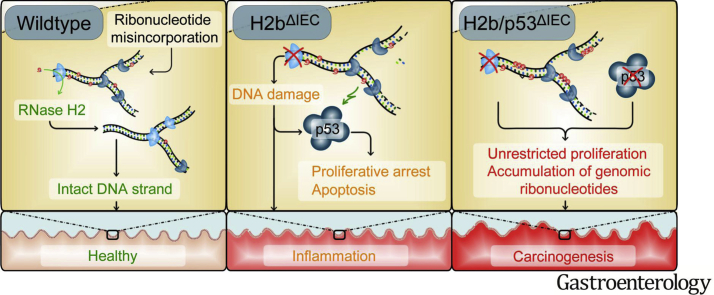
Abstract
Background & Aims
RNase H2 is a holoenzyme, composed of 3 subunits (ribonuclease H2 subunits A, B, and C), that cleaves RNA:DNA hybrids and removes mis-incorporated ribonucleotides from genomic DNA through ribonucleotide excision repair. Ribonucleotide incorporation by eukaryotic DNA polymerases occurs during every round of genome duplication and produces the most frequent type of naturally occurring DNA lesion. We investigated whether intestinal epithelial proliferation requires RNase H2 function and whether RNase H2 activity is disrupted during intestinal carcinogenesis.
Methods
We generated mice with epithelial-specific deletion of ribonuclease H2 subunit B (H2bΔIEC) and mice that also had deletion of tumor-suppressor protein p53 (H2b/p53ΔIEC); we compared phenotypes with those of littermate H2bfl/fl or H2b/p53fl/fl (control) mice at young and old ages. Intestinal tissues were collected and analyzed by histology. We isolated epithelial cells, generated intestinal organoids, and performed RNA sequence analyses. Mutation signatures of spontaneous tumors from H2b/p53ΔIEC mice were characterized by exome sequencing. We collected colorectal tumor specimens from 467 patients, measured levels of ribonuclease H2 subunit B, and associated these with patient survival times and transcriptome data.
Results
The H2bΔIEC mice had DNA damage to intestinal epithelial cells and proliferative exhaustion of the intestinal stem cell compartment compared with controls and H2b/p53ΔIEC mice. However, H2b/p53ΔIEC mice spontaneously developed small intestine and colon carcinomas. DNA from these tumors contained T>G base substitutions at GTG trinucleotides. Analyses of transcriptomes of human colorectal tumors associated lower levels of RNase H2 with shorter survival times.
Conclusions
In analyses of mice with disruption of the ribonuclease H2 subunit B gene and colorectal tumors from patients, we provide evidence that RNase H2 functions as a colorectal tumor suppressor. H2b/p53ΔIEC mice can be used to study the roles of RNase H2 in tissue-specific carcinogenesis.
Keywords: Ribonucleotide Excision Repair, Colon Cancer, Mouse Model, DNA Repair
Abbreviations used in this paper: COADREAD, RNA sequencing data of 467 patients with colorectal adenocarcinoma; COSMIC, Catalogue of Somatic Mutations in Cancer; CRC, colorectal cancer; DSS, dextran sodium sulfate; HR, homologous recombination; IEC, intestinal epithelial cell; InDel, insertion and deletion; KEGG, Kyoto Encyclopedia of Genes and Genomes; MEF, murine embryonic fibroblast; p53, tumor-suppressor protein p53; RER, ribonucleotide excision repair; SNV, single nucleotide variant; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; TCGA, The Cancer Genome Atlas
Graphical abstract
What You Need to Know.
Background and Context
Colorectal cancer arises from the gradual accumulation of mutations in intestinal epithelial stem cells. Although ribonucleotide incorporation by eukaryotic DNA polymerases represents the most frequent type of naturally occurring DNA lesions, it is not known whether this type of DNA lesion contributes to colorectal carcinogenesis.
New Findings
Intestinal epithelial RNase H2 deficiency leads to a p53-dependent proliferative exhaustion of the intestinal stem cell compartment and concomitant deletion of p53 instigates spontaneous carcinogenesis. In human CRC patients, decreased RNase H2 expression is associated with poor survival.
Limitations
A detailed delineation on the role of RNase H2 guided RER or RNA:DNA hybrid removal on cancer prevention is warranted.
Impact
This study presents a novel mouse tumor model to study the roles of RNase H2 in tissue-specific carcinogenesis.
Colorectal cancer (CRC) is the third most common type of cancer and the global burden of CRC is expected to increase by 60% to more than 2.2 million new cases and 1.1 million cancer deaths by 2030.1 Approximately 90% of CRC cases develop sporadically without genetic predisposition.2 A multi-hit genetic model of carcinogenesis has been suggested in sporadic CRC as a paradigm for solid tumor progression, and increased DNA damage and impaired DNA repair have been recognized as essential cellular properties that accompany the acquisition of mutations. CRC originates from adult stem cells that maintain lifelong proliferation and are particularly prone to accumulate hazardous mutations.3, 4 A direct correlation between the number of stem cell divisions and cancer risk over a range of tissues has been reported.5
Mis-incorporated single ribonucleotides represent the most common type of naturally occurring DNA damage (>1,000,000 per replicating mammalian genome).6, 7 Ribonucleotide excision repair (RER) is responsible for removing incorporated ribonucleotides from replicating DNA, with the RNase H2 complex providing the first key step of RER by cleaving 5′ of the DNA-embedded ribonucleotide.8 In addition, RNase H2 has been proposed to remove ribonucleotides of RNA:DNA hybrids, such as RNA oligonucleotides that prime the Okazaki fragment during replication9 or R-loops present during transcription,10 which can independently contribute to genome damage.11, 12 Despite the high frequency of such lesions, it is completely unknown whether dysfunctional RNase H2 translates into increased mutagenesis and—as a consequence—into carcinogenesis or tumor progression. Loss of RNase H2 in mice is embryonically lethal and characterized by an accumulation of genomic ribonucleotides associated with extensive DNA damage and genome instability.7, 13
A presumable implication for RNase H2 in carcinogenesis is based on the observation that community databases of mutational landscapes in human cancer (International Cancer Genome Consortium and Catalogue of Somatic Mutations in Cancer [COSMIC]) list numerous somatic RNase H2 mutations in different cancers.14 In addition, variants in the RNase H2 gene have recently been described in glioblastoma and prostate carcinoma.15 Hence, although clinical data infer a potential role for RNase H2 in cancer development, experimental evidence on the functionality of these RNase H2 cancer variants is lacking.
We report that intestinal ablation of RNase H2 results in chronic DNA damage, which, in the context of deleted tumor-suppressor protein p53 (p53), results in spontaneous manifestation of intestinal carcinomas. We present a novel link between RNase H2 protein function and intestinal epithelial carcinogenesis.
Methods
Mice
Floxed RNase H2b mice were created as described by Hiller et al,13 and p53fl/fl mice were obtained from the Jackson Laboratory (Bar Harbor, ME; catalog number 8562). Villin-Cre+, H2bfl/fl (H2bΔIEC) and p53/RNase H2bfl/fl (p53/H2bΔIEC), ERT-Cre+ (catalog number 4682; Jackson Laboratory), and RNase H2bfl/fl (RNase H2ΔTam) mice, backcrossed for at least 6 generations, were used at 8–12 weeks of age for all experiments. All mice were provided with food and water ad libitum and maintained in a 12-hour light–dark cycle under standard conditions at Kiel University (Keil, Germany). Tail or ear biopsy genomic DNA was used for genotyping of respective mouse strains. Procedures involving animal care were conducted in agreement with national and international laws and policies with appropriate permission. All experiments were carried out in accordance with the guidelines for animal care of the Christian-Albrechts-University (Kiel, Germany).
Histopathologic Analyses of Murine Small Intestinal Tissue
After sacrifice, the entire small intestine was excised and separated longitudinally into 2 equal parts. Ten centimeters of the most distal small intestine was rolled up, starting with the distal part, thereby having the distal ileum at the very inner layer and the proximal intestine at the very outer layer. The entire specimen was fixed in 10% formalin. Paraffin sections were cut and stained with hematoxylin and eosin. Histologic scoring was performed in blinded fashion by 2 independent observers. The histologic score displays the combined score of inflammatory cell infiltration and tissue damage, as described elsewhere.16
Histologic Grading of Intestinal Tumors
According to the World Health Organization’s Classification of Tumors of the Digestive System, Fourth Edition, adenocarcinomas of the small intestine were graded as “low” (≥50% gland formation) or “high” (<50% gland formation). Dysplasia of the small intestine was graded as low grade or high grade depending on the degree of architectural complexity, extent of nuclear stratification, and severity of abnormal nuclear morphology.
Exome Sequencing
DNA was isolated from the liver, small intestinal tumor, and adjacent nontumor tissue of the same mice (n = 4 mice per genotype; H2b/p53fl/fl, H2b/p53ΔIEC). DNA exome libraries were prepared using the SureSelectXT Mouse All Exon Kit (Agilent Technologies, Santa Clara, CA) and libraries were sequenced on a HiSeq3000 (Illumina, San Diego, CA). After adapter trimming and excluding bad-quality sequences and bases, all reads were mapped against the mouse genome reference mm10 using BWA 0.7.1517 for whole exome sequencing data and TopHat 2.0.8b18 for transcriptome sequences. Duplicates were marked with the Picard tool MarkDuplicates.jar 1.139 in whole exome sequencing analyses.
Single nucleotide variants (SNVs) and small insertions and deletions (InDels) were called with GATK 3.5.019 and classified using Annovar (2016-02-01)20 based on the Ensemble annotation (2014) offered by the University of California–Santa Cruz. Variants, which were not annotated in the database dbSNP build 142,21 were classified as novel. Allele counts were determined for each position using samtools mpileup 1.4.22 All called SNVs, which were covered by at least 5 reads and supported by fewer than 5% of the reads in the corresponding liver sample, were defined as somatic. The investigation of SNV patterns was performed using the R package MutationPatterns 1.0.023 and custom scripts. To determine differentially expressed genes, read counts per transcript were counted with the Python script HTSeq 0.6.1p124 using the Gencode vM6 annotation and the mode “union.” Reads with TopHat2 alignment quality lower than 20 were skipped. P values for differentially expressed genes were calculated with DeSeq2 1.14.125 after outlier replacement based on trimmed mean value. The online tool InnateDB26 was applied to test for enriched pathways. Correction for multiple testing was performed with the R function “p.adjust” using the Benjamini-Hochberg method. Networks were created with Cytoscape (https://cytoscape.org), in which connections between genes were based on entries annotated in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database with medium stringency (interaction score > 400).27 Integrated networks were produced using the R-package igraph based on the STRING database with medium stringency.28
Candidate Validation in Human CRC Samples
Paired samples of normal and tumor tissue (n = 155) were obtained during endoscopy using standard biopsy forceps and diagnostic specimens for histopathologic analysis were obtained from the interdisciplinary endoscopy unit of the University Hospital Kiel. Samples were immediately snap-frozen in liquid nitrogen, ensuring ex vivo times shorter than 30 seconds. The study protocol was approved by the institutional review board of the Medical Faculty of the Christian-Albrechts-University before commencement of the study (Ethikkommission der Medizinischen Fakultät der Christian-Albrechts-Universität zu Kiel; Az: A 110/99).
Online Database Survival Analysis
Large-cohort survival analysis was performed by using a public database with transcriptome analysis data. RNA sequencing data of 467 patients with colorectal adenocarcinoma (COADREAD) was obtained from The Cancer Genome Atlas (TCGA) database and analyzed using the SurvExpress web resource (http://bioinformatica.mty.itesm.mx/SurvExpress).29 For survival analysis, the samples of each cancer type were divided into 2 groups according to high and low expression levels to maximize risk groups according to the built-in setup of the website. The group with high RNA expression was compared with the cohort with low RNA expression. Log-rank test was used to assess association with survival.
