Abstract
Quantitative assay of microRNAs (miRNAs) with mass spectrometric detection currently suffers from two major disadvantages, i.e. being insufficient in sensitivity and requiring an extraction or chromatographic separation prior to MS detection. In this work, we developed a facile and sensitive assay of targeted miRNAs based on the combination of cyclic enzymatic amplification (CEA) with microfluidic voltage-assisted liquid desorption electrospray ionization-tandem mass spectrometry (VAL-DESI-MS/MS). The ssDNA probe was designed to have a sequence complementary to the miRNA target with an extension of a 2-base nucleotide fragment (i.e. CpC) at 3’-position as MS signal reporter, thus being easy-to-prepare and high in stability. In the proposed CEA-VAL-DESI-MS/MS assay, a ssDNA probe was added to a sample solution, forming a DNA-miRNA hybrid. Duplex-specific nuclease (DSN) was then added to cleave specifically the DNA probe in the heteroduplex strands. As the hybridization -cleavage cycle repeated itself for many rounds, a large quantity of CpC molecules was produced that was quantified by VAL-DESI-MS/MS with accuracy and specificity. miRNA-21 was tested as the model target. The assay had a linear calibration equation in the range from 2.5 pM to 1.0 nM with a limit of detection of 0.25 pM. Determination of miRNA-21 in cellular samples was demonstrated. miRNA-21 was found to be 95.3 ± 13.95 attomoles (n=3) in 100 mouse peritoneal macrophages with a recovery of 94.2 ± 2.6% (n=3). Interestingly, analysis of exosomes secreted from these cells revealed that exposure of the cells to chemical stimuli caused a 3-fold increase in exosomal level of miRNA-21. The results suggest the proposed assay may provide an accurate and cost-effective means for quantification of targeted miRNAs in biomedical samples.
Keywords: microRNA assay, cellular samples, exosomes, isothermal amplification, mass spectrometry, desorption electrospray ionization
Graphic Abstract
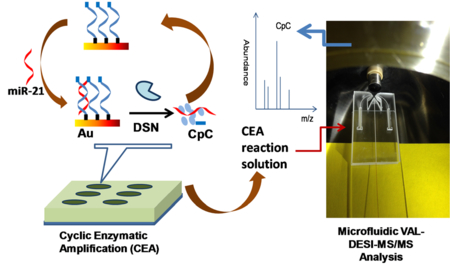
miRNA Quantification by coupling cyclic enzymatic amplification with microfluidic liquid desorption ESI-MS/MS
MicroRNAs (miRNAs) are small noncoding single-stranded RNAs and often about 22 nucleotides in length. They are known to regulate gene expression that is responsible for diverse cellular processes such as cell proliferation, migration, and death.1–3 Detection of miRNAs in biological specimens is highly significant for understanding their biological functions, early diagnosis of diseases, and discovery of new drug targets for therapeutic treatments. Up to date, analytical methods based on various techniques have been developed for miRNAs analysis. Among them, the golden standards are northern blotting,4,5 quantitative reverse transcription polymerase chain reaction (RT-qPCR),6–9 and oligonucleotide microarrays.10–12 It’s well documented, however, that methods based on northern blotting and microarrays lack the assay sensitivity needed for detection of miRNAs at low levels. Although RT-qPCR methods are highly sensitive, they normally require a delicate and tedious procedure for total RNA isolation prior to analysis. Further, due to the nature of their short strands, some miRNAs can be easily lost during this sample pretreatment procedure.
Over the past years isothermal signal amplification-based methods have been developed for miRNA analysis.13–19 In these methods no nucleic acid amplification was involved. Instead, signal amplification by a factor of 103 ~ 105 was achieved by enzyme assisted miRNA target recycling. Procedures involving various enzymes such as exonuclease (Exo)20 and duplex-specific nuclease (DSN)21 were reported. Diverse detection schemes, including chemiluminescence,22–24 fluorescence,25–28 surface-enhanced Raman spectroscopy,29–30 electrochemical,31–35 and magnetic relaxation switch36 or optomagnetic37 were deployed. Methods with fluorescence or electrochemical detection were most frequently reported. These methods require to use labeled DNA probes which are difficult to prepare and in many cases poor in stability, making the assay very costly. Moreover, as well known, fluorescence or electrochemical signals are prone to interference from biological sample matrices, producing inaccurate results. Thus, a careful isolation of miRNA targets from sample matrix is sometimes needed.
Mass spectrometry (MS) is a powerful technique for chemical characterization and measurement. A significant advantage of this detection technique lies in its applicability to a broad range of molecular classes and sizes with sensitivity and specificity.38 A few studies on miRNA assay by using mass spectrometry were recently reported. Liquid chromatography-high resolution mass spectrometric methods were developed for identification of miRNAs present in human cellular samples.39–40 These methods were useful for systematically studying miRNA modifications such as 3’-terminal variants. In other studies, different MS techniques such as MALDI and ion mobility MS were used to analyze miRNAs.41–44 In order to improve assay sensitivity, methods involving the use of DNA probes labeled with peptides45 or metal elements46 were investigated. Nevertheless, without appropriate sample pretreatments majority of these methods had difficulties to quantify miRNAs in biological samples due to either an insufficient assay sensitivity or interference from sample matrices.
