherapy.
Methods
Using four publicly available TNBC genomics datasets, we classified TNBC on the basis of the immunogenomic profiling of 29 immune signatures. Unsupervised and supervised machine learning methods were used to perform the classification.
Results
We identified three TNBC subtypes that we named Immunity High (Immunity_H), Immunity Medium (Immunity_M), and Immunity Low (Immunity_L) and demonstrated that this classification was reliable and predictable by analyzing multiple different datasets. Immunity_H was characterized by greater immune cell infiltration and anti-tumor immune activities, as well as better survival prognosis compared to the other subtypes. Besides the immune signatures, some cancer-associated pathways were hyperactivated in Immunity_H, including apoptosis, calcium signaling, MAPK signaling, PI3K–Akt signaling, and RAS signaling. In contrast, Immunity_L presented depressed immune signatures and increased activation of cell cycle, Hippo signaling, DNA replication, mismatch repair, cell adhesion molecule binding, spliceosome, adherens junction function, pyrimidine metabolism, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, and RNA polymerase pathways. Furthermore, we identified a gene co-expression subnetwork centered around five transcription factor (TF) genes (CORO1A, STAT4, BCL11B, ZNF831, and EOMES) specifically significant in the Immunity_H subtype and a subnetwork centered around two TF genes (IRF8 and SPI1) characteristic of the Immunity_L subtype.
Conclusions
The identification of TNBC subtypes based on immune signatures has potential clinical implications for TNBC treatment.
Electronic supplementary material
The online version of this article (10.1186/s13046-018-1002-1) contains supplementary material, which is available to authorized users.
Keywords: Triple-negative breast cancer, Tumor immunity, Immunogenomic profiling, Classification, Machine learning
Introduction
Triple-negative breast cancer (TNBC) is a breast cancer subtype that lacks the expression of hormone receptors (estrogen receptor (ER) and progesterone receptor (PR)) and human epidermal growth factor receptor 2 (HER2). TNBC is associated with a high risk of mortality for its aggressiveness and the lack of effective targeted therapies. Moreover, abundant evidence shows that TNBC is very heterogeneous [1–4]. Lehmann et al. identified six gene expression profile-based TNBC subtypes, including an immunomodulatory (IM) subtype that was enriched in immune cell processes [2]. Bonsang-Kitzis et al. identified six TNBC subgroups based on a biological network-driven approach, which included two immunity clusters whose stromal immune module gene signatures exhibited a strong prognostic value [3]. Burstein et al. identified four stable TNBC subgroups based on mRNA expression and DNA genomic profiling, which included Luminal/Androgen Receptor, Mesenchymal, Basal-Like Immune Suppressed, and Basal-Like Immune Activated (BLIA); furthermore, the authors identified potential therapeutic targets for these specific subtypes [4]. These efforts to classify TNBC might lay the foundation for developing targeted therapies for TNBC.
Recently, cancer immunotherapy has been successful in treating many refractory malignancies [5]. Thus, it is worth considering immunotherapy for TNBC, since the therapeutic options for this disease are significantly limited. Indeed, many experimental and clinical studies have explored the possibility of treating TNBC patients with immunotherapy [6–11]. Moreover, numerous studies have demonstrated that TNBC is more immunogenic than other breast cancer (BC) subtypes, which may warrant an immunotherapeutic approach for TNBC [12, 13]. However, currently, immunotherapeutic strategies exhibit beneficial effects in less than 20% of cancer patients. This suggests that not all TNBC patients could respond to immunotherapy. In fact, certain genetic or genomic features, such as tumor mutation burden (TMB), neoantigen load, PD-L1 expression, and deficient DNA mismatch repair, have been associated with cancer immunotherapeutic responsiveness [14–18].
In this study, we classified TNBC into three distinct subtypes by immunogenomic profiling: Immunity High (Immunity_H), Immunity Medium (Immunity_M), and Immunity Low (Immunity_L). We demonstrated the stability and reproducibility of this classification in four independent datasets by a machine learning approach. Furthermore, we identified the subtype-specific molecular features, including genes, gene ontology, pathways, and networks. The identification of immune signature-associated TNBC subtypes may facilitate the optimal selection of TNBC patients responsive to immunotherapy.
Methods
Clustering
For each TNBC dataset, we first quantified the enrichment levels of the 29 immune signatures in each TNBC sample by the single-sample gene-set enrichment analysis (ssGSEA) score [19, 20]. Based on the enrichment levels (ssGSEA scores) of the 29 immune signatures, we performed hierarchical clustering of TNBC.
Evaluation of immune cell infiltration level, tumor purity, and stromal content in TNBC
ESTIMATE [21] was used to evaluate the immune cell infiltration level (immune score), tumor purity, and stromal content (stromal score) for each TNBC sample.
Gene-set enrichment analysis
We performed gene-set enrichment analysis of the METABRIC and TCGA datasets by GSEA (R implementation) [22–24]. This analysis identified the KEGG [25] pathways that were upregulated in Immunity_H and Immunity_L (FDR < 0.05), respectively. The common pathways identified in both datasets were selected.
Correlation of pathway activities with immune cell infiltration levels in TNBC
We quantified the activity of a pathway with the ssGSEA score of the set of genes included in the pathway, and the immune cell infiltration level with the immune score. The Spearman correlation of the ssGSEA score and the immune score were used to evaluate the correlation of pathway activities with immune cell infiltration levels in TNBC.
Identification of TNBC subtype-specific gene ontology and networks
We used WGCNA [26] to identify the gene modules (gene ontology) that were significantly associated with the genes highly correlated with immune cell infiltration based on gene co-expression analysis. The gene modules specifically amplified in different TNBC subtypes were identified. On the basis of the expression correlations between the hub genes in the gene modules, we built gene–gene interaction networks. A hub gene was defined as a gene that was connected to no less than 10 other genes, with a connectedness weight greater than 0.25.
Survival analyses
We compared the survival prognosis (overall survival (OS), disease-free survival (DFS), and metastasis-free survival (MFS) of TNBC patients considering tumor subtype and the expression level of the identified genes, i.e., higher expression level (expression levels > median) versus lower expression level (expression levels < median). The log-rank test was used to calculate the significance of survival time differences using a threshold of P-value < 0.05. Kaplan–Meier curves were plotted to show the survival time differences. We performed the survival analyses using the METABRIC, TCGA, and GSE103091 datasets, where the survival data were available.