Results
Ablation of Intestinal RNase H2 in Mice Causes Age-Dependent Defects in the Crypt Epithelial Stem Cell Niche
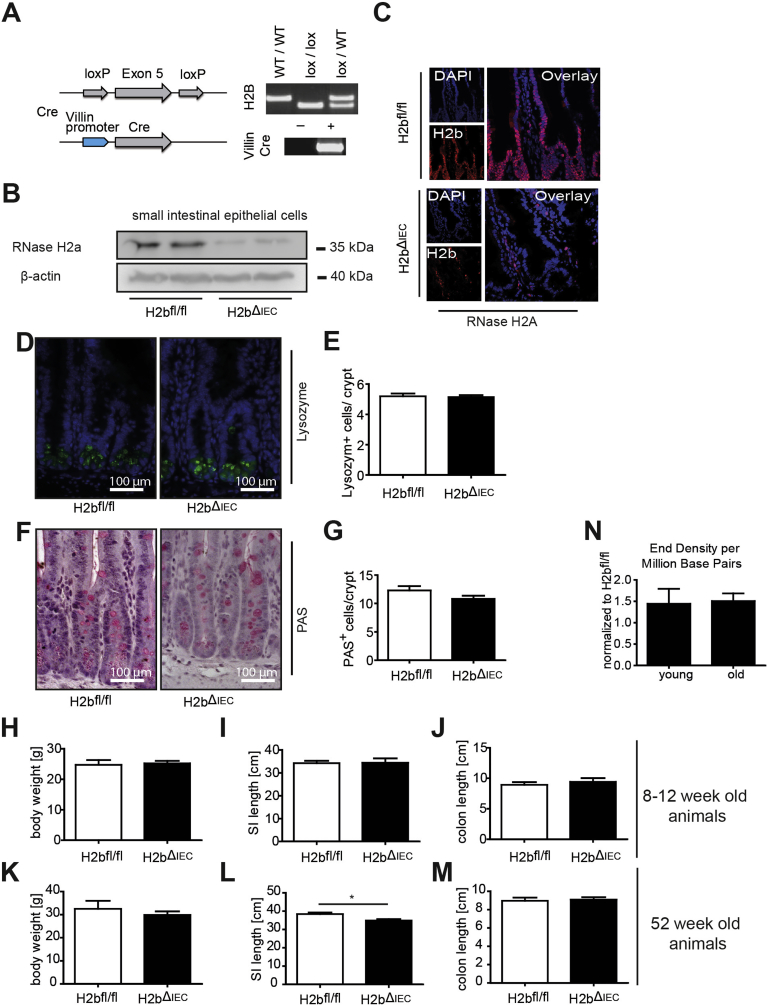
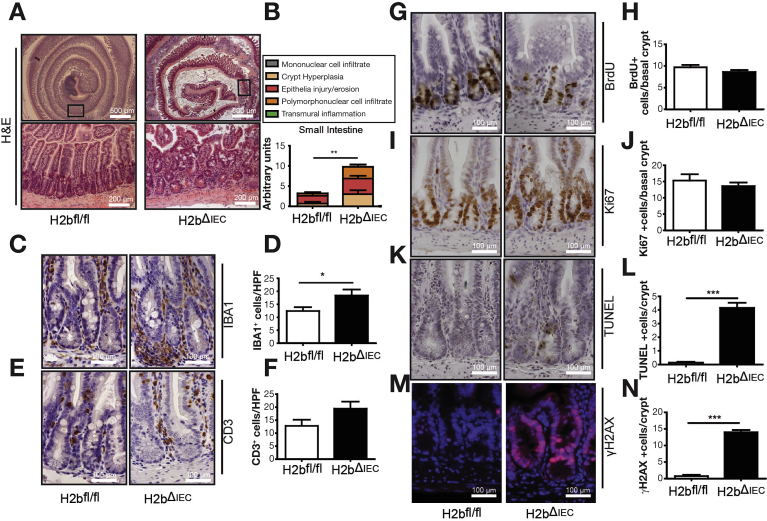
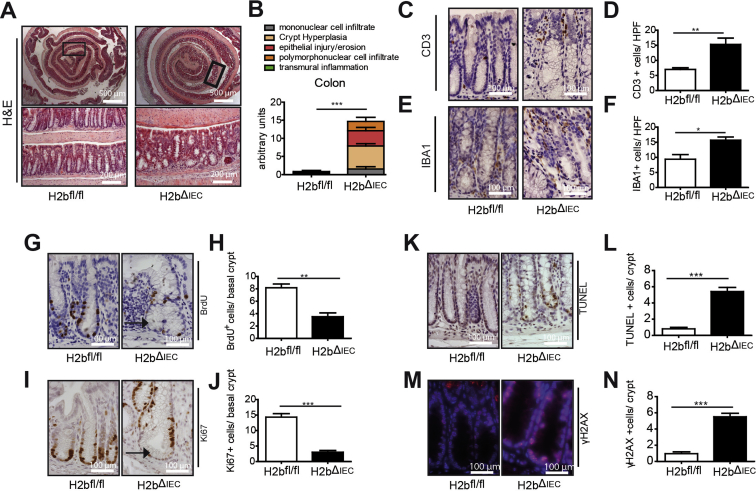
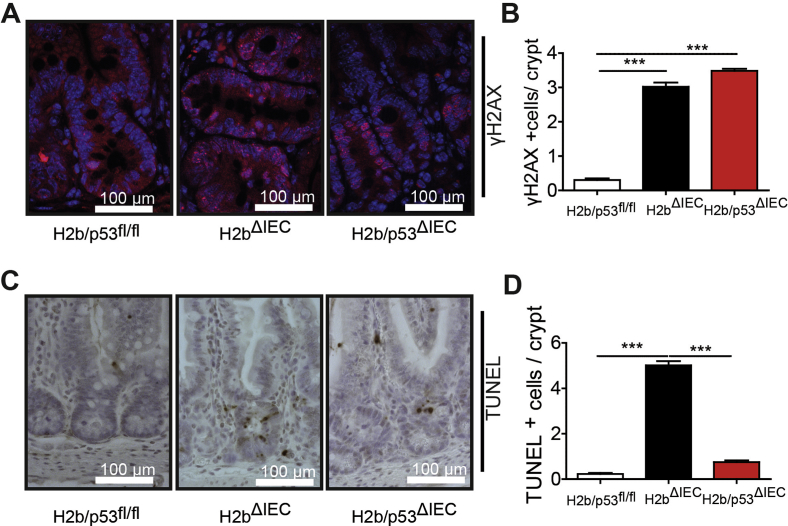
To investigate the impact of RNase H2 on the intestinal epithelium, we intercrossed floxed Rnaseh2b mice with Villin-Cre mice (hereafter H2bΔIEC) and confirmed the absence of the RNase H2 complex in H2bΔIEC mice by immunohistochemistry and western blotting (Supplementary Figure 1A–C). Of note, although the RNase H2 holoenzyme consists of subunits A (catalytic), B, and C, genetic deletion of the B subunit is sufficient to disintegrate the entire enzyme complex.7 Histopathologic analysis of 8-week-old mice showed distorted crypt architecture with modest epithelial damage (erosion and crypt hyperplasia) in the small intestinal mucosa (Figure 1A and B) of H2bΔIEC mice, which was accompanied by larger numbers of mucosal IBA1+ macrophages (Figure 1C and D) and a trend toward increased CD3+ T cells (Figure 1E and F). Further phenotyping exposed normal cellular differentiation (Supplementary Figure 1D–G) and proliferation, as shown by bromodeoxyuridine incorporation (Figure 1G and H) and Ki67 (Figure 1I and J) staining. Loss of intestinal RNase H2 led to increased epithelial apoptosis (Figure 1K and L), presumably as a consequence of the increased DNA damage (Figure 1M and N). Despite histologic evidence of intestinal inflammation, H2bΔIEC mice did not present with an overt macro-morphologic phenotype (Supplementary Figure 1H–J), indicating that constitutive intestinal epithelial DNA damage at a young age does not translate into a gross local or even systemic inflammatory response.
Supplementary Figure 1.
Generation of mice and genotype validation. (A) Generation and genotyping of H2bΔIEC and H2bfl/fl (control) mice. (B) Strongly decreased RNase H2A protein levels in western blot from isolated IECs from H2bΔIEC mice. (C) Immunohistochemistry of small intestine showing lack of RNase H2A, indicating complete RNase H2 complex destruction specifically in the intestinal epithelium of H2bΔIEC mice. Small intestines from H2bfl/fl (n = 6; 4 male and 2 female) and H2bΔIEC (n = 5; 3 male and 2 female) mice display no overt differences in epithelial differentiation markers (D, E) Lysozyme or (F, G) PAS. (H) Body weight, (I) small intestinal length, and (J) colon length in 8- to 12-week-old H2bΔIEC (n = 5; 3 male and 2 female) and H2bfl/fl control (n = 6; 4 male and 2 female) mice. (K) Body weight, (L) small intestinal length, and (M) colon length in 52-week-old H2bΔIEC (n = 10; 6 male and 4 female) and H2bfl/fl control (n = 6; 3 male and 3 female) mice. (N) Normalized end density per million base pairs of small intestinal samples from 8- to 12-week-old (young) or 52-week-old H2bΔIEC and H2bfl/fl mice. (E, G) A minimum of 100 crypts per intestine were assessed for H2bΔIEC (n = 5; 3 male and 2 female) and H2bfl/fl control (n = 6; 4 male and 2 female) mice. Data are expressed as mean ± standard error of the mean and significance was determined using nonparametric Mann-Whitney U-test. *P < .05; ***P < .001. PAS, periodic acid–Schiff.
Figure 1.
Increased DNA damage and apoptosis in young H2bΔIEC mice. Representative images (A) and histologic analysis of H&E sections (B) from small intestinal section (n = 5 for the 2 genotypes). Representative images and statistical assessment of abundance of IBA1+ (C, D) and CD3+ cells (E, F) in the lamina propria of H2bfl/fl or H2bΔIEC mice. Anti-BrdU (G, H) and anti-Ki67 (I, J) staining in small intestinal sections. TUNEL+ (K, L) and anti-γH2AX+ (M, N) staining in small intestinal sections. (D, F) A minimum of 5 HPFs per intestine were assessed in 8- to 12-week-old H2bΔIEC (n = 5; 3 male and 2 female) and H2bfl/fl control (n = 6; 4 male and 2 female) mice. (H–N) A minimum of 100 crypts per intestine were assessed for H2bΔIEC (n = 5; 3 male and 2 female) and H2bfl/fl control (n = 6; 4 male and 2 female) mice. Data are expressed as mean ± standard error of the mean and significance was determined using nonparametric Mann-Whitney U-test. *P < .05; ***P < .001. BrdU, bromodeoxyuridine; H&E, hematoxylin and eosin; HPF, high-power field; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
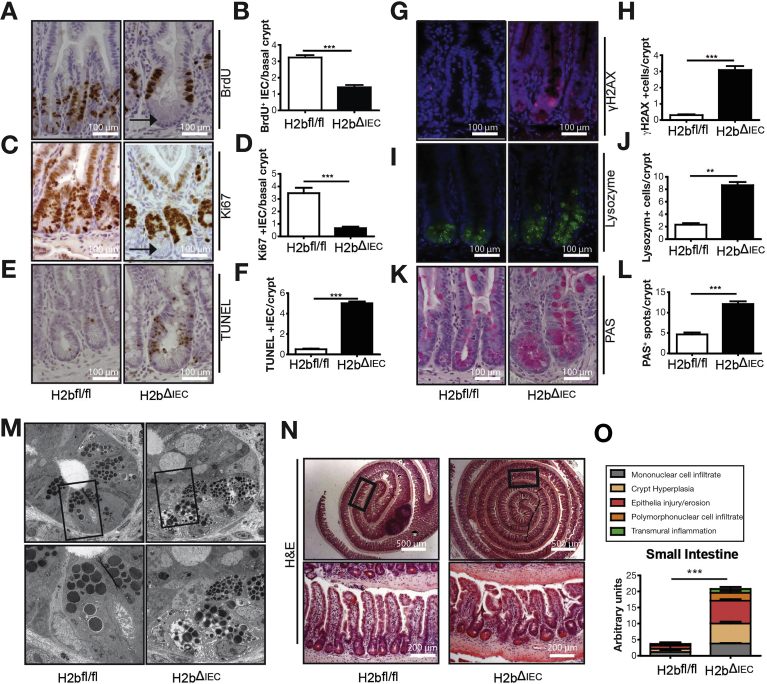
To test the consequences of long-term intestinal DNA damage, we assessed the phenotype in 52-week-old animals. At this time point, H2bΔIEC mice showed a modest increase in crypt depth and unaltered villus length (data not shown). However, we observed a proliferation deficit (by Ki67 and bromodeoxyuridine staining), specifically in the basal crypt region of the entire jejunum (data not shown) and ileum, indicative of an intestinal stem cell–specific impact of RNase H2B loss (Figure 2A–D). Proliferation in the transit-amplifying zone seemed to be retained, whereas apoptosis was increased in H2bΔIEC mice (Figure 2E and F).
Figure 2.
Impaired epithelial regeneration in aged H2bΔIEC mice. Anti-BrdU incorporation (A, B) and anti-Ki67 staining (C, D) in small intestinal sections. Note the lack of cellular proliferation in the intestinal stem cell niche located at the crypt base (arrow). TUNEL (E, F) and anti-γH2AX (G, H) staining in small intestinal sections. Lysozyme (I, J) and PAS (K, L) staining in small intestinal sections. (M) Distorted Paneth cell ultrastructure in H2bΔIEC mice (bottom pictures represent magnifications of the images above). Histologic analysis of H&E staining (N, O) in small intestinal sections (n = 5 per genotype). (B, D, F, H, J, L) A minimum of 100 crypts per intestine and (N) whole small intestinal Swiss rolls were assessed in 52-week-old H2bΔIEC (n = 10; 6 male and 4 female) and H2bfl/fl control (n = 6; 3 male and 3 female) mice. Data are expressed as mean ± standard error of the mean and significance was determined using nonparametric Mann-Whitney U-test. *P < .05; **P < .01; ***P < .001. BrdU, bromodeoxyuridine; H&E, hematoxylin and eosin; PAS, periodic acid–Schiff; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
Aged mice (52 weeks) displayed increased DNA damage (Figure 2G and H), which was associated with irregular differentiation into lysozyme-positive Paneth cells (Figure 2I and J) and goblet cells (Figure 2K and L). Paneth cells displayed a distorted ultrastructure (Figure 2M) and decreased epithelial mRNA levels of lysozyme and of cryptidin 1 (data not shown). H2bΔIEC mice presented with substantial histologic inflammation in the small intestine (Figure 2N and O) and decreased length of the small intestine, but no other significant macro-morphologic changes (Supplementary Figure 1K–M). Moreover, we did not observe age-dependent differences in genomic ribonucleotide incorporation in aged and young H2bΔIEC mice (Supplementary Figure 1N). Importantly, increased mucosal inflammation and impaired epithelial proliferation were environmentally phenotyped in the histology of the colon of aged mice (Supplementary Figure 2A–N). In conclusion, absence of intestinal RNase H2 results in epithelial DNA damage and impairment of epithelial proliferation and cell death, which manifest as spontaneous age-dependent intestinal inflammation.
Supplementary Figure 2.