In this work, we aimed to develop a label free, cost-effective, and highly sensitive mass spectrometric method for quantification of miRNAs in biological samples. The proposed assay strategy involves coupling duplex-specific nuclease (DSN)-assisted cyclic enzymatic amplification (CEA) with microfluidic voltage-assisted liquid desorption electrospray ionization-tandem mass spectrometry (VAL-DESI-MS/MS) that we recently developed.47 CEA generates a large quantity of miRNA reporter molecules, ensuring a high assay sensitivity. VAL-DESI-MS/MS allows direct analysis of CEA reaction solutions with accuracy and specificity. A label-free and easy-to-prepare DNA probe was synthesized and evaluated for the assay. Optimal CEA conditions were investigated to obtain maximum signal amplification taking miRNA-21 as a model target. Applicability of the proposed CEA-VAL-DESI-MS/MS method for quantitative assay of miRNA-21 in cellular samples was demonstrated. Finally, the assay was applied to study chemical stimulation-induced changes of intra-exosomal miRNA-21 level secreted from mouse peritoneal microphages.
EXPERIMENTAL SECTION
Materials.
miRNAs and ssDNA probes were customer synthesized by Sigma-Aldrich (St. Louis, MO) and received as lyophilized pellets in microcentrifuge tubes. The pellets were centrifuged to ensure that no residue remained on the walls or cap and were then suspended in H2O. The sequences are shown in Table 1. Duplex-specific nuclease (DSN) was purchased from Evrogen. According to the user manual, the pellet was dissolved in the DSN storage buffer provided (5 μL of buffer for each viral of 10 U DSN), gently vortexed, and incubated at 25 °C for 5 min. An equal volume of glycerol was added before the tube was gently vortexed and stored at −20 °C.
Table 1.
miRNA and ssDNA Sequences used in this work
| miRNA-21 | UAG CUU AUC AGA CUG AUG UUG A |
| miRNA single-base mismatched | UAG CUU AUC AGA CUG AUA UUG A |
| miRNA 3-base mismatched | UAG CUU AUC AGA CUA CGG UUG A |
| ssDNA probe | 5’-thiol-C6-TCAACATCAGTCTGATAAGCTACC-3’ |
Other materials used in this work, experimental procedures used for culturing and chemically stimulating mouse peritoneal microphages,48–49 isolation of exosomes from cell culture media for quantification of intra-exosomal miRNA-21 are described in supporting information.
Preparation of microtiter plates for cyclic enzymatic amplification.
Microtiter plates were in-house fabricated by bonding a PDMS sheet with holes to a glass base. The microwells had a capacity of ca. 100μL. At the bottom of each microwell a gold coating was deposited using a 108 auto sputter coater (Ted Pella, Inc.) under a vacuum pressure of 50 mTorr. The resultant gold film was about 50 nm in thickness. Before performing CEA reactions, microwells were pretreated as following: 1) thiolated DNA probe was treated with 0.10 M DTT for 20 min; 2) aliquots (25 μL) of DTT treated DNA probe solution was transferred into microwells and incubated for 5 h at room temperature; 3) after a thorough washing with water, the microwells were further incubated with 1.0 mg /mL thioglycolic acid for 30min; and then washed twice with TMD buffer.
CEA-VAL-DESI-MS/MS Assay of miRNA-21.
miRNA-containing samples (25 μL portions) were added to respective microwells. TMD buffer (50 μL) and DSN solution (1.0 μL) were added to each microwell. The mixture was incubated for 120 min at 50 °C on a shaker. A portion of the incubation solution (25 μL) was transferred into the sample reservoir on the microfluidic VAL-DESI-MS/MS platform for MS analysis.
Quantification of the signal reporter of miRNA target (i.e. CpC) was achieved by using the microfluidic VAL-DESI-MS/MS method we recently reported.47 The MS system consisted of an ion trap mass spectrometer (LCQ Deca, ThermoFinnigan, San Jose, CA), a multi-channel power supply, and a syringe pump. Xcalibur software (ThermoFinnigan) was used to acquire and to process MS data. The MS detection conditions were optimized in positive mode as follows: capillary temperature, 250°C; ion source voltage, 0V; relative collision energy, 20-30%; isolation width, 1.0 u; and activation time, 30ms. VAL-DESI conditions: ESI voltage, +4kV; sampling voltage, +75V; electrospray solvent flow rate, 250nL /min.
RESULTS AND DISCUSSION
Working Principle of the proposed miRNA Assay.
Mass spectrometry is recognized as one of the most powerful analytical tools. However, only a few MS-based methods have been reported so far for analysis of miRNAs39–46. Further, these methods have limitations associated with either insufficient assay sensitivity or signal suppression by sample matrix. In this work, a novel assay strategy based on coupling cyclic enzymatic amplification with voltage-assisted liquid desorption electrospray ionization-tandem mass spectrometry was developed to solve these problems. The strategy is illustrated by Figure 1. The experimental workflow consists of 3 steps: (1) immobilization of the DNA probe; (2) isothermal CEA based on cyclic miRNA target /DNA probe hybridization – DSN cleavage; and (3) VAL-DESI-MS/MS analysis of the CEA reaction solution.