Class prediction
We transformed each attribute (immune signature or gene set) value (ssGSEA score) xi into xi′ by the equation xi′ = (xi − xmin)/(xmax − xmin), where xmin and xmax represent the minimum and maximum of the ssGSEA scores for the gene set across all TNBC samples, respectively. The Random Forest (RF) classifier was used to classify the TNBC subtypes. We set the number of trees to 100 and all 29 immune signatures as features for the RF classifier. The classification performance was evaluated by the accuracy and the weighted F-score. We carried out the classification in Weka [27].
Comparison of the proportions of immune cell subsets between TNBC subtypes
CIBERSORT [28] was used to calculate the proportions of 22 human immune cell subsets. We set 1000 permutations and P < 0.05 as the criteria for the successful deconvolution of a sample. We compared the proportions of the immune cell subsets between TNBC subtypes using the Mann–Whitney U test.
Comparison of clonal heterogeneity between the TNBC subtypes
We used the ABSOLUTE algorithm [29] to assess the ploidy score, representing clonal heterogeneity, for each TNBC sample. We compared the ploidy scores between the TNBC subtypes using the Kruskal–Wallis test.
Comparison of biological processes between the TNBC subtypes
We compared the activities (ssGSEA scores) of stem cell-associated (marker genes ABCA8 and ALDH1A1), proliferation (MKI67), and epithelial-to-mesenchymal transition (EMT) (ZEB1, ZEB2, SNAIL, CDH2 and TGFB1) biological processes between the TNBC subtypes. The Kruskal–Wallis test was used to determine the statistical significance of the results.
Comparison of somatic copy number alteration (SCNA) levels between the TNBC subtypes
We applied GISTIC2 [30] to the SNP6 file of the SCNA data for TNBC in TCGA. We obtained arm-level SCNA frequencies for Immunity_H and Immunity_L TNBC samples and compared them. Moreover, we calculated focal SCNA levels for each TNBC samples and compared them between Immunity_H and Immunity_L.
Results
Immunogenomic profiling identifies three TNBC subtypes
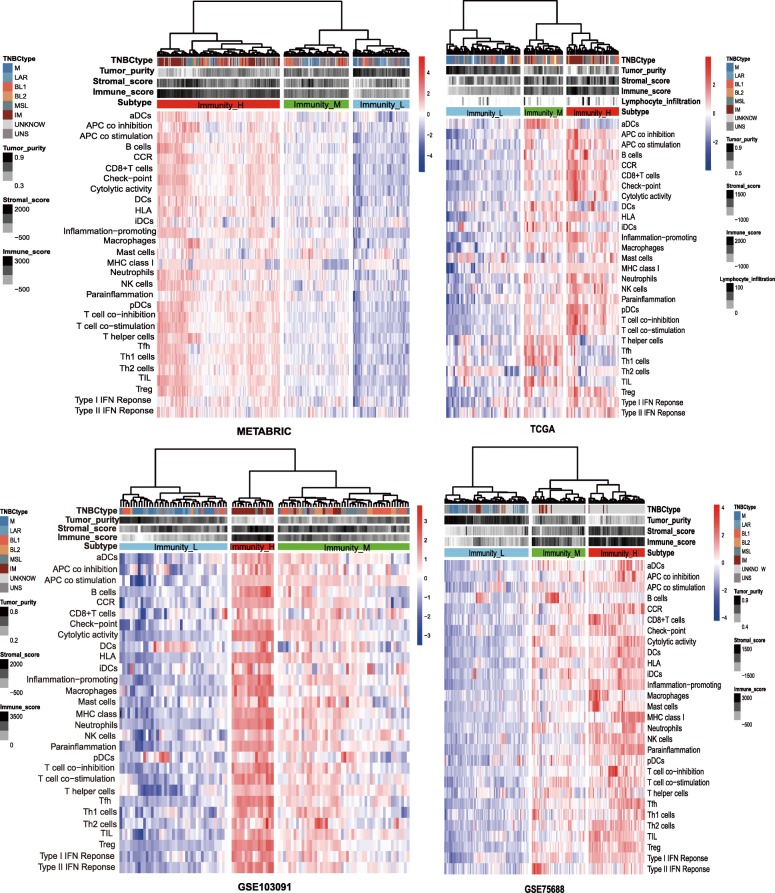
We analyzed 29 immune-associated gene sets which represented diverse immune cell types, functions, and pathways (Additional file 1: Table S1). We used the ssGSEA score [19, 20] to quantify the activity or enrichment levels of immune cells, functions, or pathways in the cancer samples. On the basis of the ssGSEA scores of the 29 gene sets, we hierarchically clustered TNBC in four BC datasets (METABRIC [31], TCGA [32], GSE75688 [33], and GSE103091 [34]). Interestingly, all four datasets showed similar clustering results, with three clusters being clearly separated (Fig. 1). We defined the three clusters as: Immunity High (Immunity_H), Immunity Medium (Immunity_M), and Immunity Low (Immunity_L). We found that the immune scores were significantly higher in Immunity_H and significantly lower in Immunity_L in all four datasets (Kruskal–Wallis test, P < 0.001) (Fig. 2a). Moreover, we found that the percentage of lymphocyte infiltration was significantly higher in Immunity_H and significantly lower in Immunity_L in TCGA based on the pathological slides data (Kruskal–Wallis test, P = 0.05). These features directed the classification. In addition, when comparing the tumor purity and stromal score of the three TNBC subtypes, we obtained opposite trends, with tumor purity increasing from Immunity_H to Immunity_L (Immunity_H < Immunity_M < Immunity_L) and stromal score decreasing from Immunity_H to Immunity_L (Immunity_H > Immunity_M > Immunity_L) (Kruskal–Wallis test, P < 0.001) (Additional file 2: Figure S1). Collectively, these results suggest that Immunity_H contains the highest number of immune cells and stromal cells, while Immunity_L contains the highest number of tumor cells.
Fig. 1.
Hierarchical clustering of triple-negative breast cancer (TNBC) yields three stable subtypes in four different datasets. Immunity_H, Immunity High; Immunity_M, Immunity Medium; Immunity_L, Immunity Low. TNBCtype, a method for classifying TBNC [2, 37]. Tumor_purity, Stromal_score, and Immune_score were evaluated by ESTIMATE [21]. Lymphocyte_infiltration, percent of lymphocyte infiltration
Fig. 2.