Colon phenotype of aged mice. (A) Representative images and (B) histologic analysis of H&E sections from small intestine show moderate intestinal inflammation in H2bΔIEC mice. Histologic evaluation and representative images of (C, D) CD3+ and (E, F) IBA1+ cells in the colonic lamina propria of 52-week-old H2bΔIEC and H2bfl/fl mice. Epithelial proliferation in the colon was significantly decreased in aged H2bΔIEC mice compared with H2bfl/fl control littermates, as evidenced by (G, H) anti-BrdU and (I, J) anti-Ki67 staining. Note the lack of cellular proliferation in the intestinal stem cell niche located at the crypt base (arrows). Increased apoptosis and DNA damage in small intestines of H2bΔIEC mice shown by (K, L) TUNEL and (M, N) γH2AX staining. (B) Small intestinal Swiss rolls, (C, E) a minimum of 5 individual high-power fields, and (G, I, K, M) a minimum of 100 crypts per intestine were assessed in 52-week-old H2bΔIEC (n = 10; 6 male and 4 female) and H2bfl/fl control mice (n = 6; 3 male and 3 female) mice. Data are expressed as mean ± standard error of the mean and significance was determined using nonparametric Mann-Whitney U-test. *P < .05; **P < .01; ***P < .001. BrdU, bromodeoxyuridine; H&E, hematoxylin and eosin; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
Ablation of Intestinal RNase H2 in Mice Confers Susceptibility to Experimental Colitis
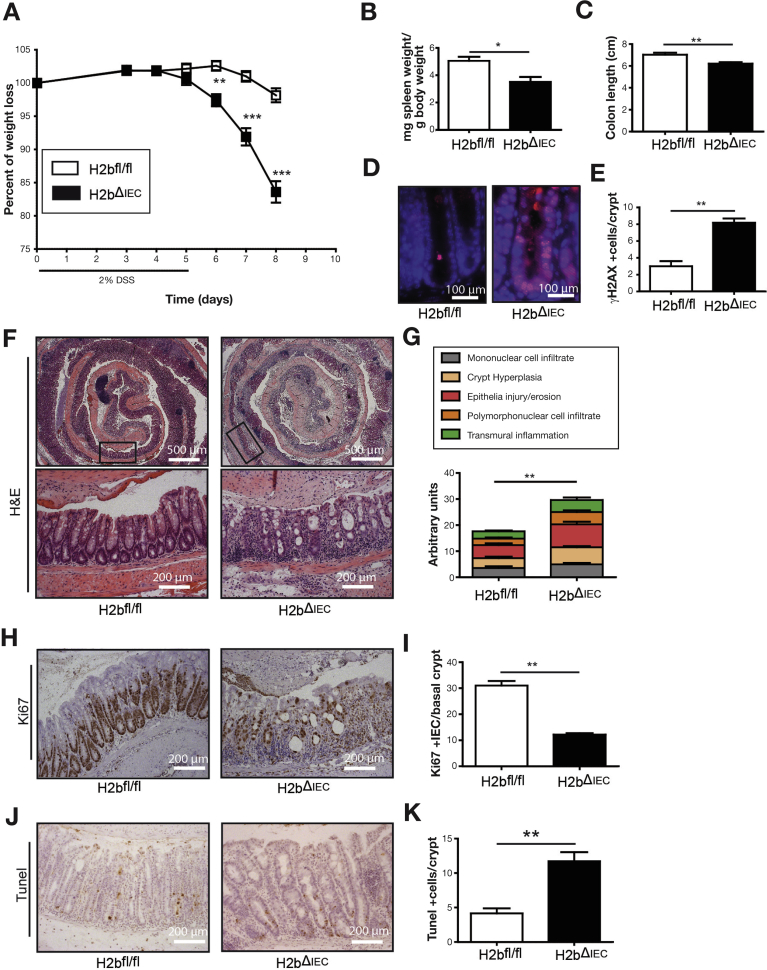
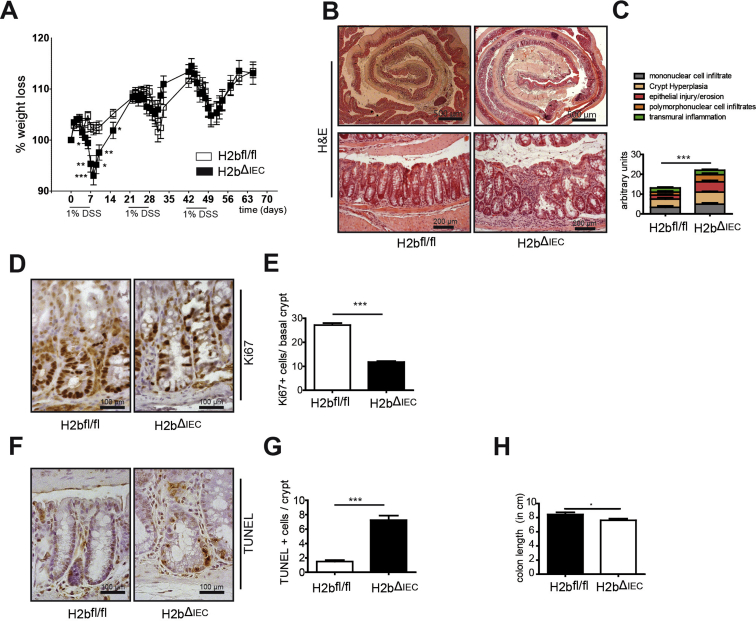
We next addressed the functional consequences of RNAse H2 deletion during forced regenerative responses of the colonic mucosa using an acute dextran sodium sulfate (DSS) model. H2bΔIEC mice developed a fulminant colitis with increased weight loss (Figure 3A), which was accompanied by decreases in spleen weight (Figure 3B) and colon length (Figure 3C). Histologic assessment exposed epithelial DNA damage (Figure 3D and E) and overall increased histopathologic inflammation (Figure 3F and G). Excessive inflammation was associated with strongly decreased epithelial proliferation (Figure 3H and I) and increased epithelial cell death in H2bΔIEC mice (Figure 3J and K). Also, in a chronic DSS colitis model (repetitive exposure of low-dose DSS over 3 cycles), H2bΔIEC mice displayed increased weight loss and histologic inflammation, decreased epithelial proliferation, and increased epithelial apoptosis (Supplementary Figure 3A–H). Notably, we did not observe any spontaneous tumor development (small intestine or colon) after chronic DSS exposure in H2bΔIEC mice.
Figure 3.
Increased susceptibility to DSS-induced colitis in H2bΔIEC mice. (A) Weight loss curve of 8- to 12-week-old H2bfl/fl (n = 12; 9 male and 3 female) and H2bΔIEC (n = 10; 5 male and 5 female) mice. Postmortem (B) spleen weight (n = 12 of 10) and (C) colon length (n = 12 of 10) of H2bfl/fl and H2bΔIEC mice. (D, E) Anti-γH2AX staining in small intestinal sections. (F) H&E staining and (G) corresponding histologic assessment from colon tissue. (H) Anti-Ki67 staining of colon Swiss rolls and (I) corresponding assessment. (J) TUNEL staining and (K) quantification of TUNEL stain of colon Swiss rolls. (E, I, K) A minimum of 100 crypts per intestine and (G) whole small intestinal Swiss rolls were assessed in H2bΔIEC (n = 10; 5 male and 5 female) and H2bfl/fl control (n = 12; 9 male and 3 female) mice. Significance was determined using 2-tailed Student t-test and expressed as mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001. H&E, hematoxylin and eosin; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
Supplementary Figure 3.
Chronic colitis in H2bfl/fl and H2bΔIEC mice. (A) Weight loss curve of RnaseH2bfl/fl (n = 8; 4 male and 4 female) and RnaseH2bΔIEC (n = 9; 5 male and 4 female) mice. (B) Representative images of colon Swiss rolls stained with H&E and (C) corresponding histologic assessment. Representative images and histologic evaluation of (D, E) Ki67- and (F, G) TUNEL-positive cells in colon crypts (≥100 crypts per intestine). (H) Postmortal colon length (n = 8 of 9) of H2bfl/fl and H2bΔIEC mice. (C) Small intestinal Swiss rolls and (E, G) a minimum of 100 crypts per intestine were assessed in H2bΔIEC (n = 8; 5 male and 3 female) and H2bfl/fl control (n = 9; 4 male and 5 female) mice. Significance was determined using 2-tailed Student t-test and expressed as mean ± standard error of the mean *P < .05; **P < .01; ***P < .001. H&E, hematoxylin and eosin; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
Impaired Epithelial Regeneration in Aged H2bΔIEC Mice Depends on Functional p53
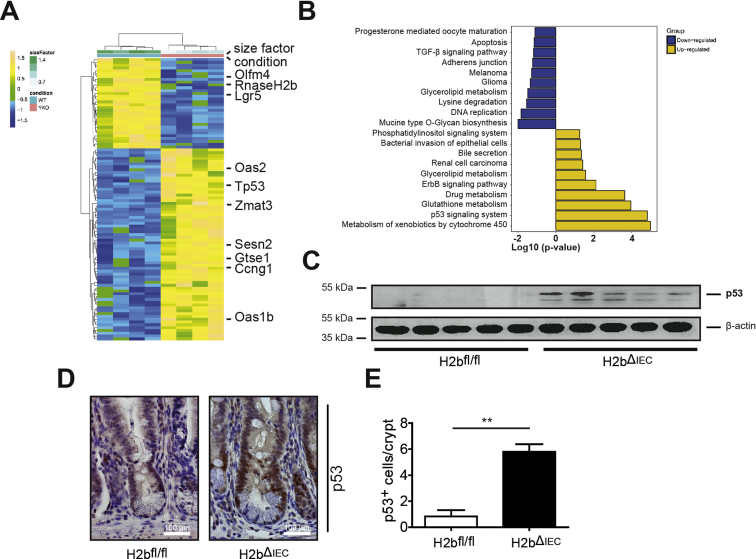
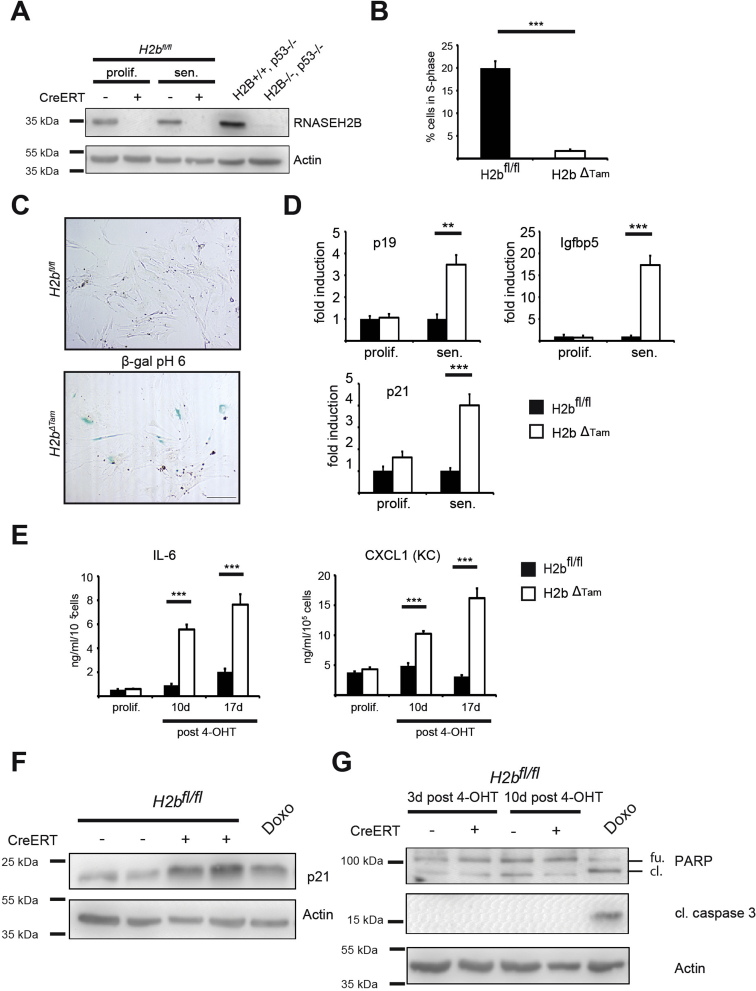
To understand the underlying molecular principle of intestinal growth arrest in H2bΔIEC mice, we subjected purified small intestinal epithelial cells (IECs) from aged mice (n = 4 mice; 2 female and 2 male per genotype) to RNA sequencing. Hierarchical clustering of the top 100 regulated genes (up and down) displayed numerous p53-related genes up-regulated in the transcriptome of H2bΔIEC epithelial cells (Supplementary Figure 4A and Supplementary Tables 1 and 2). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the top 250 regulated genes showed that “p53” was among the top regulated terms in H2bΔIEC epithelial cells (Supplementary Figure 4B). Increased p53 protein stabilization was further confirmed using western blot with protein lysates from isolated H2bΔIEC IECs and immunohistochemistry in the small intestine (Supplementary Figure 4C–E). To confirm a link between Rnaseh2b deletion and p53 activation, we generated intestinal organoids from mice carrying a tamoxifen-inducible knockout of RNase H2b (H2bΔTam). After tamoxifen treatment H2bΔTam intestinal organoids rapidly developed epithelial cell death and activation of the p53 signaling pathway (Supplementary Figure 5). The molecular nexus between RNase H2 deficiency and p53 activation was further confirmed in murine embryonic fibroblasts (MEFs) from H2bΔTam mice.30 Tamoxifen-induced Rnaseh2b ablation in MEFs led to up-regulation of p21, a canonical downstream target of p53, and induction of cellular senescence. However, we did not observe detectable activation of caspase-3, indicating a difference of the biological impact of RNase H2 deficiency between MEFs and IECs (Supplementary Figure 6).
Supplementary Figure 4.
RNA sequencing of isolated IECs. (A) Heatmap showing clustering of top 100 differentially expressed genes in isolated IECs from H2bfl/fl (n = 4; 2 male and 2 female) and H2bΔIEC (n = 4; 2 male and 2 female) mice. (B) Gene Ontology term analysis of differentially up- and down-regulated genes between H2bfl/fl (n = 4) and H2bΔIEC (n = 4). (C) Western blot from isolated IECs from aged H2bfl/fl (n = 5) and eH2bΔIEC (n = 5) mice probed against anti-p53 antibody. (D) Representative images and (E) statistical evaluation of p53+ cells per crypt. (E) A minimum of 100 crypts per intestine were assessed in 52-week-old H2bΔIEC (n = 5; 3 male and 2 female) and H2bfl/fl control (n = 6; 3 male and 3 female) mice. Significance was determined using 2-tailed Student t-test and expressed as mean ± standard error of the mean. **P < .01.
Supplementary Figure 5.
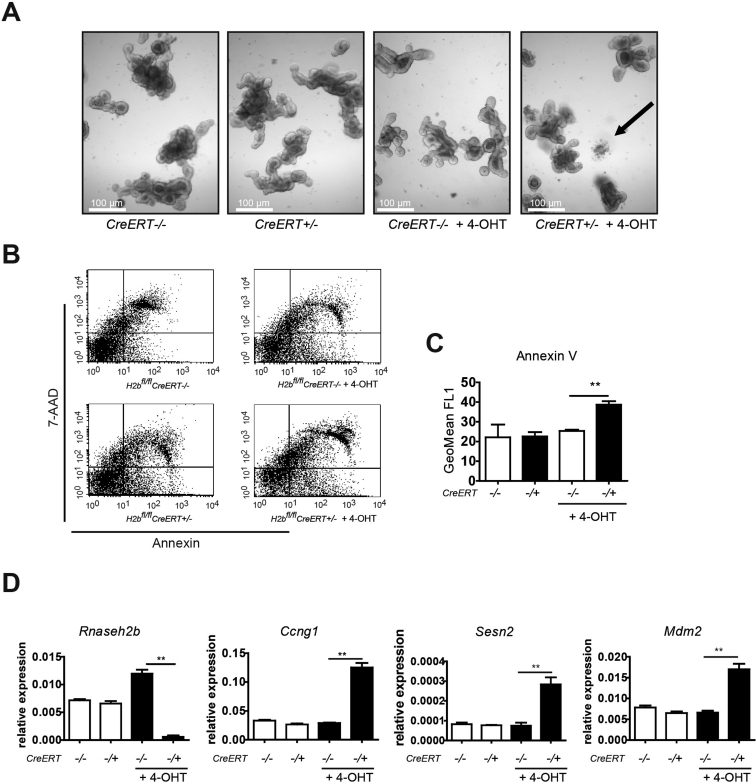
Phenotyping of H2bΔTam organoids. Administration of a low dose of 4-OHT (100 nmol/L) for 3 days resulted in increased cell death and induction of apoptosis and induction of canonical p53 target genes in primary intestinal organoids derived from H2bfl/fl mice bearing a ubiquitous Cre recombinase (CreERT). (A) Representative images of intestinal organoids 3 days after 4-OHT treatment (white bars, 100 μm; black arrow, dying organoid). Representative (B) fluorescence-activated cell sorter plots and (C) statistical analysis of GeoMean intensity of annexin V and fluorescein isothiocyanate staining. (D) Gene expression of intestinal organoids 3 days after 4-OHT assessing Rnaseh2b, Ccng1, Sesn2, Mdm2 by quantitative polymerase chain reaction. Significance was determined using Student t-test and expressed as mean ± standard error of the mean. **P < .01. 4-OHT, 4-hydroxytamoxifen; 7-AAD, 7-aminoactinomycin D.
Supplementary Figure 6.