Figure 1.
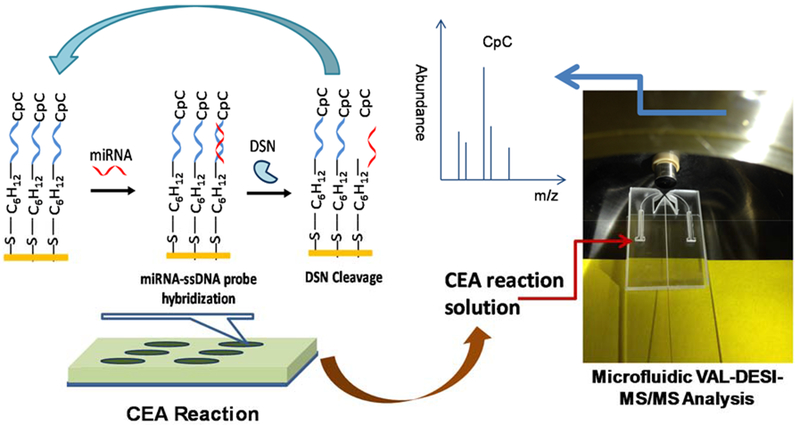
Schematic illustration of the proposed miRNA assay: ssDNA probe is immobilized on gold-coated bottom of microwells; miRNA target is introduced for hybridization; DSN is added to cleave the DNA probe hybridized, producing CpC (the signal reporter of miRNA target) and releasing the miRNA target for next round of hybridization-cleavage; and the CEA reaction solution is analyzed by VAL-DESI-MS/MS to quantify CpC.
A CEA reaction can be carried out either in a homogeneous solution using a free DNA probe or in a heterogeneous environment using a DNA probe immobilized on a solid surface. Our preliminary study showed that with using immobilized DNA probes a more effective CEA (in terms of the quantity of reporter molecules produced by unit amount of miRNA target) could be obtained. This might be because a more favorable 3-dimensional structure and a higher microenvironment concentration of DNA probes could be obtained for the CEA reaction when the probes were immobilized. A thiolated DNA probe was, therefore, utilized. The probe could be readily immobilized on the gold coated bottom of microwells via Au-S bond. For assaying miRNA-21 (5’-UAG CUU AUC AGA CUG AUG UUG A-3’) selected as the model miRNA target in this study, a 24-nucleotide oligomer (5’-SH-C6-TCA ACA TCA GTC TGA TAA GCT ACC-3’) was synthesized and used as the DNA probe. It’s worth noting that synthesis of this probe is very easy compared with the synthesis of those with a fluorescent or electrochemical label. The probe also has a better stability than those fluorescently or electrochemically labeled probes. Various enzymes such as Exo III, T7 Exo, and DSN have been used in CEA of miRNA.13 Duplex-specific DSN selectively cleaves DNA strands in DNA-DNA and DNA-RNA duplexes with little activity toward single-stranded nucleic acids and RNA-RNA duplexes.21 In the proposed assay, DSN cleaves the DNA probes in DNA probe-miRNA-21 duplex strands. The cleavage releases a nucleotide fragment (i.e. CpC, the MS signal reporter) and miRNA-21 that initiates a next round of hybridization, cleavage and release of CpC, thus achieving an isothermal signal amplification.
CpC, a 2-nucleotide fragment at 3’-terminal of the DNA probe was for the first time evaluated as the MS signal reporter for miRNA assay in this work. An advantage of using CpC as a reporter lies in that it makes the miRNA assay label free. CpC can be effectively released as one unit by DSN enzyme cleavage from the DNA probe/miRNA duplex strand because it extrudes from the duplex strand. Further, CpC can be sensitively detected by VAL-DESI-MS/MS. Figure 2 shows the MS2 spectrum of CpC (ion m/z 597.3) obtained from a CEA reaction solution containing 0.2nM miRNA-21, confirming the identity of CpC. The fragment pathways, producing major ions m/z 579.3, m/z 306.0, and m/z 485.0 have been elucidated and illustrated by the inset. In further experiments, ion transition m/z 597 →579 was monitored for quantification of CpC. It was noted that CpC in a CEA reaction solution was stable for at least 24 h at 4°C.
Figure 2.