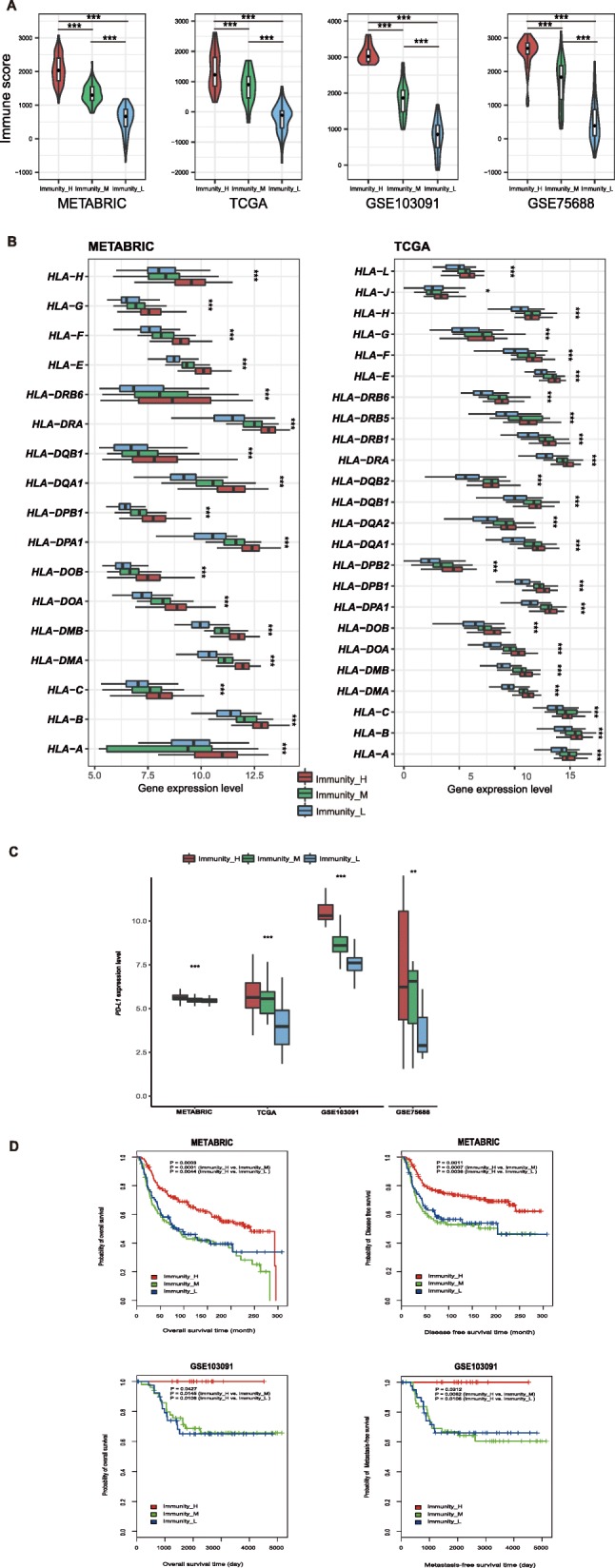
Three TNBC subtypes show differential phenotypes. a. Comparison of the immune cell infiltration levels between TNBC subtypes (Mann–Whitney U test). b. Comparison of the expression levels of HLA genes between TNBC subtypes (ANOVA test). c. Comparison of PD-L1 expression levels between TNBC subtypes (ANOVA test). d. Comparison of survival prognosis between TNBC subtypes (log-rank test). *P < 0.05, **P < 0.01, ***P < 0.001. It also applies to following figures
Notably, most HLA genes showed significantly higher expression levels in Immunity_H and significantly lower expression levels in Immunity_L (ANOVA test, P < 0.05) (Fig. 2b, Additional file 3: Figure S2A). Moreover, the expression levels of various immune cell subpopulation marker genes [35] were the highest in Immunity_H and the lowest in Immunity_L, such as CD8A (cytotoxic T cell), CD45RO (memory T cell), CD20 (B cell), CXCR5 (Tfh cell), FOXP3 (Treg), IL-17 (Th17 cell), CD1A (iDC), and IL3RA (pDC) (Additional file 3: Figure S2B).
We examined the expression of PD-L1 (programmed cell death 1 ligand) in the three TNBC subtypes and found that Immunity_H had the highest PD-L1 expression levels and Immunity_L had the lowest PD-L1 expression levels (ANOVA test, P < 0.05) (Fig. 2c). This suggest that the TNBC subtype Immunity_H might better respond to anti-PD-L1 immunotherapy than the other TNBC subtypes, since PD-L1 expression tends to be positively associated with immunotherapeutic responsiveness [36].
Survival analyses showed that these TNBC subtypes had distinct clinical outcomes. The Immunity_H subtype likely had a better survival prognosis than the Immunity_M and Immunity_L subtypes, but there was no significant survival difference between the Immunity_M and the Immunity_L subtypes (Fig. 2d). This is consistent with previous studies showing that TNBC with elevated immune activity were associated with more favorable clinical outcomes [4, 12, 34].