Phenotyping of H2bΔTam MEF. Administration of a low dose of 4-OHT (100 nmol/L) for 3 days resulted in abrogation of RNASEH2B protein expression in primary MEFs derived from H2bfl/fl embryos bearing a ubiquitous Cre recombinase (CreERT). H2bfl/fl MEFs without CreERT were used as control cells. For conciseness, 4-OHT–treated H2bfl/fl plus CreERT MEFs are termed H2bΔTam MEFs. Rnaseh2b/p53 double-knockout MEFs2 served as a negative control. The RNase H2 holoenzyme was detected by western blotting in proliferating (3 days after 4-OHT) and senescent (10 days after 4-OHT) MEFs using a specific rabbit antiserum raised against the whole murine enzyme complex.2 (A) Actin as loading control. H2bΔTam MEFs at 10 days after 4-OHT ceased proliferation. Proliferation was assessed by measuring 5-ethynyl-2′-deoxyuridine incorporation into replicating DNA using flow cytometry. (B) Percentage of cells in S-phase after a 4-hour 5-ethynyl-2′-deoxyuridine pulse is depicted. Error bars represent standard error of the mean; ***P < .001 by t-test (n = 5). (C) H2bΔTam MEFs at 17 days after 4-OHT stained positive for senescence-associated β-galactosidase. Cells also showed an altered morphology with an enlarged and flattened appearance (scale bar, 200 μmol/L). H2bΔTam MEFs in prolonged culture exhibited increased expression of senescence-associated genes p19, Cdkn1a (p21), and Igfbp5. (D) Transcript levels were gauged by quantitative polymerase chain reaction (p19 10 days and Igfbp5 and Cdkn1a 17 days after 4-OHT). Error bars represent standard error of the mean. ***P < .001; **P < .01 by 2-way analysis of variance (n = 3–4). Senescent H2bΔTam MEFs displayed a senescence-associated secretory phenotype and secreted the proinflammatory cytokines IL-6 and CXCL1 (KC). (E) MEF supernatant 10 and 17 days after 4-OHT was harvested for 24 hours and cytokine levels were analyzed by enzyme-linked immunosorbent assay. Note that secreted cytokine levels tended to increase over time. Error bars represent standard error of the mean. ***P < .001 by 2-way analysis of variance (n = 3). (F) Increased expression of p21 protein in H2bΔTam MEFs 3 days after Cre induction by a low dose of 4-OHT (100 nmol/L). Doxorubicin treatment at a concentration of 1 μmol/L for 24 hours was used as a positive control. (G) Deletion of Rnaseh2b does not induce apoptosis in primary MEFs. Administration of 4-OHT 100 nmol/L for the indicated time points does not lead to increased cell death, as assessed by western blot analysis of cleaved PARP-1 (full length, 116 kDa; cleaved, 89 kDa) and caspase 3. Apoptosis in control cells was induced by doxorubicin at 25 μmol/L for 24 hours. 4-OHT, 4-hydroxytamoxifen; β-gal, β-galactosidase; cl., cleaved; CXCL1, chemokine (C-X-C motif) ligand 1; Doxo, doxorubicin; fu., full length; IL-6, interleukin-6; PARP, poly(adenosine diphosphatase ribose)polymerase; prolif., proliferating; sen., senescent.
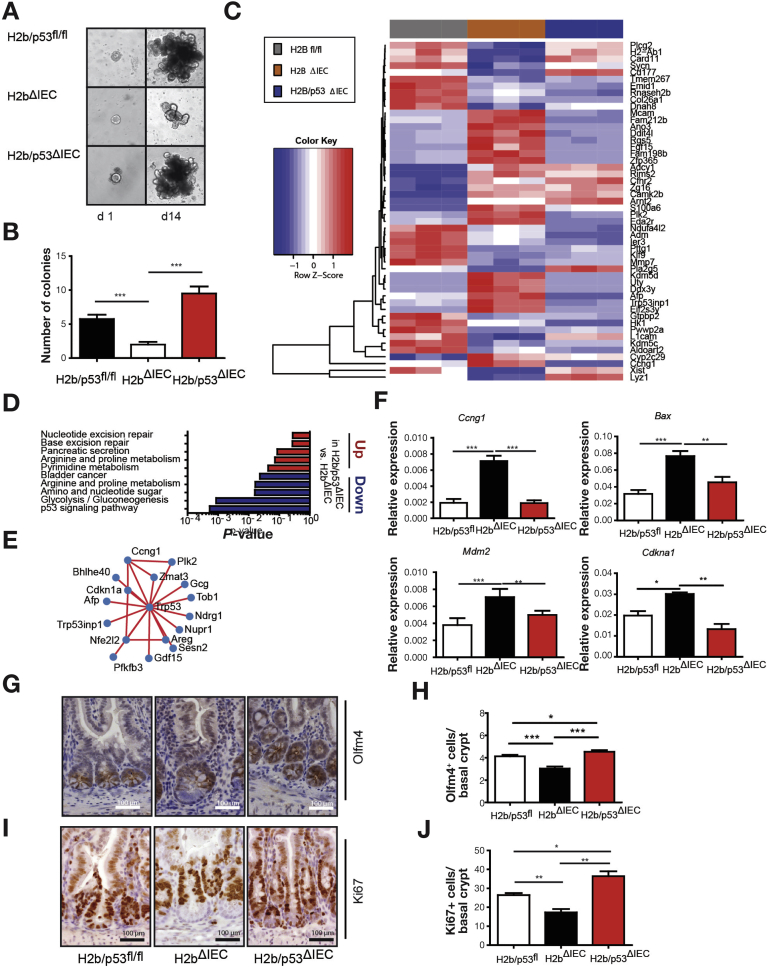
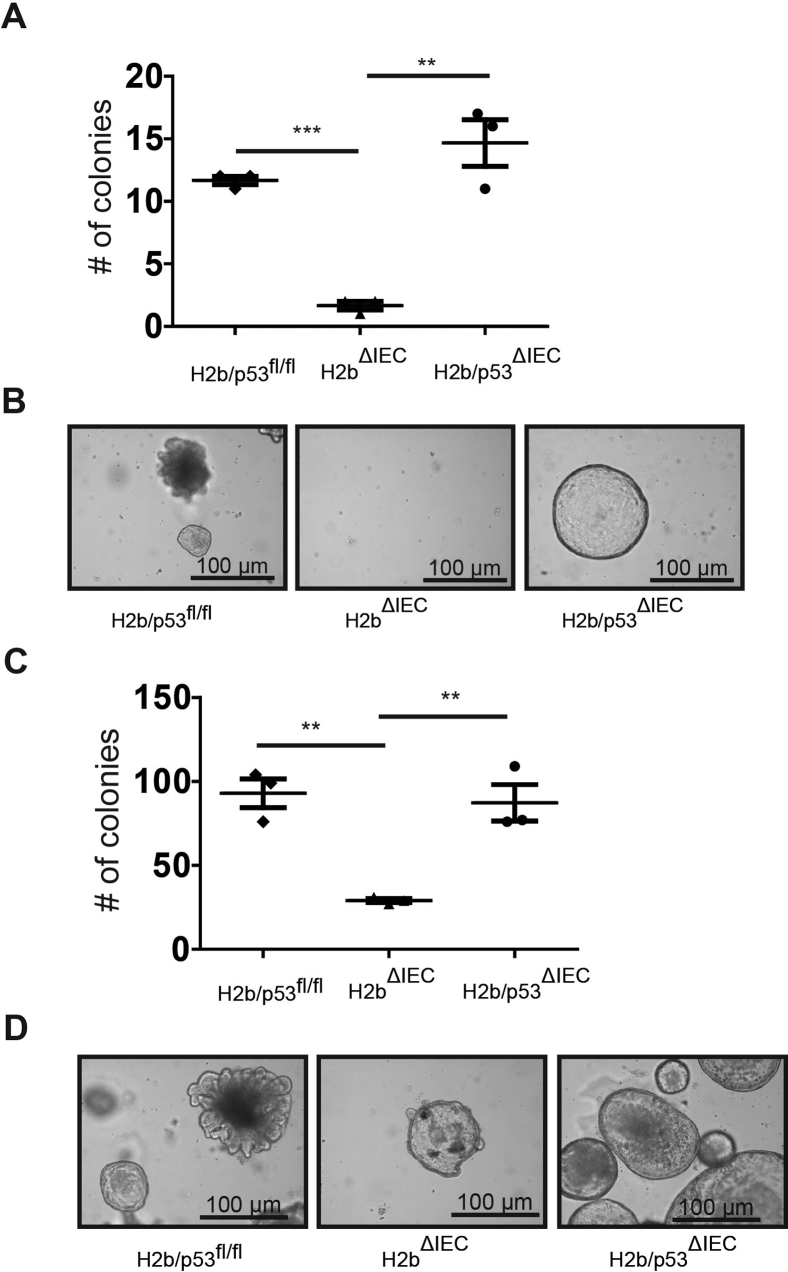
To further corroborate the hypothesis that p53 activation is responsible for the proliferation deficit in the absence of RNase H2, we generated mice lacking intestinal Rnaseh2b and p53 (H2b/p53ΔIEC). By assessing the growth capacity in small intestinal organoids from 8-week-old mice, we observed that H2bΔIEC organoids displayed severe growth impairment, which was restored with simultaneous p53 deletion (Figure 4A and B). This effect was environmentally phenotyped in colon organoids from 52-week-old mice, suggesting the presence of the molecular effect throughout the intestinal epithelial lining (Supplementary Figure 7).
Figure 4.
Epithelial regeneration failure in aged H2bΔIEC mice depends on p53. (A) Representative images and (B) statistical analysis of small intestinal growth after 14 days of cultivation. (C) Heatmap showing clustering of top 50 up- and down-regulated genes in H2bΔIEC, H2bfl/fl, and H2b/p53ΔIEC intestinal organoids. (D) Gene set enrichment (KEGG) analysis of top 250 up- and down-regulated genes in H2b/p53ΔIEC vs H2bΔIEC intestinal organoids. STRING-based network analysis of top 50 down-regulated genes in H2b/p53ΔIEC vs H2bΔIEC. (E) Genes not being connected to the central p53 hub were manually removed. (F) Transcript levels in small intestinal epithelial crypts from 20-week-old H2bfl/fl (n = 8; 5 male and 3 female), H2bΔIEC (n = 5; 2 male and 3 female), and H2b/p53ΔIEC (n = 9; 4 male and 5 female) mice were measured by quantitative polymerase chain reaction. (G, H) Anti-Olfm4 and (I, J) anti-Ki67 immunohistochemical assessments in small intestinal sections. (G–J) A minimum of 100 crypts per intestine were assessed (n = 5 per genotype). Significance was determined using nonparametric Mann-Whitney U-test and expressed as mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001.
Supplementary Figure 7.
Phenotyping of 35-week-old colon intestinal organoids. Statistical evaluation and representative images of colon organoids were derived from 35-week-old H2b/p53fl/fl, H2bΔIEC, or H2b/p53ΔIEC mice. Statistical evaluation and representative images of obtained colon organoid colonies at day 4 after passaging and seeding in the (A, B) absence or (C, D) presence of the anoikis inhibitor Y-27632. Significance was determined using nonparametric Mann-Whitney U-test and expressed as mean ± standard error of the mean. **P < .01; ***P < .001.
Further RNA sequencing of small intestinal organoids from H2bfl/fl, H2bΔIEC, and double-deficient H2b/p53ΔIEC intestinal organoids confirmed the up-regulation of p53-dependent genes in H2bΔIEC, which was abolished in H2b/p53ΔIEC intestinal organoids (Figure 4C and D and Supplementary Tables 3 and 4). Notably, KEGG gene set enrichment analysis comparing H2bΔIEC with H2b/p53ΔIEC transcriptomes showed that up-regulated pathways were enriched for alternative DNA damage repair pathways (nucleotide excision repair, base excision repair) in H2b/p53ΔIEC organoids, indicating the induction of a compensatory DNA repair mechanism in the absence of functional RNase H2 (Figure 4D). Interaction analysis using the STRING database31 of top 50 down-regulated genes in H2b/p53ΔIEC showed a densely connected network of p53-dependent genes (Figure 4E). Thus, we conclude that restoration of epithelial proliferation in H2b/p53ΔIEC organoids is essentially orchestrated by p53. This finding was confirmed in vivo, because isolated intestinal crypts of 20-week-old H2b/p53ΔIEC mice specifically displayed the absence of p53-dependent gene induction (Ccng1, Bax, Mdm2, Cdkna1; Figure 4F). Most importantly, we observed the reappearance of Olfm4+ stem cells, excessively (compared with H2b/p53fl/fl animals) restored epithelial proliferation (Figure 4G–J), similar levels of DNA damage, and decreased epithelial apoptosis in H2b/p53ΔIEC mice (Supplementary Figure 8A–D).
Supplementary Figure 8.
Phenotyping of 20-week-old H2b/p53ΔIEC mice. Immunohistochemistry of (A, B) γH2AX and (C, D) TUNEL staining in the small intestine shows restoration of background (H2b/p53fl/fl) apoptosis levels in H2b/p53ΔIEC double-knockout mice, albeit with a similar degree of DNA damage. (A, C) A minimum of 100 crypts per intestine were assessed in 20-week-old H2bfl/fl (n = 8; 5 male and 3 female), H2bΔIEC (n = 5; 2 male and 3 female), and H2b/p53ΔIEC (n = 9; 4 male and 5 female) mice. Significance was determined using nonparametric Mann-Whitney U-test and expressed as mean ± standard error of the mean. ***P < .001. TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling.