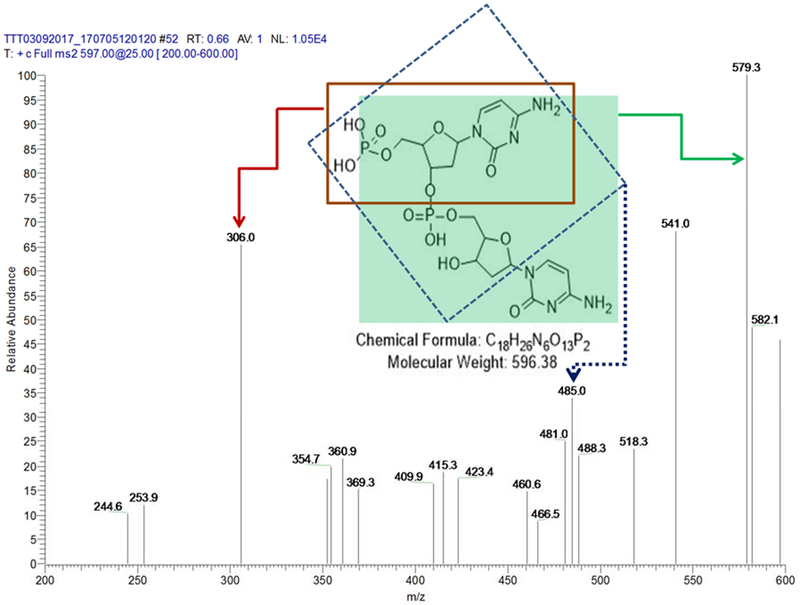
MS/MS Spectrum of CpC ([M+H]+ m/z 597) obtained from a CEA reaction solution of 0.2 nM miRNA-21, indicating that CpC can be effectively released from the DNA probe by DSN cleavage and sensitively detected by VAL-DESI-MS/MS as the MS signal reporter of miRNA-21.
Study of Experimental Conditions for the Assay.
To achieve an effective CEA, effects of reaction conditions were investigated. A 1.0 nM standard miRNA-21 solution prepared in water was assayed as the test sample in these experiments. After CEA incubation, CpC concentrations in the reaction solutions were measured to assess the CEA efficiency. DNA probe concentration in the solution used for coating the bottom of microwells was studied in the range from 0.5 to 10 μM. The results are shown in Figure S2A. As shown, DNA probe concentration ranging from 5.0 to 7.0 μM produced similar results. Using a solution at a higher concentration range increases the probe density on the gold layer surface meanwhile it causes a rise in steric hindrance to DNA probe-miRNA-21 hybridization and DSN cleavage. Therefore, a 6.0 μM DNA probe solution was used for further tests. CEA incubation time strongly affected the CEA results as expected. As the incubation time increased, CpC concentration in the CEA reaction solution increased until becoming constant after 180 min when all DNA probes were consumed. It should be pointed out that these results were obtained when miRNA-21 concentration was at 1.0 nM which was relatively high compared with those in real clinical samples. An incubation time of 120 min was selected for further tests. Effects of DSN amount used in CEA reaction were also investigated. The results are shown in Figure S2B. In 75 μL CEA reaction solution 0.8 U DSN was enough to produce reproducible results at an optimal temperature of 50°C. It was noted that DSN enzyme was stable at a wide range of pH (from 4 to 12) at 50°C in the presence of 5 mM Mg2+. However, DSN cleavage decreased sharply when temperature raised to 80°C. The decrease in activity may be attributable to double-strand substrate denaturation.
Under the CEA reaction conditions selected, analytical figures of merit were assessed. A set of 10 water solutions containing miRNA-21 at various concentrations ranging from 2.5 pM to 10.0 nM were prepared. Triplicate analyses were performed for each sample. To improve MS measurement accuracy and reproducibility, 8-bromoadenosine was added to the samples at 0.50 μM as internal standard (IS). For quantification of miRNA-21, ion transitions m/z 597 →579 for CpC and m/z 348 →216 for IS were monitored using SRM detection mode. A plot of abundance ratio of CpC /IS versus miRNA concentration is shown in Figure S2C. As shown, abundance of the signal reporter (i.e. CpC) increased with the increase in miRNA-21 concentration. A good linear relationship between the abundance ratio and logarithm of miRNA-21 concentration was obtained in the concentration range from 2.5pM to 1.0nM:
where C was miRNA-21 concentration in pM (Figure S2D). The limit of detection was estimated to be 0.25pM based on 3S/N. The assay sensitivity is notably higher than those of the mass spectrometric methods previously reported39–46, in which MS techniques, including liquid chromatography-tandem mass spectrometry, ion mobility MS, high-resolution MS, and ICP-MS were deployed. The significantly improved assay sensitivity can be attributed to: (1) a reasonable design of the CEA reaction, which include utilizing CpC as MS signal reporter and immobilized DNA probes for enhancing hybridization with miRNA target and then DSN cleavage; and (2) deploying microfluidic VAL-DESI-MS/MS measurement with nearly zero background signal and little sample dilution.
Assay repeatability was assessed by analyzing a standard solution containing 10.0 pM miRNA-21 five times. The relative standard deviation (RSD) as found to be 4.2% (n=5). The assay specificity was assessed by measuring the MS signal response to three types of miRNA sequences, i.e. miRNA-21, single-base mismatched miRNA-21 (SBM-21), and three bases mismatched miRNA-21 (TBM-21). As shown in Figure 3, TBM-21 produced a MS signal that was about ~5% of that produced by miRNA-21. In response to merely a single base mismatch the MS signal drastically decreased by 85%, indicating an excellent discrimination capability for sequences with base-mismatch. Good detection sensitivity and specificity of the proposed assay allowed its application to analysis of “real-life” biological samples.