Comparisons of the immunogenomic profiling-based TNBC classification with other TNBC classification methods
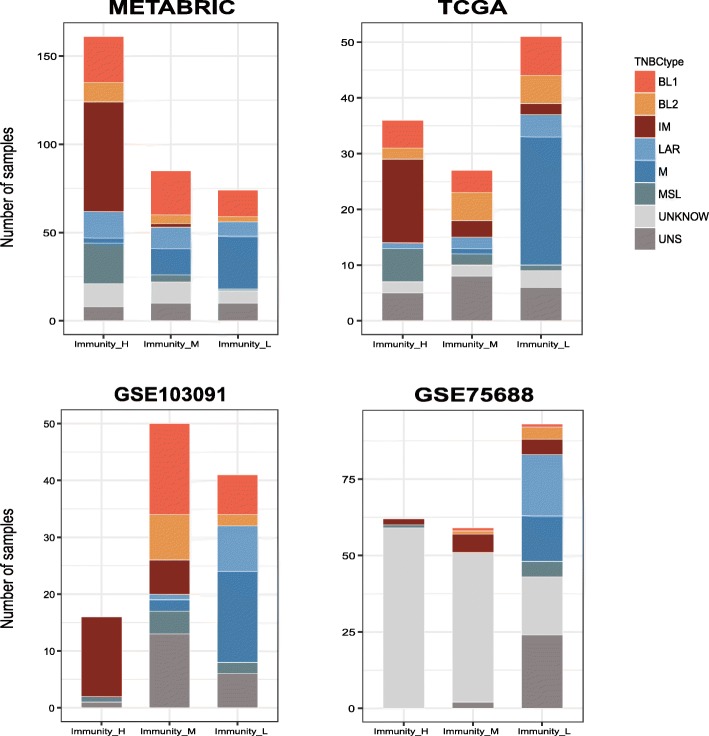
We used the TNBCtype method [2, 37] to classify the four TNBC datasets. We found that the immunomodulatory (IM) subtype of TNBCs was most frequently associated with Immunity_H and least frequently associated with Immunity_L (Fisher’s exact test, P < 0.001) (Fig. 3). This is consistent with the enrichment of immune cell processes in the IM subtype [2]. In contrast, the mesenchymal (M) subtype of TNBCs was mostly detected in Immunity_L and least frequently detected in Immunity_H (Fisher’s exact test, P < 0.001) (Fig. 3). The M subtype is mainly characterized by pathways involved in cell motility, ECM receptor interaction, and cell differentiation, such as Wnt, ALK, and TGF-β signaling [2]. Our results suggest that the activities of these pathways may be associated with reduced tumor immunity in TNBC.
Fig. 3.
Comparison of the immune signature-based TNBC classification results with the results by TNBCtype shows that Immunity_H is most enriched in IM while Immunity_L is most enriched in M. IM, immunomodulatory; M, mesenchymal
Identification of TNBC subtype-specific pathways, gene ontology, and networks
Identification of TNBC subtype-specific pathways
GSEA identified a number of KEGG [25] pathways enriched in Immunity_H and Immunity_L (Fig. 4a, Additional file 4: Figure S3A). Typically, the immune-associated pathways were highly active in Immunity_H and included antigen processing and presentation pathways, B and T cell receptor signaling, chemokine signaling, cytokine–cytokine receptor interactions, IL-17 signaling, Jak–STAT signaling, natural killer cell-mediated cytotoxicity, NF-kappa B signaling, NOD-like receptor signaling, TNF signaling, and Toll-like receptor signaling (Fig. 4a, Additional file 4: Figure S3A). This result confirmed the elevated immune activity in Immunity_H. Besides, we identified various cancer-associated pathways that were hyperactivated in Immunity_H, including apoptosis, calcium signaling, MAPK signaling, PI3K–Akt signaling, and RAS signaling (Fig. 4a, Additional file 4: Figure S3A). This suggests that the activities of these cancer-associated pathways are positively associated with TNBC immunity. In contrast, the TNBC subtype Immunity_L was enriched in pathways related to Hippo signaling, DNA replication, mismatch repair, spliceosome, adherens junctions, pyrimidine metabolism, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, and RNA polymerase (Fig. 4a, Additional file 4: Figure S3A). This indicates that the activities of these pathways could be negatively associated with TNBC immunity. In fact, a previous study has shown that the activities of MAPK and PI3K–Akt cascades positively correlated with the activation of various immune pathways, while the activity of the mismatch repair pathway showed a negative correlation with immune activation in TNBC [12]. Furthermore, we confirmed that all the cancer-associated pathways hyperactivated in Immunity_H were positively associated with the immune scores, whereas the pathways hyperactivated in Immunity_L likely showed a negative correlation (Spearman’s correlation test, P < 0.05) (Fig. 4b).
Fig. 4.
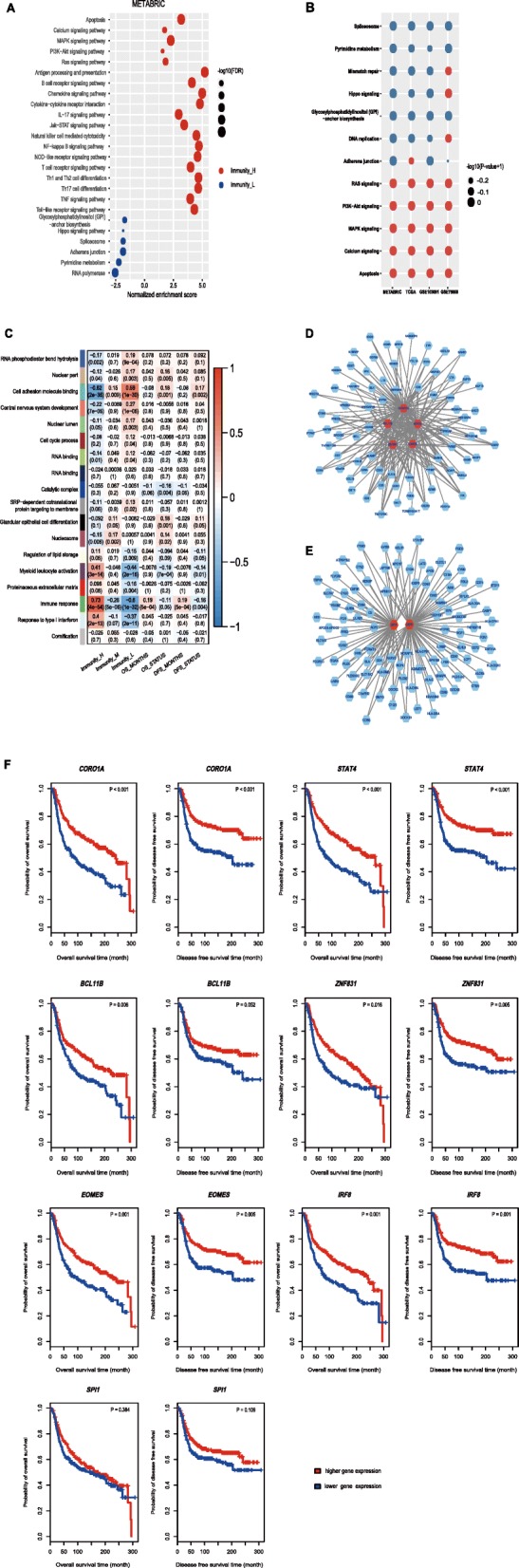
Identification of TNBC subtype-specific pathways, gene ontology, and networks. a. KEGG pathways enriched in Immunity_H and Immunity_L. b. The cancer-associated pathways upregulated in Immunity_H positively correlated with the immune scores, and the pathways upregulated in Immunity_L negatively correlated with the immune scores in TNBC (Spearman’s correlation test, P < 0.05). c. Gene modules significantly differentiating TNBC by subtype, survival time, or survival status. d. A network significantly active in Immunity_H, centered on five TFs (highlighted in red). e. A network significantly active in Immunity_L, centered on two TFs (highlighted in red). f. Kaplan–Meier curves showing that the expression of the hub TF genes is positively associated with survival prognosis in TNBC (log-rank test, P < 0.05). TF, transcription factor; FDR, false discovery rate
Identification of TNBC subtype-specific gene ontology