Spontaneous Tumor Development in H2b/p53ΔIEC Mice
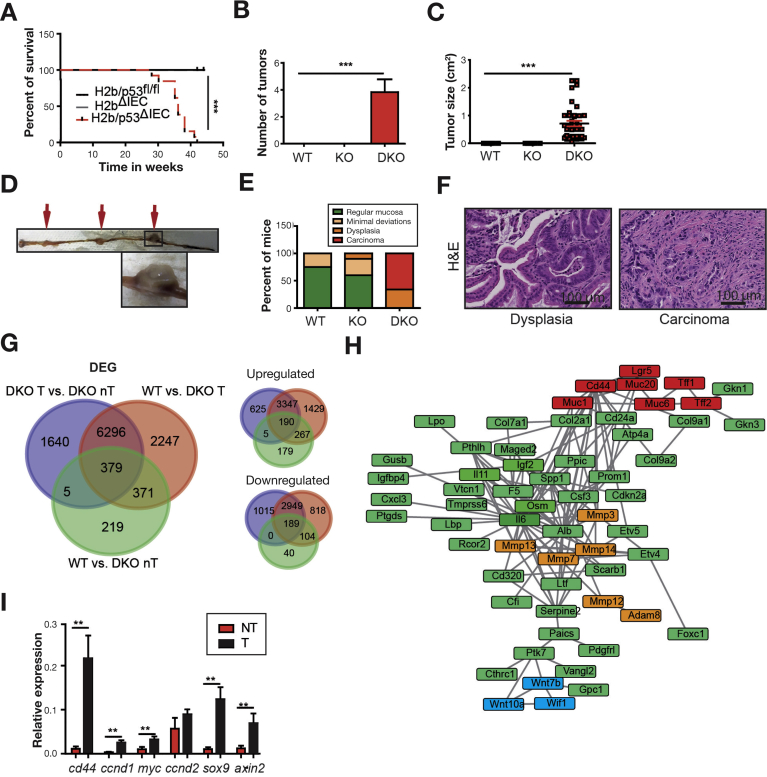
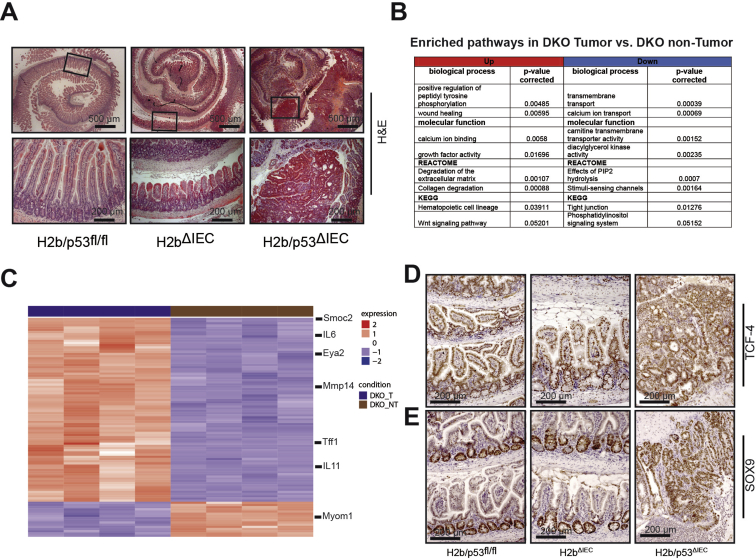
Starting at 30 weeks of age, H2b/p53ΔIEC mice died or had to be withdrawn from the study because of their deteriorating physical condition (Figure 5A). Postmortem analysis showed the development of small intestinal (n = 17/17) and colon (n = 2/17) tumors (Figure 5B–D). Interestingly, male vs female mice showed slightly shorter overall survival despite comparable tumor burden (data not shown). Histopathologic examination graded tumors as adenomas with high-grade dysplasia (33%) or low-grade epithelial carcinomas (66%; Figure 5E and F and Supplementary Figure 9A). Importantly, p53ΔIEC mice do not display spontaneous intestinal tumor formation up to 14 months,32 even under persistent DNA damage.33 RNA sequencing of tumors and adjacent nontumor tissue from H2b/p53ΔIEC mice showed differential expression of 1640 genes (625 up, 1015 down; Figure 5G and Supplementary Tables 5 and 6). Up-regulated transcripts were enriched for the Wnt signaling pathway (Wnt7b, Wnt10a, Wif1) and pathways associated with epithelial-to-mesenchymal transition (matrix metalloproteinases 3, 7, 13, and 14; Figure 5H and Supplementary Figure 9B and C). In line with this, we found nuclear accumulation of TCF-4 and SOX-9 (Supplementary Figure 9D and E) and up-regulation of canonical Wnt target genes (Cd44, Axin2, Sox9) in tumor vs adjacent nontumor tissue (Figure 5I), although we did not observe increased nuclear accumulation of active β-catenin (data not shown). Altogether our data show that concomitant deletion of RNase H2 and p53 drives spontaneous intestinal tumorigenesis in H2b/p53ΔIEC mice.
Figure 5.
Spontaneous intestinal tumor development in H2b/p53ΔIEC mice. (A) Survival curve for H2b/p53ΔIEC (n = 17; 9 male and 6 female), H2b/p53fl/fl (n = 13; 7 male and 6 female), and H2bΔIEC (n = 9; 3 male and 6 female) mice. (B) Total number of tumors per small intestine, (C) average tumor size in small intestinal tumors and (D) representative image of a H2b/p53ΔIEC small intestine bearing a total of 3 tumors (arrows). (E) Histologic grading of intestinal mucosa with or without tumors. (F) Representative image of a dysplasia or low-grade carcinoma in small intestine from H2b/p53ΔIEC mice. (G) Venn diagram of DEGs from small intestinal tissues derived from H2b/p53ΔIEC tumors (DKO T; n = 4; 3 male and 1 female) or adjacent nontumor tissue from the same mice (DKO nT, n = 4; 3 male and 1 female) or H2b/p53fl/fl (WT, n = 4; 2 male and 2 female). Total number of DEGs (left) and number of up- and down-regulated genes (right) are displayed. (H) STRING-based network analysis of top 100 up-regulated genes in tumor vs nontumor intestinal tissue from H2b/p53ΔIEC mice. Manual coloring was used to highlight genes involved in extracellular matrix degradation (orange), cellular differentiation (red), and Wnt signaling (blue). (I) Quantitative polymerase chain reaction of Wnt target genes (relative to adjacent nontumor small intestinal tissue, n = 8). Data are expressed as mean ± standard error of the mean and significance was determined using log-rank Mantel Cox test (A) or nonparametric Mann-Whitney U-test. *P < .05; **P < .01; ***P < .001. DEG, differentially expressed gene; DKO, double knockout; KO, knockout; NT, nontumor; T, tumor; WT, wild type.
Supplementary Figure 9.
RNA sequencing of intestinal tumor and nontumor tissue from H2b/p53ΔIEC mice. (A) Representative image of small intestinal tumors in H2b/p53ΔIEC, but not in H2b/p53fl/fl or H2bΔIEC, mice. RNA sequencing was performed from the RNA of small intestinal sections of H2b/p53fl/fl (n = 4, 3 female and 1 male) and paired tumor and nontumor sections of H2b/p53ΔIEC (n = 4; 2 male and 2 female) mice. (B) Overview of selected signaling pathways (Gene Ontology terms and KEGG pathways) enriched in H2b/p53ΔIEC tumors compared with adjacent nontumor tissue. (C) Heatmap of the top 100 differentially expressed genes, ranked according to adjusted P value. Representative immunohistochemical staining of small intestinal tissue stained against (D) TCF-4 and (E) SOX9. Images indicate increased nuclear translocation (TCF-4) or overall expression (SOX9) in tumors from H2b/p53ΔIEC mice. DKO, double knockout; DKO_NT, double-knockout nontumor; DKO_T, double-knockout tumor; H&E, hematoxylin and eosin.
H2b/p53ΔIEC Mice Display a Distinct Mutational Signature
Next, we investigated the genomic events related to this malignant transformation and assessed the ribonucleotide content of tumor-derived DNA by gauging its alkaline sensitivity.34
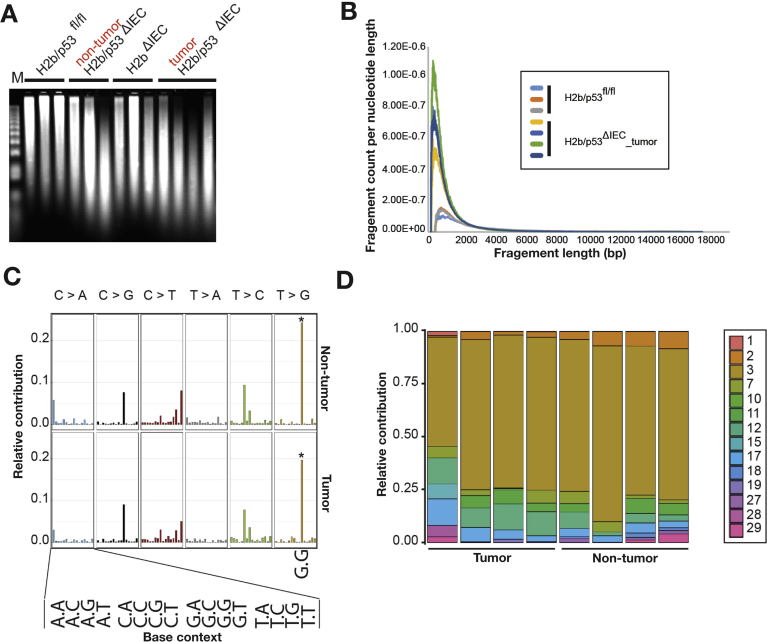
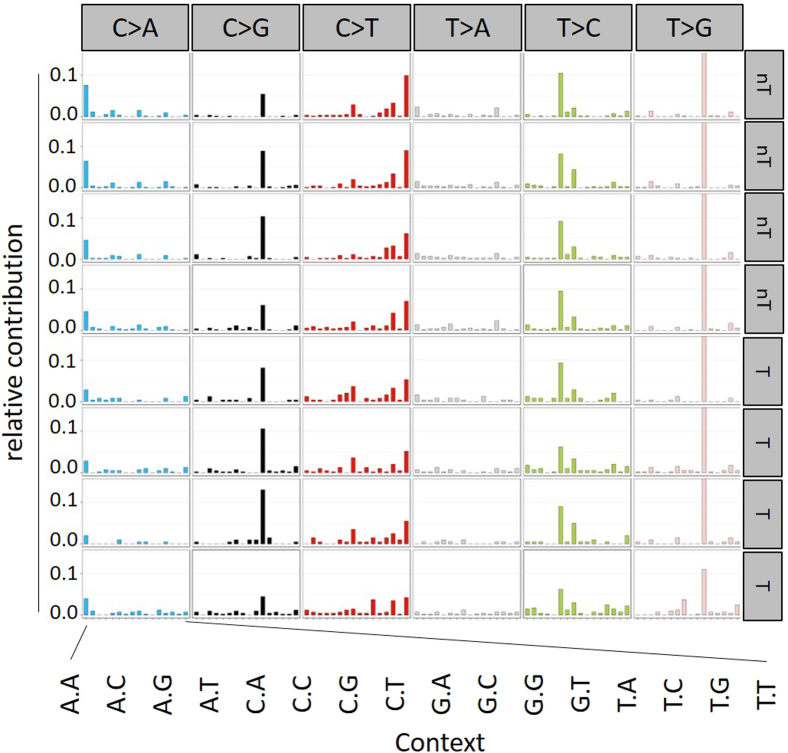
We observed a strong increase in genomic ribonucleotides in tumor DNA and to a lesser extent in nontumor H2b/p53ΔIEC and H2bΔIEC, but not in H2b/p53fl/fl, intestinal DNA (Figure 6A and B and Supplemental Figure 10A and B). In contrast to H2bΔIEC mice (Supplementary Figure 1K), we observed a gradual age-dependent increase of genomic ribonucleotide incorporation in intestines from H2b/p53ΔIEC mice (Supplemental Figure 10C). To investigate whether genomic ribonucleotides resulting from defective RNase H2 are mutagenic, we exome-sequenced tumors of individual mice (n = 4 mice; 2 female and 2 male per genotype) and compared them with (1) adjacent nontumor H2b/p53ΔIEC epithelium and (2) RNase H2-proficient liver DNA from the same animal. We used an established approach to describe the observed somatic variants in their 5′ and 3′ base context, thus resulting in 96 possible mutation types.35 Tumor and nontumor tissues shared a remarkable degree of similarity and individual samples shared a high degree of conformity of enriched mutation types with a striking over-representation of T>G transversions in the context of a 5′ and 3′G (Figure 6C and Supplementary Figure 11), which were observed in all 8 samples from H2b/p53ΔIEC mice in tumor and nontumor tissues. Importantly, the definition of mutation types has been used in the COSMIC database36 from more than 7000 human tumor genomes to infer specific mutational signatures (ie, combinations of mutation types) describing the heterogeneity of mutational processes operative during tumorigenesis.37 We tested to which extent mutational patterns in our mouse samples shared similarity with those previously established mutational signatures in humans. We found that mutational signature 3 (according to the COSMIC nomenclature) contributed up to 75% of the observed patterns of mutation types in H2b/p53ΔIEC mice (Figure 6D). This signature has been associated with failure of DNA double-strand break–repair by homologous recombination (HR).35
Figure 6.
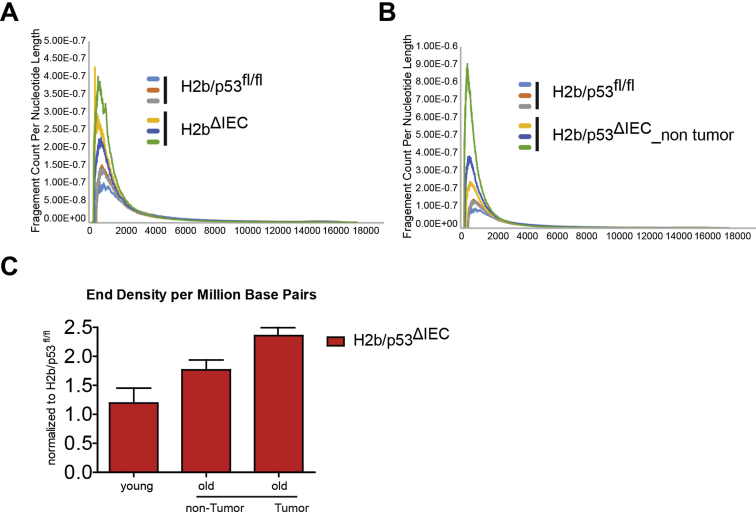
A mutational signature associated with genomic ribonucleotides. Ribonucleotide content of genomic intestinal DNA was assessed by alkaline hydrolysis and subsequent electrophoresis of fragmented DNA. Increased DNA fragmentation indicated a higher genomic ribonucleotide load. (A) Gel image shows intestinal DNA samples from individual mice (n = 3–4). (B) Quantification of fragment count per nucleotide length based on electrophoresis gel shown in A. Graph shows comparison of H2b/p53ΔIEC tumor with H2b/p53fl/fl control DNA. (C) Relative contribution of mutational signatures described in the COSMIC database to individual tumor and H2b/p53ΔIEC nontumor signatures. Note the high contribution of signature 3 to all H2b/p53ΔIEC mutational signatures, which is independent of tumor status. A distinct T>G substitution at GpTpG trinucleotides is marked (asterisk). (D) Average mutational signatures extracted from tumor and H2b/p53ΔIEC nontumor intestinal DNA exhibit a striking degree of similarity.
Supplementary Figure 10.
Alkali hydrolysis of intestinal samples. Quantification of fragment count per nucleotide length based on electrophoresis gel shown in Figure 6A. Graphs show the comparison of (A) H2bΔIEC vs H2b/p53fl/fl control DNA with (B) H2b/p53ΔIEC nontumor vs H2b/p53fl/fl. (C) End density per million base pairs of small intestinal samples from young H2b/p53ΔIEC or aged H2b/p53ΔIEC mice (tumor vs adjacent normal), normalized to control DNA from H2b/p53fl/fl.
Supplementary Figure 11.
Mutational signature in intestinal tumor and nontumor tissue from H2b/p53ΔIEC mice. The figure shows the contribution of each SNV type including the base context to the somatic mutational signature of each sample. Base context is magnified and shown for C>A conversion, but applies to all depicted nucleotide SNVs. nT, nontumor; T, tumor.