Figure 3.
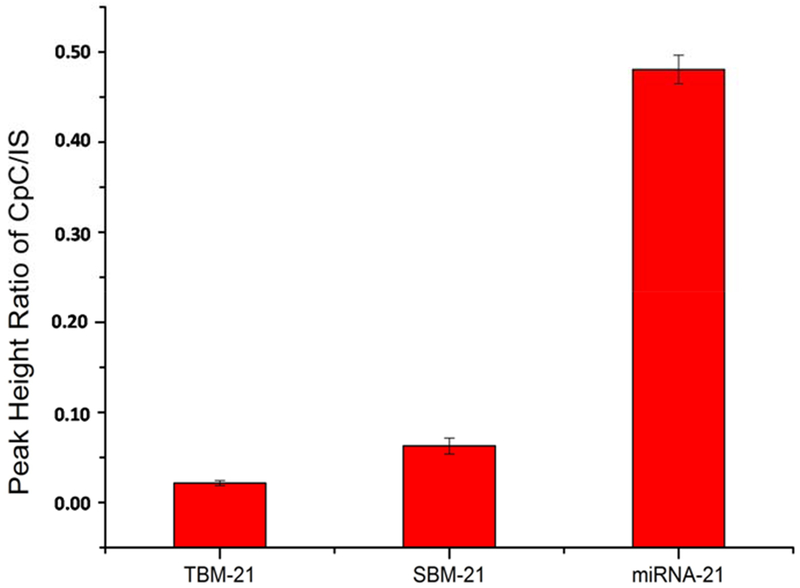
Specificity of the proposed miRNA assay: comparison of MS signal responses to complementary target (miRNA-21), single-base mismatch strand (SBM-21), and three-base mismatch strand (TBM-21) (all were at 65pM).
Quantification of miRNA-21 in Cells.
Sensitive and specific detection of miRNA biomarkers in cellular samples such as circulating tumor cells is highly significant for disease diagnosis and therapeutic evaluation. miRNA-21 is recognized as a versatile biomarker of disease. In this work, we investigated the feasibility of the proposed CEA-VAL-DESI-MS/MS method to quantification of miRNAs in cells. Mouse peritoneal macrophages were analyzed as the model cellular system. A set of cellular samples containing different numbers of cells were prepared in PBS from a cell suspension of known cell density. After lysing the cells, miRNA-21 concentration was determined to evaluate the assay sensitivity and applicability with this sample matrix. The results obtained are illustrated by Figure 4. As shown, miRNA-21 concentration measured increased with the increase of cell number. In a range from 100 to 500 cells, a linear correlation was obtained between the abundance ratio of CpC to IS and cell number, indicating there was no interference with the assay from this sample matrix. It’s worth noting that no isolation of total RNAs was needed for the assay. By using the calibration equation shown above, miRNA-21 content in 100 mouse peritoneal macrophages was measured to be 95.3 ± 13.95 attomoles (n=3). To assess the assay accuracy, a cell homogenate sample was spiked with miRNA-21 at 50.0pM and quantified against the calibration curve shown above. The recovery was found to be 94.2 ± 2.6% (n=3), indicating the assay was accurate and there was no interference from this sample matrix. It’s worth mentioning that there was no need for total RNA isolation in order to quantify miRNA-21 at trace levels in these samples by using the present assay. This is a significant advantage because total RNA isolation is not only tedious and lab skill-demanding, but also may cause a loss of miRNAs due to the nature of their short sequences during the isolation. Although the present assay involves CEA incubation of 120 min multiple samples can be treated at the same time in a microtiter plate. The analysis of CEA reaction solutions takes less than 1 min per sample on a VAL-DESI-MS/MS platform with two sample reservoirs (as shown in Figure 1). The proposed assay can be, therefore, high throughput and suitable for analysis of large batches of samples.
Figure 4.
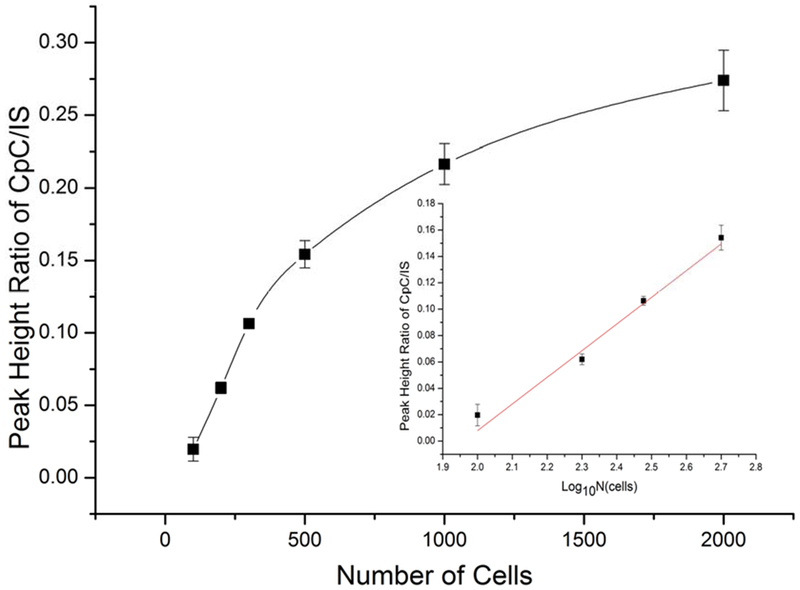
Quantification of miRNA-21 in mouse peritoneal macrophages by the proposed CEA-VAL-DESI-MS/MS method: abundance ratio of CpC /IS versus number of peritoneal macrophages. Inset: abundance ratio has a linear correlation with the number of cells in the range of 100-500 cells. Error bars indicate the standard deviation of three measurements.