We performed a weighted gene co-expression network analysis of the METRABRIC dataset by WGCNA [26] and identified a set of gene modules (gene ontology) associated with the highly expressed genes previously determined. We found several gene modules that significantly differentiated TNBC by subtype, survival time, or survival status (Fig. 4c). As expected, the immune response was significantly elevated in Immunity_H (P = 4.0*10− 54), while was depressed in Immunity_L (P = 1.0*10− 32). Moreover, a high immune response was associated with a better survival prognosis in TNBC patients (P = 5.0*10− 4). This finding is in line with the previous observation that the subtype Immunity_H is associated with better clinical outcomes than the other subtypes. Similar results were observed for the TCGA dataset (Additional file 4: Figure S3B). The other two immune-associated gene modules, i.e., myeloid leukocyte activation and response to type I interferon, were also enriched in Immunity_H (P = 3.0*10− 14 and 2.0*10− 13, respectively), and were reduced in Immunity_L (P = 2.0*10− 16 and 2.0*10− 11, respectively). In contrast, cell adhesion molecule (CAMD) binding activity was significantly increased in Immunity_L (P = 1.0*10− 30) and decreased in Immunity_H (P = 2.0*10− 35). This suggests that CAMD activity has a strong inverse correlation with tumor immunity in TNBC. Interestingly, CAMD activity correlated with reduced survival (P = 0.001 for OS, and P = 0.002 for DFS). Cell cycle process was also increased in Immunity_L (P = 0.04), suggesting that the cell cycle signature correlates with reduced tumor immunity. This finding is consistent with results from previous studies [38, 39].
Identification of TNBC subtype-specific networks
WGCNA generated a gene module (green color, Fig. 4c) that was specifically significant in Immunity_H. We identified 98 hub genes from the gene module, including five transcription factor (TF) genes, i.e., CORO1A, STAT4, BCL11B, ZNF831, and EOMES. The five TFs interact with each other and form a subnetwork with diverse immune and cancer-related genes that they regulate (Fig. 4d). Typically, CD247 (the marker gene for a T cell subpopulation) was regulated by all these TFs, and the cytotoxic T cell marker gene CD8A was co-regulated by CORO1A, STAT4, and EOMES. MAP4K1 (Mitogen-Activated Protein Kinase Kinase Kinase Kinase 1), which is involved in multiple immune and cancer-related pathways including B cell receptor signaling, JNK, EGF/EGFR, TGF-β, and MAPK signaling, was also regulated by the five TFs. CORO1A encodes a member of the WD repeat protein family which is involved in diverse cellular processes including cell cycle, apoptosis, signal transduction, and gene regulation. The main pathways related to CORO1A include cytoskeletal signaling and phagosome function, and its relatedness with immune regulation has been revealed [40, 41]. The association of the other TFs STAT4 [42], BCL11B [43], and EOMES [44] with immunity has been examined, whereas the role of ZNF831 in immune regulation remains unexplored.
WGCNA also generated a gene module (turquoise color, Fig. 4c) that was more enriched in Immunity_L. This module included 112 hub genes, two of which encode the TFs IRF8 and SPI1. A subnetwork of the hub genes centered on IRF8 and SPI1is shown in Fig. 4e. IRF8 (interferon regulatory factor 8) has been shown to play a negative role in immune cell regulation [45]. Thus, the IRF8-centered regulatory network may be responsible for the depressed immunity of the TNBC subtype Immunity_L. SPI1 (Spi-1 proto-oncogene) encodes a transcription factor that activates gene expression during immune cell development. As a result, the deregulation of SPI1 may affect immunity. In fact, SPI1 showed significantly lower expression levels in Immunity_L than in Immunity_H (Student’s t test, P = 9.1*10− 28, fold change > 2). Therefore, the down regulation of SPI1 may contribute to the decreased immunity of the Immunity_L subtype. The contribution of the IRF8- and SPI1-centered regulatory network to the depressed immunity of Immunity_L is evidenced by a previous study showing that IRF8 and SPI1 together negatively regulated immune cell differentiation [45].
Interestingly, survival analyses showed that elevated expression levels of these TF genes (except SPI11) were consistently associated with better survival prognosis in TNBC (Fig. 4f), suggesting the pivotal role of these TFs in TNBC immunity and prognosis.
Class prediction of TNBC subtypes based on immunogenomic profiling
We first used 10-fold cross validation (CV) to evaluate the classification performance in METABRIC and then predicted the TNBC subtypes in the other three datasets using the METABRIC dataset as the training set. The 10-fold CV accuracy was 89% in classifying the METABRIC dataset. The classification accuracies were 70, 84, and 63% in TCGA, GSE75688, and GSE103091, respectively. The weighted F-scores in these classifications were 89, 71, 83, and 63% for METABRIC, TCGA, GSE75688, and GSE103091, respectively (Fig. 5). These results demonstrate that the immunogenomic profiling-based classification of TNBC is stable and predictable.
Fig. 5.