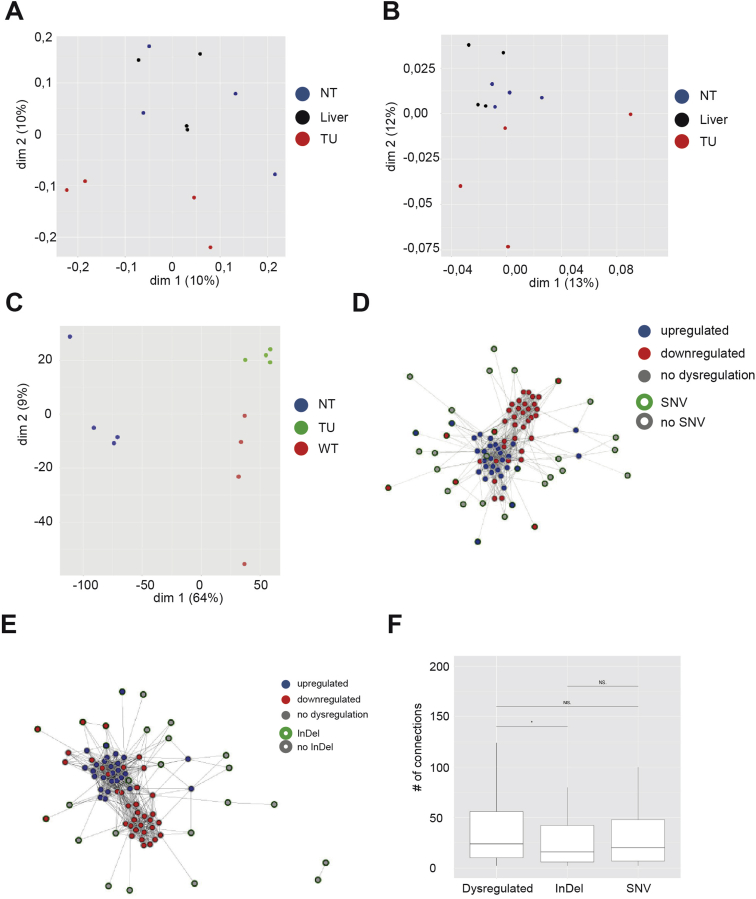
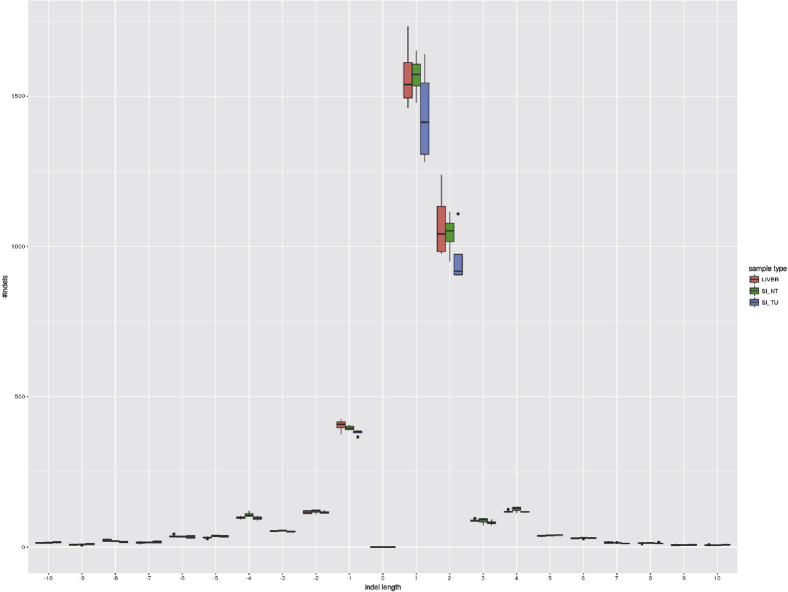
Despite displaying highly similar mutational signatures, tumor and corresponding nontumor samples clustered separately in gene expression and observed mutations, which included InDels and SNVs (Supplementary Figure 12A–C). To identify a potential mutation hotspot involved in tumorigenesis in H2b/p53ΔIEC mice, we compiled a list of tumor-specific mutations (SNVs and small InDels) containing only genetic alterations that were not present in the corresponding nontumor H2b/p53ΔIEC epithelium of the same mouse (Supplementary Tables 7 and 8). With the exception of a hotspot of small InDels of all tumors within the Nlrp1a gene, which has been causally linked to skin cancer in humans,38, 39 we could not detect recurrent mutations, which would indicate a specific mechanism downstream of RNase H2 deficiency. In protein interaction network reconstruction (using dysregulated and mutated genes as input), genes affected by somatic SNVs or small InDels showed only very few connections among one another but were individually linked to a central network of dysregulated genes (Supplementary Figure 12D–F). Interestingly and in contrast to RNase H2-null yeast,6 no global increase in small deletions in the 2 RNase H2-deficient experimental groups was found (Supplementary Figure 13).
Supplementary Figure 12.
Gene expression and InDel count. Multidimensional scaling plots based on (A) InDels, (B) SNVs, or (C) RNA sequencing data. Network analysis integrating somatic (E) SNV or (F) InDels with genes differentially expressed between tumor and nontumor H2b/p53ΔIEC epithelium samples. Note that most dysregulated genes are clustered in the center, whereas most genes affected by an InDel or an SNV are localized to the periphery, with little connectivity between the 2 datasets. (E, F) For better readability, exclusively dysregulated genes having more than 200 connected dysregulated genes or a cancer proliferation indices sum of connected genes higher than 60 are displayed. (G) Network-based analysis of the difference between dysregulated and mutated genes. As input, all significantly dysregulated genes (tumor vs nontumor; P < .001) and all genes affected by a somatic SNV or a somatic InDel were considered. As shown in the boxplot, dysregulated genes were characterized by a larger number of connections than mutated genes (P = .023 for dysregulated vs InDel; P = .152 for dysregulated vs SNV; P = .011 for dysregulated vs SNV + InDel). NT, nontumor; T, tumor; WT, wild type.
Supplementary Figure 13.
InDel count. The figure displays the number of InDels for each InDel length subdivided into sample type groups. Negative InDel lengths indicate deletions, and positive values describe insertions.
RNase H2 Acts as a Tumor Suppressor in Human Colorectal Carcinogenesis
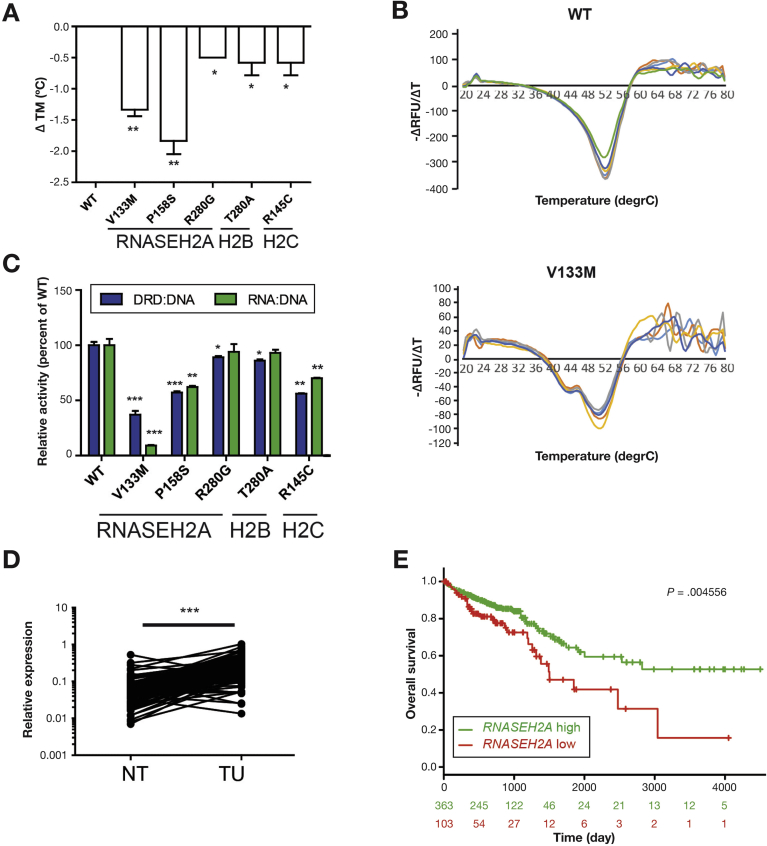
To explore whether impaired RNase H2 function is linked to colorectal carcinogenesis in humans, we examined the impact of the 5 most common somatic mutations found in 1 of the 3 RNase H2 subunits in human cancers, deposited in the COSMIC cancer database on RNase H2 function. Notably, 3 of 5 mutations were found in adenocarcinoma of the large intestine (Supplementary Table 9). Although the strength of the effect was variable, all 5 mutations showed a significant decrease in nuclease activity toward DNA duplexes containing single ribonucleotides and RNA:DNA hybrids and had a decreased thermostability, indicating that the cancer-associated RNase H2 variants are indeed hypomorphs (Figure 7A–C).
Figure 7.
Functional characterization of somatic RNase H2 cancer variants and RNase H2 expression in human colorectal tumors. Thermal stability of mutant RNase H2 complexes vs WT (represented as ΔTm; negative values represent a less stable complex). (A) Mean ΔTm ± standard error of the mean displayed for 6 technical replicates. (B) Representative graphs of thermostability of WT and V133M mutant. Decreased enzymatic activity of somatic RNase H2 cancer variants toward a DNA duplex containing a single ribonucleotide (DRD:DNA) or RNA–DNA hybrid (RNA:DNA). (C) Initial activity measured at RNase H2 15.6 pmol/L and substrate 250 μmol/L, with mean ± standard error of the mean displayed for 3 independent experiments. (D) Relative expression of RNASEH2A in paired tumor and adjacent normal tissue from patients with CRC (n = 155). Kaplan-Meier Plots for high vs low RNASEH2A expression in 467 patients with colorectal adenocarcinoma, retrieved from the TCGA COADREAD cohort. (E) Low expression levels of RNASEH2A were correlated with poor overall survival (P = .004). Data are expressed as mean ± standard error of the mean and significance was determined using 2-sided t-test (A, C), nonparametric Mann-Whitney U-test (D), or log-rank Mantel Cox Test (E). *P < .05; **P < .01; ***P < .001. NT, nontumor; TU, tumor; WT, wild type.
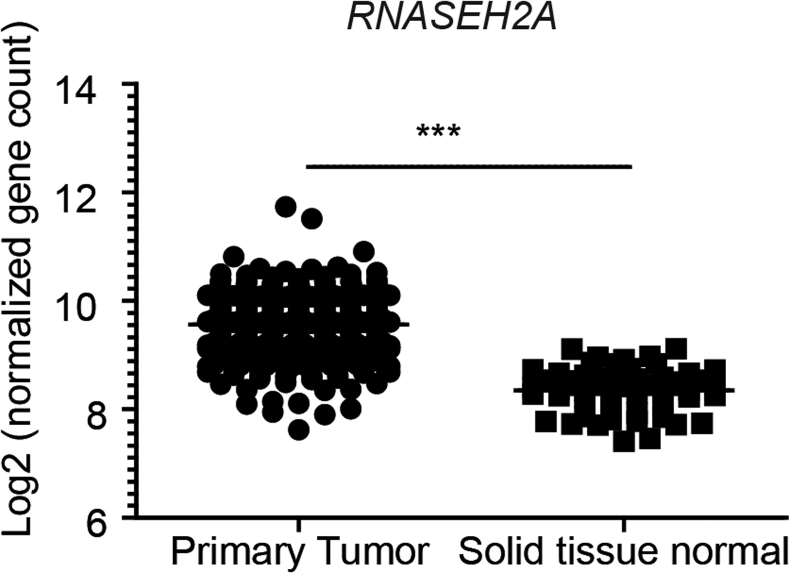
Next, we monitored RNase H2 expression in a cohort of patients with CRC (n = 155) and compared RNASEH2A transcript levels in paired tumor and adjacent normal tissues. Clinical data were retrieved from 122 patients and no significant correlation between RNASEH2A expression and disease subtype was observed (Supplementary Table 10). Expression of RNASEH2A was significantly increased in tumor samples vs adjacent normal colon tissue (Figure 7D). We independently validated tumor-specific overexpression of RNASEH2A in non-paired samples of the colorectal adenocarcinoma cohort (COADREAD; n = 467) of TCGA consortium40 (Supplementary Figure 14). We used publically accessible databases to address whether RNASEH2A expression affects the outcome of CRC. For this purpose, we analyzed RNA sequencing and clinical survival data from the COADREAD cohort using the SurvExpress web resource.29 Interestingly, low RNASEH2A tumor expression significantly correlated with poor overall survival compared with patients with high RNASEH2A tumor expression (Figure 7E).
Supplementary Figure 14.
In silico validation in COADREAD cohort. RNASEH2A expression in primary tumor (n = 380) or normal solid tissue (n = 51) based on normalized read counts from RNA sequencing (Illumina HiSeq). Data were retrieved from the COADREAD cohort of the TCGA and made publically available (www.xenabrowser.net). Significance was determined using Mann-Whitney test. ***P < .001.
Discussion
In this study, we investigated the role of RNase H2 in intestinal epithelial genome integrity and tumor formation by generating an intestinal epithelial-specific knockout mouse. Young H2bΔIEC mice displayed epithelial DNA damage and increased apoptosis, which manifested over time (1 year) into substantial inflammation and structural damage of the intestinal epithelium. Importantly, epithelial proliferation in aged H2bΔIEC mice was specifically impaired at the base of the crypts, with virtually no cycling intestinal stem cells, whereas proliferation (and accompanying apoptosis) in aged H2bΔIEC mice was largely restricted to the transit-amplifying cell region. This might indicate that fast cycling transit-amplifying cells react to chronic DNA damage predominantly by cell death, whereas intestinal stem cells appear to favor proliferative arrest as response to persistent DNA damage.
Using RNA sequencing we identified p53 as a key suppressor of epithelial proliferation at RNase H2 loss in IECs. Vice-versa proliferation in H2b/p53ΔIEC mice was excessively restored and associated with persistent DNA damage, which likely resulted in the accumulation of hazardous mutations and intestinal tumor development. Interestingly, the observation that H2b/p53ΔIEC mice predominantly develop small intestinal and only few colonic tumors is similar to Apc/Min+ mice, a classic murine model of human CRC. This model, although being largely reminiscent of human CRC with regard to β-catenin deregulation, also develops large numbers of small intestinal tumors compared with fewer colonic tumors.41 In contrast, adenomatous polyposis coli mutations are rarely associated with small intestinal tumors in humans, which indicates a potential species-specific difference between mice and humans in the propensity of their small intestinal epithelium to give rise to tumors.
H2b/p53ΔIEC tumors and, to a lesser extent, H2b/p53ΔIEC nontumor intestines contained large numbers of genomic ribonucleotides. Of note, incorporated ribonucleotides themselves cannot be regarded as a heritable mutation, because they will not be copied to the nascent strand in the following rounds of DNA replication. Therefore, it is likely that other mechanisms (eg, degree of epithelial proliferation, age-dependent decline of compensating repair mechanisms42) might affect the amount of potentially hazardous genome-embedded ribonucleotides in H2b/p53ΔIEC mice. Altogether, our findings suggest that genomic ribonucleotides are the products of dysfunctional RNase H2 that constitute the mutagenic DNA lesions and consequently drive tumorigenesis in this model.