Detection of changes in exosomal miRNA-21 expression caused by chemical stimulation.
Exosomes are biologically active vesicles secreted by cells. Cargos of exosomes include proteins, DNAs, and RNAs.50–52 In this work, exosomes secreted from mouse peritoneal macrophages were analyzed. To study the effects of chemical stimulation on exosomal miRNA-21 expression, macrophages were cultured in serum-free media containing interferon (IFN)-γ (at 20 ng/mL) and lipopolysaccharide (LPS, at 1 μg/mL) for 8 h to prime the cells to a pro-inflammatory M1 phenotype.48–49 The culture media were collected and span to remove cells and cell debris. Exosomes were isolated from the media by using anti CD-63 magnetic beads (obtained from Invitrogen). To investigate the affinity binding based isolation, exosomes were first labeled with a lipophilic fluorescence dye, DiO (obtained from Molecular Probe). After incubation with a fluorescently labeled exosome sample the magnetic beads were separated and their fluorescence microscopic images were taken against a blank (neat cell culture media). As shown in Figures S3a and 3b, fluorescently labeled exosomes were enriched on the surface of magnetic beads, confirming that exosomes were successfully isolated from the solution. Further, effects of magnetic beads amount used for the isolation was investigated. As shown in Figure S3c, for 5.0 mL culture media 50 μL bead suspension was enough to achieve the optimal isolation results. Exosomes isolated from conditioned culture media were lysed for miRNA-21 quantification. Since the VAL-DESI-MS/MS platform had two sample reservoirs, CEA solutions obtained from both the control and the chemically stimulated cellular sample were analyzed in parallel (i.e. in an alternating manner as we previously described44). Figure 5 shows the typical MS data obtained from the analysis. As shown, miRNA-21 was detected in both groups of exosomal samples, but exosomal miRNA-21 expression in the chemically stimulated sample was found significantly elevated. Calculated from the abundance ratios of CpC /IS in these samples, the chemical stimulation caused an increase of 3.0 ± 0.82 (n=3) times in exosomal miRNA-21 content. To our knowledge, this is the first report on increased exosomal miRNA expression due to exposure of parent cells to chemical stimuli. The increase can be due to either an elevated secretion of exosomes from the cells under chemical stimulation or an increase in intra-exosomal concentration of miRNA-21 caused by chemical stimulation or both.
Figure 5.
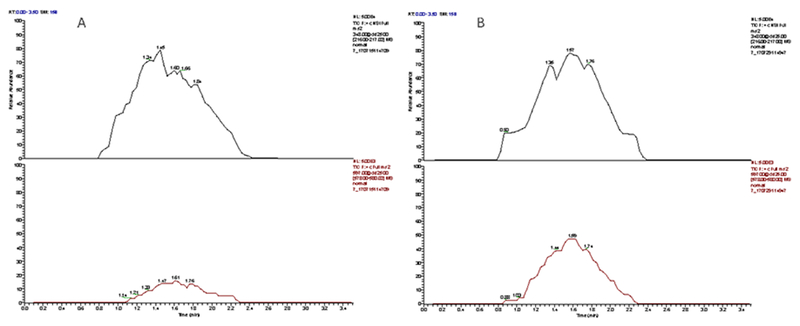
Extracted ion abundance traces for internal standard (upper traces) and miRNA-21 (lower traces) obtained from analysis of exosomes secreted from mouse peritoneal macrophages cultured in serum-free medium (A); and in serum-free medium containing chemical stimulants (i.e. IFN-γ at 20 ng/mL and lipopolysaccharide at 1 μg/mL) (B). Both were cultured for 8 hrs.