Performance in the classification of TNBC subtypes based on immune signatures. F-score, weighted average of F-scores
Discussion
A number of prior studies have identified TNBC subtypes on the basis of genomic profiling [2–4, 34]. However, very few studies have investigated the classification of TNBC specifically based on immune signatures. To fill this knowledge gap, we focused on identifying immune-related TNBC subtypes using immunogenomic profiling. Our results show that TNBC could be classified into three stable subtypes: Immunity High, Immunity Medium, and Immunity Low. Furthermore, we demonstrated that this classification was reproducible and predictable. The Immunity High TNBC subtype was enriched not only in immune signatures, but also in many cancer-associated pathways including apoptosis, calcium signaling, MAPK signaling, PI3K–Akt signaling, and RAS signaling (Fig. 4a). This is in line with our previous study showing that diverse immune signatures positively correlated with the MAPK and PI3K–Akt signaling pathways in TNBC [12]. In contrast, the Immunity Low TNBC subtype was impoverished in immune signatures but enriched in Hippo signaling, DNA replication, mismatch repair, spliceosome, adherens junction, pyrimidine metabolism, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, and RNA polymerase pathways (Fig. 4a). It is rational that the mismatch repair pathway activity was significantly negatively correlated with immune signatures in cancer, since deficient mismatch repair often results in elevated tumor immunity [18]. Interestingly, we found that the Hippo signaling pathway had a significantly negative correlation with immune signatures in TNBC. This observation is in agreement with findings from previous studies showing that the Hippo signaling pathway plays a key role in regulating tumor immunity [46–48]. Deficiency of Hippo pathway components such as kinases LATS1/2 (large tumor suppressor 1 and 2) [46], effector YAP (Yes-associated protein) [47], and transcriptional co-activator TAZ (WW domain-containing transcription regulator 1) [48] could promote anti-tumor immunity. Overall, these results revealed potential positive or negative associations between pathway activities and immune activities in TNBC.
Immunity_H had stronger immune cell infiltration and anti-tumor immune activities, e.g., high levels of cytotoxic T cells and B cells infiltration (Fig. 1). When we used CIBERSORT [28] to calculate the proportions of 22 immune cell subsets in TNBC, we found that CD8 T cells, CD4 T cells, NK cells, and M1 macrophages tended to be present in significantly higher numbers in Immunity_H than in Immunity_L (Mann–Whitney U test, P < 0.05) (Fig. 6). This analysis again confirmed elevated anti-tumor immune activity in Immunity_H. The high anti-tumor immune activation could explain why Immunity_H had more favorable clinical outcomes compared to the other subtypes (Fig. 2d). In fact, numerous studies have demonstrated that the density of tumor-infiltrating lymphocytes (TILs) is positively associated with survival prognosis in various cancers [35, 49, 50]. Immunity_H more highly expressed most HLA genes, which is indicative of stronger immunogenicity compared to the other subtypes. However, Immunity_H did not show higher TMB or neoantigen load than the other subtypes. This suggests that the differential immunogenicity between the TNBC subtypes cannot be attributed to TMB and neoantigens. In addition, we did not find lower clonal heterogeneity in Immunity_H, as estimated by ABSOLUTE [29], than in the other subtypes, although, in some studies, clonal heterogeneity was shown to have a significant negative correlation with tumor immunity [13, 51]. Interestingly, Immunity_H exhibited more active stem cell-associated biological processes than the other subtypes (Additional file 5: Figure S4), while it showed no significant differences in proliferation and epithelial-to-mesenchymal transition (EMT) processes.
Fig. 6.
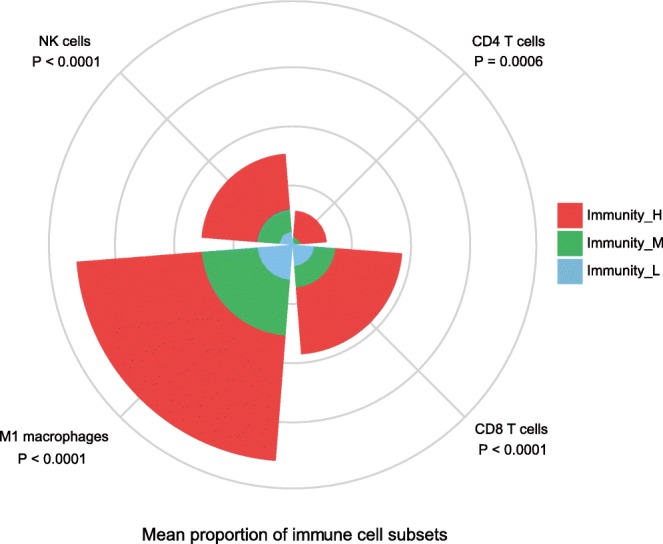
Comparison of the proportions of immune cell subsets between TNBC subtypes. Kruskal-Wallis test, P values are shown
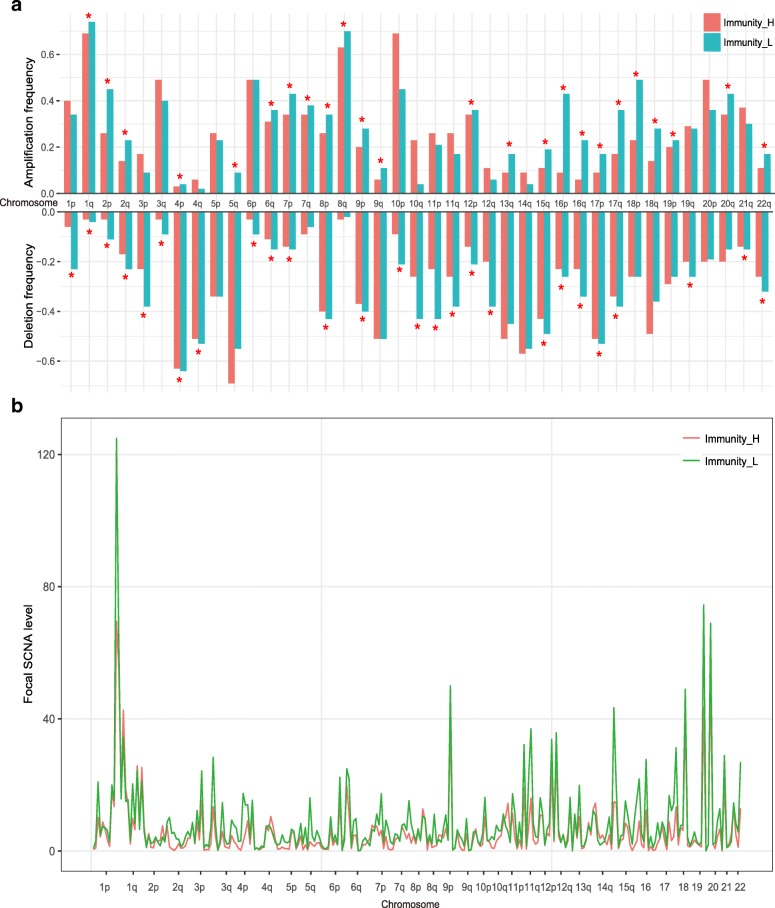
Furthermore, we compared SCNA levels between Immunity_H and Immunity_L subtypes. We found that Immunity_H had significantly lower arm-level SCNAs than Immunity_L (Wilcoxon signed-rank test, P = 0.04, 0.001, 0.0006 for comparisons of amplification, deletion, and total alteration frequencies, respectively) (Fig. 7a). Moreover, Immunity_H had significantly lower focal SCNA levels than Immunity_L (Mann–Whitney U test, P = 0.01, 0.02, 0.01 for comparisons of amplification, deletion, and total alteration levels, respectively) (Fig. 7b). These findings demonstrated that Immunity_H had lower levels of SCNAs compared to Immunity_L, supporting the notion that high tumor aneuploidy correlates with reduced tumor immune infiltration [52].