To decipher the mutational consequences of genomic ribonucleotides, we used exome sequencing to establish a mutational signature specific for defective RNase H2 function. Mutational patterns from RNase H2-negative tissues (tumor, nontumor) invariantly contained distinct T>G transversions at GTG trinucleotides. T>G transversion has been described in mutational signature 3 (according to the COSMIC nomenclature), which has been linked to a failure of DNA double-strand break–repair through HR.
The high prevalence of signature 3 in H2b/p53ΔIEC tumors (and H2b/p53ΔIEC nontumor tissue for that matter) could indicate that an alternative DNA repair mechanism (eg, nonhomologous end joining) might occur at increased frequency alongside HR to allow RNase H2-deficient cells to cope with the high level of DNA damage. This would be consistent with the increased HR and synthetic lethality with HR deficiency observed in RNase H2-null human cells.43 Interestingly and in contrast to previous reports on the function of RNase H2 in yeast,6 our exome sequencing data did not show increased InDel frequency in H2b/p53ΔIEC tissue. Because specific 2- to 5-bp deletions observed in yeast resulted from positive selection in the experimental setting, discovering this specific mutation type is unlikely in the described in vivo setting of H2b/p53ΔIEC mice at a high frequency.
To assess the impact of RNase H2 on human tumorigenesis, we recombinantly expressed 5 recurrent somatic tumor variants of human RNase H2 and found that enzymatic activity and complex stability were decreased in all tested RNase H2 tumor variants, albeit to a varying degree. Although our data intriguingly point to a specific role for RNase H2 in malignant transformation of the intestinal epithelium, it must be noted that the degree of activity impairment varied across cancer mutations and even a significant decrease of enzymatic activity might not be equivalent to the total knockout of RNase H2 as described in our murine model. Therefore, further molecular delineation of the impact of loss-of-function mutations in RNase H2 on maintenance of genome integrity is needed. Moreover, we cannot differentiate in our murine model whether accumulating mutations and DNA damage in RNase H2-deficient IECs result from impaired RER or from a failure to resolve RNA:DNA hybrids (eg, R-loops) because the 2 processes are known to influence genome integrity. Given the sheer number of mis-incorporated ribonucleotides per replication round, the fact that RNase H2 mutations lead to poly(adenosine diphosphate ribose)polymerase trapping because of impaired RER43 and that impaired RER has been linked to skin cancer,44 it is tempting to speculate that defective RER plays a dominant role in our model.
In apparent contrast to this, we established that RNASEH2A expression was increased in CRC. This finding indicates that increased RNase H2 expression displays a physiologic adaption of cells with high proliferative turnover. This interpretation is substantiated by our finding that RNase H2B deficiency leads to a dramatic decrease of cellular proliferation and by the finding that rapidly proliferating tissues display significantly increased RNASEH2A expression levels.7 Therefore, increased RNASEH2A mRNA expression in tumor specimens from patients with CRC could represent a futile attempt of cells to maintain genomic integrity despite uncontrolled proliferation. It must be noted that we did not observe any significant association of tumor RNASEH2A expression with several clinical features of CRC (eg, microsatellite instability vs stability or histologic subtype) and a more thorough correlation analysis is clearly warranted in larger prospective studies. When stratifying according to RNASEH2A expression status in a survival analysis, patients with lower tumor RNASEH2A expression levels had a significantly shorter survival time, which is in line with previous findings from RNASEH2A expression in cervical cancer.45
Hence, although the exact impact of altered RNase H2 activity (eg, by loss-of-function mutations) during tumorigenesis of human CRC remains elusive, this seeming paradox could be well explained by the findings in our murine model. Therefore, our mouse model provides an excellent tool to decipher the potential roles of RNase H2 function (RER, R-loop resolution) and its therapeutic exploitation (increased poly[adenosine diphosphatase ribose]polymerase trapping in RNase H2-deficient tumor cells43) in intestinal carcinogenesis.
Acknowledgments
We gratefully appreciate the technical assistance of Maren Reffelmann, Dorina Oelsner, Tanja Klostermeier, Katharina Göbel, Karina Greve, Melanie Nebendahl, Sabine Kock, Tatjana Schmidtke, and Stefanie Baumgarten. We are deeply grateful for constructive and encouraging insight from Andrew P. Jackson.
Author contributions: Konrad Aden, Björn Rabe, and Philip Rosenstiel designed the study. Konrad Aden, Kareen Bartsch, Daniela Esser, Joseph Dahl, Felix Wottawa, Martin A.M. Reijns, Raheleh Sheibani-Tezerji, Anupam Sinha, Katharina Knittler, Adam Burkholder, Lina Welz, Johan van Es, Florian Tran, Simone Lipinski, Nassim Kakavand, Lennart Lenk, Christine Boeger, Ralph Lucius, Witigo von Schoenfels, Clemens Schafmayer, and Athena Chalaris performed experiments and analyzed the data. Hans Clevers, Christoph Röcken, Christoph Kaleta, Stefan Rose-John, Stefan Schreiber, Thomas Kunkel, Björn Rabe, and Philip Rosenstiel planned the project and supervised the experiment. Konrad Aden, Björn Rabe, and Philip Rosenstiel wrote the manuscript.
Footnotes
Conflicts of interest Authors disclose no conflicts.
Funding This work was supported by the DFG Excellence Cluster Inflammation at Interfaces (Philip Rosenstiel and Björn Rabe); the DFG Research Training Group 1743 (Philip Rosenstiel), the CRC877 B9 project (Philip Rosenstiel) and DFG project grant RA 2404/1-1 (Björn Rabe), and the DGIM Clinician Scientist Programme (Konrad Aden); the Medical Research Council (MRC, U127580972) attributed to (Martin Reijns) and CRC1182 C2 (Philip Rosenstiel).
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2018.09.047.
Supplementary Materials
Methods
cDNA Synthesis and Gene Expression Analysis
The mRNA isolation of cells washed in phosphate buffered saline (PBS), snap-frozen tissue, and PBS-washed Matrigel containing organoids was performed using the RNEasy Kit (Qiagen, Hilden, Germany). The cDNA synthesis was performed using RevertAid Premium cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada) according to the manufacturer’s protocol. Gene expression was subjected to the cDNA samples using SYBR Green quantitative reverse transcription polymerase chain reaction or TaqMan assays, which were purchased from Applied Biosystems (Foster City, CA). Reactions were carried out on the PRISM Sequence 7700 Detection System (Applied Biosystems), and relative transcript levels were determined using β-actin (TaqMan and SYBR Green) and glyceraldehyde 3-phosphate dehydrogenase (SYBR Green) as housekeepers. A list of all TaqMan probes and primers is below.
TaqMan Probes and Primers
| Gene name | Symbol | Species | TaqMan probe ID |
|---|---|---|---|
| Axin2 | Axin2 | murine | 00443610 |
| BCL2-associated X protein | Bax | murine | 00432051 |
| Bmi1 polycomb ring finger oncogene | Bmi1 | murine | 03053308 |
| Ccnd2 | Ccnd2 | murine | 00438070 |
| Cyclin D1 | Ccng1 | murine | 00432359 |
| Cd44 | Cd44 | murine | 01277161 |
| Cyclin-dependent kinase inhibitor 1A (P21) | cdkn1a | murine | 04205640 |
| CHGA | Chromogranin A | murine | 00154441 |
| HOP homeobox | Hopx | murine | 00558630 |
| leucine rich repeat containing G-protein–coupled receptor 5 | Lgr5 | murine | 00438905 |
| Leucine-rich repeats and immunoglobulin-like domains 1 | Lrig1 | murine | 00456116 |
| Lysozym | LysZ | murine | 00657323 |
| Transformed mouse 3T3 cell double minute 2 | Mdm2 | murine | 01233138 |
| Male-specific lethal 1 homolog | Msl1 | murine | 01303585 |
| Mucin 2 | MUC2 | murine | 00458299 |
| Myelocytomatosis oncogene | Myc | murine | 00487804 |
| Olfactomedin 4 | Olfm4 | murine | 00197437 |
| SPARC-related modular calcium binding 2 | Smoc2 | murine | 00491553 |
| SRY (sex determining region Y)-box 9 | Sox9 | murine | 00448840 |
| β-Actin | Actb | murine | 007393.1 |
Tamoxifen-Induced Knockout of RNaseH2 in RNaseH2ΔTam MEFs
MEFs derived from H2bfl/fl; ERT-Cre+ (RNase H2ΔTam) embryos were generated as previously described.1 Early-passage MEFs were cultivated in Dulbecco’s modified Eagle’s Medium (high glucose [4.5 g/L] with stable glutamine) supplemented with 10% fetal calf serum (v/v), 1% (v/v) penicillin, and 1% (v/v) streptomycin at 37°C. MEFs were held at 5% CO2 and physiologic O2 levels (3%). For conditional depletion of the RnaseH2b gene, H2bΔTam MEFs were treated for 3 days with 4-hydroxytamoxifen 100 nmol/L (Sigma-Aldrich, St Louis, MO; H7904, stock: 500 μmol/L in dimethyl sulfoxide). Three days after 4-hydroxytamoxifen treatment, the medium was changed and cells were further cultured until reaching senescence. MEFs derived from H2bfl/fl littermate embryos served as controls.
Assessment of Cellular Senescence
Senescence-associated activity of acidic β-galactosidase was measured using the Senescence Detection Kit (BioVision, Milpitas, CA) according to the manufacturer’s instructions. Lack of proliferation in senescent cells was assessed by 5-ethynyl-2′-deoxyuridine (EdU) incorporation into replicating DNA. To that end, 10 days after 4-hydroxytamoxifen treatment, RNase H2ΔTam MEFs were subjected to a 4-hour EdU pulse and EdU content of genomic DNA was quantified with the Click-iT EdU Flow Cytometry Assay Kit (Invitrogen, Waltham, MA). Stained cells were analyzed using a FACS Canto flow cytometer and FlowJo 10 (BD Biosciences, Heidelberg, Germany). To assess cytokine concentration in cell supernatants, MEFs were plated in 6 wells and incubated with fresh medium 600 μL for 24 hours. Removed cell-free supernatant was analyzed for murine interleukin-6 and C-X-C motif chemokine 10 by enzyme-linked immunosorbent assay according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN). For quantification of enzyme-linked immunosorbent assay data, the amount of secreted protein was normalized to the number of proliferating and senescent cells.
Transmission Electron Microscopy
For electron microscopy, tissue was fixed at 4°C with 3% glutaraldehyde, washed with PBS, exposed to 2% osmium tetroxide for 30 minutes, dehydrated in a series of increasing ethanol concentrations, and embedded in Araldite (Huntsman Advanced Materials GmbH, Basel, Switzerland). Ultrathin (60-nm) sections were cut, mounted on carbon-coated copper grids (Science Service GmbH, München, Germany), and contrasted with a saturated solution of uranyl acetate (Merck KGaA, Darmstadt, Germany) in H2O. The grids were examined with a JEOL (Freising, Germany) 1400 plus transmission electron microscope at 120-kV operating voltage.
Immunohistochemistry and Immunofluorescence
For immunohistochemical staining, 5-μm sections of paraffin-embedded colon–ileum Swiss rolls were deparaffinized with Xylol substitute (Carl Roth, Karlsruhe, Germany), incubated in citrate buffer for 3 minutes, and subsequently blocked with blocking serum (Vectastain; Vector Laboratories, Burlingame, CA) for 20 minutes.
For anti-bromodeoxyuridine staining, mice were pulsed with bromodeoxyuridine 10 mg/kg bodyweight 1.5 hours before sacrifice. Primary bromodeoxyuridine (BD Pharmingen, San Diego, CA) was incubated overnight. Antibodies used were mouse anti-Ki67 (1:100 dilution; Novocastra, Sheffield, UK), rabbit anti-SOX9 (1:600 dilution; Chemicon, Temecula, CA), mouse anti-Tcf4 (1:250 dilution; Millipore, Beverly, MA), and β-catenin (BD Transduction, BD Biosciences). Incubation of antibodies was performed overnight in bovine serum albumin in PBS at 4°C for antibodies directed against bromodeoxyuridine and Tcf4 and for 1 hour at room temperature for antibodies directed against Ki67, β-catenin, SOX9, and lysozyme. In all cases, the Envision+ kit (Dako, Agilent, Santa Clara, CA) was used as a secondary reagent. Stainings were developed with 3,3′-diaminobenzidine. Slides were counterstained with hematoxylin and mounted. Sections were washed and incubated with secondary antibodies and 3,3′-diaminobenzidine substrate (Vectastain ABC Kit). For periodic acid–Schiff staining, slides were dehydrated, rinsed for 5 minutes with aqua, and 0.5% periodic acid, respectively, and incubated with Schiff reagent for 1 minute. Schiff reagent was rinsed off for 5 minutes and slides were counterstained with hematoxylin. For the terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling assay, slides were subjected to the Apop Tag Plus Peroxidase In Situ Apoptosis Detection Kit (Merck Millipore) according to the manufacturer’s protocol. Slides were visualized by an AxioImager Z1 microscope (Zeiss, Jena, Germany). Pictures were captured by a digital camera system (AxioCam HrC/HrM; Zeiss). Measurements were made using semiautomated image analysis software (AxioVision 08/2013).
In Situ Hybridization
Freshly isolated intestines were flushed with formalin (4% formaldehyde in PBS) and fixed by incubation in a 10-fold excess of formalin overnight at room temperature. The formalin was removed and the intestines washed twice in PBS at room temperature. Then, the intestines were transferred to a tissue cassette and dehydrated by serial immersion in 20-fold volumes of 70%, 96%, and 100% EtOH for 2 hours, each at 4°C. Excess ethanol was removed by incubation in xylene for 1.5 hours at room temperature and then the cassettes immersed in liquid paraffin (56°C) overnight. Paraffin blocks were prepared using standard methods. For in situ hybridization, 8-μm-thick sections were rehydrated as described earlier. Afterward, the sections were treated with sodium chloride 0.2 mol/L and proteinase K. Slides were post-fixed, and then sections were demethylated with acetic anhydride and pre-hybridized. Hybridization was performed in a humid chamber with freshly prepared digoxigenin-labeled RNA probe 500 ng/mL of Olfm4 (image clone 1078130) Sections were incubated for at least 48 hours at 68°C. The slides were washed and incubation of the secondary anti-digoxigenin antibody (Hoffman-La Roche, Basel, Switzerland) was performed at 4°C overnight. The next day, sections were washed and developed using nitro blue tetrazolium chloride/5-brom-4-chlor-3-indolyl-phosphat. Slides were visualized by an AxioImager Z1 microscope (Zeiss). Images were captured by a digital camera system (Axiocam HrC/HrM; Zeiss). Measurements were made using semiautomated image analysis software (Axiovision 08/2013).
Antibodies and Reagents
Antibodies targeting γH2A.X (catalog number 2577; Cell Signaling Technology, Danvers, MA), murine RNase H2 holo enzyme,2 p53 (NCL-p53-CM5p; Leica Biosystems, Buffalo Grove, IL), and β-Actin (catalog number 4967; Cell Signaling Technology) were used for immunoblot assay. Antibodies used for immunohistochemistry are listed separately in the Immunohistochemistry and Immunofluorescence section.