CONCLUSIONS
In this study it’s shown that coupling duplex-specific nuclease-based cyclic enzymatic amplification (CEA) with microfluidic voltage-assisted liquid desorption electrospray ionization-tandem mass spectrometry (VAL-DESI-MS/MS) is an effective assay strategy for quantification of targeted miRNAs in biological samples. A novel label free, easy-to-prepare, and highly stable ssDNA probe is prepared and proven effective. The assay has a detection limit of 0.25 pM miRNA-21 and is sensitive enough for quantification of miRNA-21 present in 10 living cells. Also, importantly, there is no need for total RNA isolation prior to the assay. Since multiple samples can be treated at the same time in a microtiter plate to complete the CEA reaction and VAL-DESI-MS/MS analysis takes <1min per sample, the assay is well suited for high throughput screening. Compared with fluorescence signal measurement, mass spectrometric measurement is far more selective and thus less likely to be interfered from sample matrix. Considering the high assay sensitivity, specificity and simple implement features, the proposed assay has a potential to become a cost-effective, accurate, and reliable means for quantification of targeted miRNAs in biomedical and clinical samples.
Supplementary Material
ACKNOWLEDGMENTS
Financial support from U.S. National Institutes of Health (GM089557 to YML and partially G12MD007581 to PBT) and American Heart Association (partially 15SDG22930009 to MY) is gratefully acknowledged.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:
Materials; Preparation and treatments of mouse peritoneal macrophages; Isolation of exosomes from cell culture media; Results of studying experimental conditions for the assay; Results of isolation of exosomes from cell culture media using anti CD-63 magnetic beads. (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1).Volinia S; Calin GA; Liu C-G; Ambs S; Cimmino A; Petrocca F; Visone R; Iorio M; Roldo C; Ferracin M A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A 2006, 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Shah IM; Macrae IM; Di Napoli M Neuroinflammation and neuroprotective strategies in acute ischaemic stroke-from bench to bedside. Curr. Mol. Med 2009, 9, 336–354. [DOI] [PubMed] [Google Scholar]
- 3).Davies BP; Arenz C A homogenous assay for micro RNA maturation. Angew. Chem. Int. Ed 2006, 45, 5550–5552. [DOI] [PubMed] [Google Scholar]
- 4).Lagos-Quintana M; Rauhut R; Lendeckel W; Tuschl T Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 5).Várallyay É; Burgyán J; Havelda Z MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc 2008, 3, 190–196. [DOI] [PubMed] [Google Scholar]
- 6).Chen C; Ridzon DA; Broomer AJ; Zhou Z; Lee DH; Nguyen JT; Barbisin M; Xu NL; Mahuvakar VR; Andersen MR Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005, 33, e179–e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Benes V; Castoldi M Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 2010, 50, 244–249. [DOI] [PubMed] [Google Scholar]
- 8).Redshaw N; Wilkes T; Whale A; Cowen S; Huggett J; Foy CA A comparison of miRNA isolation and RT-qPCR technologies and their effects on quantification accuracy and repeatability. BioTechniques 2013, 54 (3), 155–164. [DOI] [PubMed] [Google Scholar]
- 9).Curry E; Ellis S; Pratt S Detection of porcine sperm microRNAs using a heterologous microRNA microarray and reverse transcriptase polymerase chain reaction. Mol. Reprod. Dev 2009, 76, 218–219. [DOI] [PubMed] [Google Scholar]
- 10).Barad O; Meiri E; Avniel A; Aharonov R; Barzilai A; Bentwich I; Einav U; Gilad S; Hurban R; Karov Y MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 2004, 14, 2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Thomson JM; Parker J; Perou CM; Hammond SM A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 2004, 1, 47–53. [DOI] [PubMed] [Google Scholar]
- 12).Li W; Ruan K MicroRNA detection by microarray. Anal. Bioanal. Chem 2009, 394, 1117–1124. [DOI] [PubMed] [Google Scholar]
- 13).Deng R; Zhang K; Li J Isothermal amplification for microRNA detection: from the test tube to the cell. Acc. Chem. Res 2017, 50, 1059–1068. [DOI] [PubMed] [Google Scholar]
- 14).Tang X; Deng R; Sun Y; Ren X; Zhou M; Li J Amplified Tandem Spinach-Based Aptamer Transcription Enables Low Background miRNA Detection. Anal. Chem 2018, 90 (16), 10001–10008. [DOI] [PubMed] [Google Scholar]
- 15).Deng R; Tang L; Tian Q; Wang Y; Lin L; Li J Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angewandte Chemie 2014, 126(9), 2421–2425. [DOI] [PubMed] [Google Scholar]
- 16).Hu J; Kuang T DNA-Coded Fluorescence for Color Painting RNAs. Chem 2018, 4(6), 1194–1196. [Google Scholar]