Fig. 7.
Comparison of the somatic copy number alteration (SCNA) levels between TNBC subtypes. a. Comparison of the arm-level SCNAs between Immunity_H and Immunity_L. The red asterisks indicate the chromosome arms in which Immunity_H presents higher amplification or deletion frequency than Immunity_L. b. Comparison of the focal SCNA levels between Immunity_H and Immunity_L
Currently, immunotherapy for TNBC is an active field of investigation [53], and the stronger immunogenicity exhibited by TNBC compared to other breast cancer subtypes suggests that immunotherapy could be a viable option for TNBC patients [12]. However, some preliminary TNBC immunotherapy clinical trials have not shown significant patients’ improvement (personal communication). Thus, the immune signature-based classification of TNBC may aid the stratification of TNBC patients to identify those responsive to immunotherapy. It is imaginable that patients with an Immunity_H subtype of TNBC would be more likely to respond to anti-PD-1/PD-L1 therapy than patients with other TNBC subtypes, since PD-L1 is more highly expressed in Immunity_H TNBC, and PD-L1 expression is a predictive biomarker for the response to PD-1/PD-L1-directed immunotherapy [36, 54].
Conclusions
The identification of TNBC subtypes based on immune signatures has potential clinical implications for TNBC treatment.
Additional files
Table S1. The 29 immune signatures represented by 29 different gene sets. (XLSX 41 kb)
Figure S1. Comparisons of the stromal content and tumor purity between TNBC subtypes (Mann–Whitney U test). (PDF 368 kb)
Figure S2. Comparisons of the expression levels of immune-related genes between TNBC subtypes. A. Comparisons of the expression levels of HLA genes between TNBC subtypes. B. Comparisons of the expression levels of immune cell subpopulation marker genes between TNBC subtypes. ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. (PDF 168 kb)
Figure S3. Identification of TNBC subtype-specific pathways and gene ontology. A. KEGG pathways enriched in Immunity_H and Immunity_L. B. Gene modules significantly differentiating TNBC by subtype, survival time, or survival status. (PDF 164 kb)
Figure S4. Stem cell-associated activity is higher in Immunity_H than in the other subtypes. (PDF 110 kb)
Acknowledgements
We thank Dr. Agnese Mariotti for editing the manuscript.
Funding
This study was funded by the China Pharmaceutical University (grant number 2632018YX01, 3150120001).
Availability of data and materials
We used four publicly available TNBC genomic datasets: METABRIC [31], TCGA [32], GSE75688 [33] and GSE103091 [34]. The METABRIC dataset was obtained from cBioPortal (http://www.cbioportal.org/study?id=brca_metabric#summary). The TCGA dataset was obtained from the TCGA data portal (https://portal.gdc.cancer.gov/). The GSE75688 and GSE103091 datasets were downloaded from the NCBI gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/). We obtained 29 immune signatures (represented by 29 different gene sets, respectively) from the publications [12, 35] (Additional file 1: Table S1).
Abbreviations
- aDCs
Activated dendritic cells
- BC
Breast cancer
- BLIA
Basal-like immune activated
- CAMD
Cell adhesion molecule binding
- CCR
Cytokine and cytokine receptor
- CV
Cross validation
- DCs
Dendritic cells
- DFS
Disease-free survival
- EMT
Epithelial-to-mesenchymal transition
- ER
Estrogen receptor
- FDR
False discovery rate
- GSEA
Gene-set enrichment analysis
- HLA
Human leukocyte antigen
- HER2
Human epidermal growth factor receptor 2
- iDCs
Immature dendritic cells
- IM
Immunomodulatory
- Immunity_H
Immunity high
- Immunity_M
Immunity medium
- Immunity_L
Immunity low
- M
Mesenchymal
- MFS
Metastasis-free survival
- MHC
Major histocompatibility complex
- NK cells
Natural killer cells
- OS
Overall survival
- pDCs
Plasmacytoid dendritic cells
- PR
Progesterone receptor
- RF
Random forest
- ssGSEA
Single-sample gene-set enrichment analysis
- SCNA
Somatic copy number alteration
- TCGA
The Cancer Genome Atlas
- TF
Transcription factor
- Tfh cells
Follicular helper T cells
- Th17 cells
T helper 17 cells
- TILs
Tumor-infiltrating lymphocytes
- TME
Tumor microenvironment
- TNBC
Triple-negative breast cancer
- Treg
Regulatory T cells
- WGCNA
Weighted gene co-expression network analysis
Authors’ contributions
YH performed data analyses and helped prepare for the manuscript. ZJ performed data analyses and helped prepare for the manuscript. CC performed data analyses. XW conceived the research, designed the analyses strategies, and wrote the manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval and consent to participate was waived since we used only publicly available data and materials in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yin He, Email: yin.he@stu.cpu.edu.cn.
Zehang Jiang, Email: zehang.jiang@stu.cpu.edu.cn.
Cai Chen, Email: cac005@eng.ucsd.edu.
Xiaosheng Wang, Email: xiaosheng.wang@cpu.edu.cn.