Immunoblot Analysis
Cells were lysed using sodium dodecylsulfate–based DLB buffer plus 1% Halt Protease inhibitor cocktail (ThermoFischer Scientific, Steineich, Germany) before heating at 95°C for 5 minutes and followed by ultra-sonication for 5 seconds twice. To remove cell remnants, lysates were centrifuged at 16,000g for 15 minutes at 4°C. For protein extraction of organoids, Matrigel was removed by several centrifugation steps at 4°C followed by lysis as described earlier. Afterward, equal amounts of lysates containing Laemmle buffer were heated at 95°C and electrophoresed on 12% polyacrylamide gels under standard sodium dodecylsulfate polyacrylamide gel electrophoresis conditions before being transferred onto a polyvinylidene fluoride membranes (GE Healthcare, Freiburg, Germany). Protein-loaded membranes were blocked with 5% milk in tris buffered saline and Tween 20 and incubated with primary antibody overnight and with horseradish peroxidase–conjugated secondary antibody for 1 hour at the indicated concentrations. Proteins were detected using the Amersham ECL Prime Western Blot Detection Reagent (GE Healthcare).
Cultivation of Intestinal Organoids
Mouse intestinal organoids were cultivated as described previously.3 In brief, the small intestine was removed and cleared of intestinal content by flushing the intestine with Hank’s Balanced Saline Solution (without calcium and magnesium). After removal of residual fat and Peyer patches, the intestine was cut longitudinally and then laterally in pieces of 0.5 cm. Intestinal pieces were incubated in ice-cold PBS plus EDTA 10 nmol/L for 10 minutes for 4 times, with intermittent vigorous shaking and replacement with fresh PBS and EDTA 10 nmol/L after every shaking process. The crypt suspension was strained through a 1000μm strainer, followed by a spin at 1200 rpm at +4°C. Pure epithelial crypts were resuspended in BD Matrigel (BD Bioscience, Heidelberg, Germany) to a concentration of 5–10 crypts per 1 μL of Matrigel, embedded in 24-well plates, and cultivated in intestinal stem cell medium (IntestiCult Organoid Growth Medium [Mouse]; STEMCELL Technologies, Cambridge, MA) based on previously described organoid medium containing murine epidermal growth factor, murine Noggin, and human R-spondin 1.3 The medium was changed every other day and organoids were stimulated after 7 days of cultivation. For murine colon organoids, pure epithelial crypts were suspended in BD Matrigel (BD Bioscience) to a concentration of 5–10 crypts per 1 μL of Matrigel, embedded in 24-well plates, and cultivated in L-WRN conditioned medium (L-WRN cells were kindly provided by Markus Tschurtschenthaler, Munich) in the presence or absence of anoikis inhibitor Y-27632. The medium was changed every day or every other day and passage was performed every 3–5 days. For the colon colony formation assay, 10,000 cells were seeded in Matrigel 40 μL and the number of grown colonies was assessed at day 4 and day 10 after seeding.
Transcriptome Analysis
Total RNA samples from isolated intestinal epithelial crypts of H2bfl/fl (n = 4) and H2bΔIEC (n = 4) or whole small intestinal pieces from nontumorous mucosa H2bfl/fl (n = 4) and p53/H2bΔIEC (n = 4), or small intestinal tumor tissue from p53/H2bΔIEC (n = 4) mice were sequenced on Illumina HiSeq4000 using Illumina stranded TruSeq protocol (GEO Project Accession number GSE111318). An average of ∼40 million 75-nt paired-end reads was sequenced for each sample. Raw reads were preprocessed using cutadapt4 to remove adapter and low-quality sequences and then aligned to the mm10 (GrCm38) reference genome with the Ensemble gene annotation using TopHat2.5 Gene expression values of the transcripts were computed by HTSeq.6 Differential gene expression levels were analyzed and visualized by the Bioconductor package DESeq2.7 To interpret the biological significance of the differentially expressed genes (up- and down-regulated), Gene Ontology enrichment analysis was performed to investigate their functional distribution using the InnateDB database (www.innatedb.com).8 Transcription factor binding sites that were over-represented in the promoter region of differentially expressed genes (up- and down-regulated) were identified by innateDB,8 integrating predicted transcription factor binding site data from the CisRED database (www.cisred.org).9
Method for Statistical Treatment of End Density for 8- vs 52-Week-Old Mice
Fluorescence intensity as a function of migration distance was determined for each SybrGold-stained alkali-treated DNA sample using ImageQuant. These results were blank subtracted and fragment length was determined with 1-kb and 100-bp ladders as standards. The median fragment length for each sample was calculated and then averaged for isogenic samples. IEC samples were normalized to the isogenic paired fl/fl sample (un-induced; ie, nominally wild type for Rnase H2) to account for the background of alkali-sensitive sites that are due to IEC H2b or H2b/p53 knockout. Error bars represent compounded standard deviations of the normalized end densities.
RNase H2 Purification and Activity Assay
Recombinant RNase H2 was expressed in Rosetta-2 cells using a polycistronic construct (pMAR22) based on pGEX6P1 and purified by affinity purification as previously described.10 Site-directed mutagenesis to introduce the following mutations was performed using the Quikchange method: RNASEH2A-V133M, P158S and R280G, RNASEH2b-T280A, and RNASEH2C-R145C. Substrate was formed by annealing a 3′-fluorescein–labeled oligonucleotide (GATCTGAGCCTGGGaGCT; uppercase, DNA; lowercase, RNA) to a complementary 5′ DABCYL-labeled DNA oligonucleotide (Eurogentec, Liege, Belgium) by heating for 5 minutes at 95°C followed by slow cooling to room temperature.10 To measure enzyme activity, purified recombinant RNase H2 (concentration range, 0.001–1 nmol/L) was incubated with substrate 250 nmol/L in 100-μL reactions (KCl 60 mmol/L, Tris-HCl [pH 8] 50 mmol/L, MgCl2 10 mmol/L, 0.01% bovine serum albumin, and 0.01% Triton X-100) at 24 ± 2°C for 90 minutes in 96-well flat-bottomed plates. Fluorescence was read every 5 minutes for 100 ms using a VICTOR2 1420 multi-label counter (PerkinElmer, Waltham, MA), with a 480-nm excitation filter and a 535-nm emission filter. Relative activities were determined by comparing the slopes of the initial linear substrate conversion rates for wild-type and mutant RNase H2.
Thermofluor Protein Stability Assay
Thermal stability assays were performed using the fluorescence-based thermal shift (Thermofluor) method11 as previously described,10 with minor modifications. Proteins were diluted in 50-μL volumes to 2 μmol/L in Tris-HCl (pH 7.5) 50 mmol/L, NaCl 150 mmol/L, EDTA 1 mmol/L, dithiothreitol 1 mmol/L, and 5× SYPRO Orange (Invitrogen, Waltham, MA). Assays were carried out in a Bio-Rad iQ5 rtPCR Thermocycler (Bio-Rad, Hercules, CA) by heating the samples from 20°C to 80°C at 0.5°C increments and holding each temperature for 30 seconds. Fluorescence intensity was measured in relative fluorescence units (RFUs) using 485-nm excitation and 575-nm emission wavelengths. Thermal denaturation graphs were plotted as a function of the gradient of protein unfolding [d(RFU)/dT] against the temperature gradient, with the melting temperature defined as the temperature at the maximum of d(RFU)/dT.
Induction of Colitis, Determination of Clinical Scores, and Histology
For acute colitis induction, mice were supplied with 2% of DSS (molecular mass, 40 kDa; TdB Consultancy, Uppsala, Sweden) dissolved in drinking water for 5 days followed by 5 days of regular drinking water. The Disease Activity Index was obtained as described previously.12 Consumption of drinking water was measured daily. For induction of chronic DSS colitis, mice were supplied with 1% DSS for 3 cycles of 7 days followed by 14 days of regular drinking water. Histopathologic and analyses were performed according to standard methods. Histologic scoring was performed in blinded fashion by 2 independent observers and displays a combination of inflammatory cell infiltration, epithelial cell damage, and transmural inflammation, as described elsewhere.13
Online Database Expression Analysis
A publically available web-based tool (https://xenabrowser.net/) was used to (1) assess tissue (tumor vs solid normal)-specific expression of RNASEH2A and (2) for association analysis of RNASEH2A expression in CRC tumor samples with clinical and molecular denominators of CRC in subtypes in the COADREAD cohort, as described previously.14 Tumor RNASEH2A expression was compared in non-paired tumor vs solid normal tissue or was associated with (1) microsatellite instability, (2) histologic cancer type (rectal adenocarcinoma, rectal mucinous adenocarcinoma, colon adenocarcinoma, colon mucinous adenocarcinoma), (3) loss of mismatch repair protein expression (MLH1, MSH2, MSH6, PMS2), as defined by immunohistochemistry, and (4) pathologic TNM stage. Statistical significance was determined using 1-way analysis of variance.
References
- 1.Arnold M., Sierra M.S., Laversanne M. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Melo FdSe, Kurtova A.V., Harnoss J.M. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 4.Tomasetti C., Durrett R., Kimmel M. Role of stem-cell divisions in cancer risk. Nature. 2017;548:E13. doi: 10.1038/nature23302. [DOI] [PubMed] [Google Scholar]
- 5.Tomasetti C., Li L., Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355:1330–1334. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McElhinny S.A.N., Kumar D., Clark A.B. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reijns Martin A.M., Rabe B., Rigby Rachel E. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reijns Martin A.M., Jackson Andrew P. Ribonuclease H2 in health and disease. Biochem Soc Trans. 2014;42:717–725. doi: 10.1042/BST20140079. [DOI] [PubMed] [Google Scholar]
- 9.Liu B., Hu J., Wang J. Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J Biol Chem. 2017;292:4777–4788. doi: 10.1074/jbc.M116.758599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelio D.A., Sedam H.N.C., Ferrarezi J.A. Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability. DNA Repair. 2017;52:110–114. doi: 10.1016/j.dnarep.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucherlapati M., Yang K., Kuraguchi M. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci U S A. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groh M., Gromak N. Out of Balance: R-loops in Human Disease. PLoS Genet. 2014;10:e1004630. doi: 10.1371/journal.pgen.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiller B., Achleitner M., Glage S. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes S.A., Bindal N., Bamford S. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beyer U., Brand F., Martens H. Rare ADAR and RNASEH2B variants and a type I interferon signature in glioma and prostate carcinoma risk and tumorigenesis. Acta Neuropathol. 2017;134:905–922. doi: 10.1007/s00401-017-1774-y. [DOI] [PubMed] [Google Scholar]
- 16.Adolph T.E., Tomczak M.F., Niederreiter L. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D., Pertea G., Trapnell C. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna A., Hanna M., Banks E. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry S.T., Ward M.H., Kholodov M. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Handsaker B., Wysoker A. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blokzijl F., Janssen R., Van Boxtel R. MutationalPatterns: an integrative R package for studying patterns in base substitution catalogues. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breuer K., Foroushani A.K., Laird M.R. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen L.J., Kuhn M., Stark M. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csardi G., Nepusz T. The igraph software package for complex network research. https://www.researchgate.net/profile/Gabor_Csardi/publication/221995787_The_Igraph_Software_Package_for_Complex_Network_Research/links/0c96051d301a30f265000000/The-Igraph-Software-Package-for-Complex-Network-Research.pdf Available at:
- 29.Aguirre-Gamboa R., Gomez-Rueda H., Martínez-Ledesma E. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartsch K., Knittler K., Borowski C. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet. 2017;26:3960–3972. doi: 10.1093/hmg/ddx283. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D., Franceschini A., Wyder S. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwitalla S., Ziegler Paul K., Horst D. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 2013;23:93–106. doi: 10.1016/j.ccr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Begus-Nahrmann Y., Lechel A., Obenauf A.C. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet. 2009;41:1138. doi: 10.1038/ng.426. [DOI] [PubMed] [Google Scholar]
- 34.Kim N., Huang S.N., Williams J.S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrov L.B., Nik-Zainal S., Wedge D.C. Signatures of mutational processes in human cancer. Nature. 2013;500:415. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbes S.A., Bhamra G., Bamford S. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;57 doi: 10.1002/0471142905.hg1011s57. 10.11.1–10.11.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandrov L.B., Stratton M.R. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong F.L., Mamaï O., Sborgi L. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell. 2016;167:187–202.e17. doi: 10.1016/j.cell.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Zhai Z., Liu W., Kaur M. NLRP1 promotes tumor growth by enhancing inflammasome activation and suppressing apoptosis in metastatic melanoma. Oncogene. 2017;36:3820. doi: 10.1038/onc.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moser A., Pitot H., Dove W. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 42.Vaidya A., Mao Z., Tian X. Knock-in reporter mice demonstrate that DNA repair by non-homologous end joining declines with age. PLoS Genet. 2014;10:e1004511. doi: 10.1371/journal.pgen.1004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmermann M., Murina O., Reijns M.A.M. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 2018;559:285–289. doi: 10.1038/s41586-018-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiller B., Hoppe A., Haase C. Ribonucleotide excision repair is essential to prevent squamous cell carcinoma of the skin. Cancer Res. 2018;78:5917–5926. doi: 10.1158/0008-5472.CAN-18-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Tian R., Gao H. Identification of significant gene signatures and prognostic biomarkers for patients with cervical cancer by integrated bioinformatic methods. Technol Cancer Res Treat. 2018;17 doi: 10.1177/1533033818767455. 1533033818767455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Bartsch K., Knittler K., Borowski C. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet. 2017;26:3960–3972. doi: 10.1093/hmg/ddx283. [DOI] [PubMed] [Google Scholar]
- 2.Reijns M.A., Rabe B., Rigby R.E. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T., Vries R.G., Snippert H.J. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 4.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17 [Google Scholar]
- 5.Trapnell C., Roberts A., Goff L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breuer K., Foroushani A.K., Laird M.R. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson G., Bilenky M., Lin K. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;34:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reijns M.A., Bubeck D., Gibson L.C. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J Biol Chem. 2011;286:10530–10539. doi: 10.1074/jbc.M110.177394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nettleship J.E., Brown J., Groves M.R. Methods for protein characterization by mass spectrometry, thermal shift (ThermoFluor) assay, and multiangle or static light scattering. Methods Mol Biol. 2008;426:299–318. doi: 10.1007/978-1-60327-058-8_19. [DOI] [PubMed] [Google Scholar]
- 12.Siegmund B., Lehr H.-A., Fantuzzi G. IL-1β–converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci U S A. 2001;98:13249–13254. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adolph T.E., Tomczak M.F., Niederreiter L. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehrer S., Rheinstein P.H., Rosenzweig K.E. Glioblastoma multiforme: fewer tumor copy number segments of the SGK1 gene are associated with poorer survival. Cancer Genomics Proteomics. 2018;15:273–278. doi: 10.21873/cgp.20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.