References
- 1.Lehmann BD, et al. Refinement of triple-negative breast Cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11(6):e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonsang-Kitzis H, et al. Biological network-driven gene selection identifies a stromal immune module as a key determinant of triple-negative breast carcinoma prognosis. Oncoimmunology. 2016;5(1):e1061176. doi: 10.1080/2162402X.2015.1061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burstein MD, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Paggio JC. Immunotherapy: Cancer immunotherapy and the value of cure. Nat Rev Clin Oncol. 2018;15(5):268–270. [DOI] [PubMed]
- 6.Salatino M, Girotti MR, Rabinovich GA. Glycans pave the way for immunotherapy in triple-negative breast Cancer. Cancer Cell. 2018;33(2):155–157. doi: 10.1016/j.ccell.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, et al. Combination immunotherapy of MUC1 mRNA Nano-vaccine and CTLA-4 blockade effectively inhibits growth of triple negative breast Cancer. Mol Ther. 2018;26(1):45–55. doi: 10.1016/j.ymthe.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, et al. Targeting tissue factor for immunotherapy of triple-negative breast Cancer using a second-generation ICON. Cancer Immunol Res. 2018;6(6):671–684. doi: 10.1158/2326-6066.CIR-17-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi AH, et al. Novel chimeric parapoxvirus CF189 as an oncolytic immunotherapy in triple-negative breast cancer. Surgery. 2018;163(2):336–342. doi: 10.1016/j.surg.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernier C, et al. DZ-2384 has a superior preclinical profile to taxanes for the treatment of triple-negative breast cancer and is synergistic with anti-CTLA-4 immunotherapy. Anti-Cancer Drugs. 2018. [DOI] [PMC free article] [PubMed]
- 11.Nanda R, et al. Pembrolizumab in patients with advanced triple-negative breast Cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, et al. A comprehensive immunologic portrait of triple-negative breast Cancer. Transl Oncol. 2018;11(2):311–329. doi: 10.1016/j.tranon.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safonov A, et al. Immune gene expression is associated with genomic aberrations in breast Cancer. Cancer Res. 2017;77(12):3317–3324. doi: 10.1158/0008-5472.CAN-16-3478. [DOI] [PubMed] [Google Scholar]
- 14.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witten IH, et al. Data Mining: Practical Machine Learning Tools and Techniques. 4th ed: Morgan Kaufmann, San Francisco; 2016.
- 28.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas, N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jezequel P, et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015;17:43. doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindea G, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in Cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, et al. TNBCtype: a subtyping tool for triple-negative breast Cancer. Cancer Inform. 2012;11:147–156. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goel S, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Z, et al. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric Cancer. Transl Oncol. 2018;11(5):1171–1187. doi: 10.1016/j.tranon.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pick R, et al. Coronin 1A, a novel player in integrin biology, controls neutrophil trafficking in innate immunity. Blood. 2017;130(7):847–858. doi: 10.1182/blood-2016-11-749622. [DOI] [PubMed] [Google Scholar]
- 41.Pieters J, Muller P, Jayachandran R. On guard: coronin proteins in innate and adaptive immunity. Nat Rev Immunol. 2013;13(7):510–518. doi: 10.1038/nri3465. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, et al. CD4+ T-cell-mediated anti-tumor immunity can be uncoupled from autoimmunity via the STAT4/STAT6 signaling axis. Eur J Immunol. 2009;39(5):1252–1259. doi: 10.1002/eji.200839152. [DOI] [PubMed] [Google Scholar]
- 43.Avram D, Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J Immunol. 2014;193(5):2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knox JJ, et al. Characterization of T-bet and eomes in peripheral human immune cells. Front Immunol. 2014;5:217. doi: 10.3389/fimmu.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carotta S, et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014;211(11):2169–2181. doi: 10.1084/jem.20140425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moroishi, T., et al., The hippo pathway kinases LATS1/2 suppress Cancer immunity. Cell, 2016. 167(6): p. 1525–1539 e17. [DOI] [PMC free article] [PubMed]
- 47.Guo X, et al. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 2017;31(3):247–259. doi: 10.1101/gad.294348.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janse van Rensburg HJ, et al. The hippo pathway component TAZ promotes immune evasion in human Cancer through PD-L1. Cancer Res. 2018;78(6):1457–1470. doi: 10.1158/0008-5472.CAN-17-3139. [DOI] [PubMed] [Google Scholar]
- 49.Ali HR, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 50.Garnelo M, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66(2):342–351. doi: 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorsson V, et al. The immune landscape of Cancer. Immunity. 2018;48(4):812–830 e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davoli T, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322). [DOI] [PMC free article] [PubMed]
- 53.Wang X, et al. Immunological therapy: A novel thriving area for triple-negative breast cancer treatment. Cancer Lett. 2018;442:409–428. [DOI] [PubMed]
- 54.Aguiar PN, Jr, et al. PD-L1 expression as a predictive biomarker in advanced non-small-cell lung cancer: updated survival data. Immunotherapy. 2017;9(6):499–506. doi: 10.2217/imt-2016-0150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The 29 immune signatures represented by 29 different gene sets. (XLSX 41 kb)
Figure S1. Comparisons of the stromal content and tumor purity between TNBC subtypes (Mann–Whitney U test). (PDF 368 kb)
Figure S2. Comparisons of the expression levels of immune-related genes between TNBC subtypes. A. Comparisons of the expression levels of HLA genes between TNBC subtypes. B. Comparisons of the expression levels of immune cell subpopulation marker genes between TNBC subtypes. ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. (PDF 168 kb)
Figure S3. Identification of TNBC subtype-specific pathways and gene ontology. A. KEGG pathways enriched in Immunity_H and Immunity_L. B. Gene modules significantly differentiating TNBC by subtype, survival time, or survival status. (PDF 164 kb)
Figure S4. Stem cell-associated activity is higher in Immunity_H than in the other subtypes. (PDF 110 kb)
Data Availability Statement
We used four publicly available TNBC genomic datasets: METABRIC [31], TCGA [32], GSE75688 [33] and GSE103091 [34]. The METABRIC dataset was obtained from cBioPortal (http://www.cbioportal.org/study?id=brca_metabric#summary). The TCGA dataset was obtained from the TCGA data portal (https://portal.gdc.cancer.gov/). The GSE75688 and GSE103091 datasets were downloaded from the NCBI gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/). We obtained 29 immune signatures (represented by 29 different gene sets, respectively) from the publications [12, 35] (Additional file 1: Table S1).