- 17).Zhao Y; Chen F; Li Q; Wang L; Fan C Isothermal amplification of nucleic acids Chem. Rev 2015, 115, 12491–12545. [DOI] [PubMed] [Google Scholar]
- 18).Shen Y; Tian F; Chen Z; Li R; Ge Q; Lu Z Amplification-based method for microRNA detection. Biosens. Bioelectron 2015, 71, 322–331. [DOI] [PubMed] [Google Scholar]
- 19).Chen Y-X; Huang K-J; Niu K-X Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens. Bioelectron 2018, 99, 612–624. [DOI] [PubMed] [Google Scholar]
- 20).Ge J; Zhang L; Liu S; Yu R; Chu X A highly sensitive target-primed rolling circle amplification (TPRCA) method for fluorescent in situ hybridization detection of microRNA in tumor cells. Anal. Chem 2014, 86, 1808–1815. [DOI] [PubMed] [Google Scholar]
- 21).Yin B-C; Liu Y-Q; Ye B-C One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J. Am. Chem. Soc 2012, 134, 5064–5067. [DOI] [PubMed] [Google Scholar]
- 22).Feng Q-M; Shen Y-Z; Li M-X; Zhang Z-L; Zhao W; Xu J-J; Chen H-Y Dual-wavelength electrochemiluminescence ratiometry based on resonance energy transfer between Au nanoparticles functionalized g-C3N4 nanosheet and Ru (bpy) 32+ for microRNA detection. Anal. Chem 2015, 88, 937–944. [DOI] [PubMed] [Google Scholar]
- 23).Wang Q; Yin B-C; Ye B-C A novel polydopamine-based chemiluminescence resonance energy transfer method for microRNA detection coupling duplex-specific nuclease-aided target recycling strategy. Biosens. Bioelectron 2016, 80, 366–372. [DOI] [PubMed] [Google Scholar]
- 24).Bi S; Zhang J; Hao S; Ding C; Zhang S Exponential amplification for chemiluminescence resonance energy transfer detection of microRNA in real samples based on a cross-catalyst strand-displacement network. Anal. Chem 2011, 83, 3696–3702. [DOI] [PubMed] [Google Scholar]
- 25).Degliangeli F; Kshirsagar P; Brunetti V; Pompa PP; Fiammengo R Absolute and direct microRNA quantification using DNA-gold nanoparticle probes. J. Am. Chem. Soc 2014, 136, 2264–2267. [DOI] [PubMed] [Google Scholar]
- 26).Xi Q; Zhou D-M; Kan Y-Y; Ge J; Wu Z-K; Yu R-Q; Jiang J-H Highly sensitive and selective strategy for microRNA detection based on WS2 nanosheet mediated fluorescence quenching and duplex-specific nuclease signal amplification. Anal. Chem 2014, 86, 1361–1365. [DOI] [PubMed] [Google Scholar]
- 27).Lv W; Zhao J; Situ B; Li B; Ma W; Liu J; Wu Z; Wang W; Yan X; Zheng L A target-triggered dual amplification strategy for sensitive detection of microRNA. Biosens. Bioelectron 2016, 83, 250–255. [DOI] [PubMed] [Google Scholar]
- 28).Yan H; Bhattarai U; Guo Z-F; Liang F-S Regulating miRNA-21 biogenesis by bifunctional small molecules. J. Am. Chem. Soc 2017, 139, 4987–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Pang Y; Wang C; Wang J; Sun Z; Xiao R; Wang S, Fe3O4@ Ag magnetic nanoparticles for microRNA capture and duplex-specific nuclease signal amplification based SERS detection in cancer cells. Biosens. Bioelectron. 2016, 79, 574–580. [DOI] [PubMed] [Google Scholar]
- 30).Chiadò A; Novara C; Lamberti A; Geobaldo F; Giorgis F; Rivolo R Immobilization of oligonucleotides on metal-dielectric nanostructures for miRNA detection. Anal. Chem 2016, 88, 9554–9563. [DOI] [PubMed] [Google Scholar]
- 31).Jou AF-J; Chen Y-J; Li Y; Chang Y-F; Lee J.-j.; Liao AT; Ho J.-a.A. Target-Triggered, Dual Amplification Strategy for Sensitive Electrochemical Detection of a Lymphoma-associated MicroRNA. Electrochim. Acta 2017, 236, 190–197. [Google Scholar]
- 32).Yang C; Dou B; Shi K; Chai Y; Xiang Y; Yuan R Multiplexed and amplified electronic sensor for the detection of microRNAs from cancer cells. Anal. Chem 2014, 86, 11913–11918. [DOI] [PubMed] [Google Scholar]
- 33).Hao N; Dai P-P; Yu T; Xu J-J; Chen H-Y A dual target-recycling amplification strategy for sensitive detection of microRNAs based on duplex-specific nuclease and catalytic hairpin assembly. Chem. Commun 2015, 51, 13504–13507. [DOI] [PubMed] [Google Scholar]
- 34).Zhang J; Wu D-Z; Cai S-X; Chen M; Xia Y-K; Wu F; Chen J-H An immobilization-free electrochemical impedance biosensor based on duplex-specific nuclease assisted target recycling for amplified detection of microRNA. Biosens. Bioelectron 2016, 75, 452–457. [DOI] [PubMed] [Google Scholar]
- 35).Labib M; Khan N; Ghobadloo SM; Cheng J; Pezacki JR; Berezovski MV Three-mode electrochemical sensing of ultralow microRNA levels. J. Am. Chem. Soc 2013, 135, 3027–3038. [DOI] [PubMed] [Google Scholar]
- 36).Lu W; Chen Y; Liu Z; Tang W; Feng Q; Sun J; Jiang X, Quantitative detection of microRNA in one step via next generation magnetic relaxation switch sensing. ACS Nano, 2016, 10, 6685–6692. [DOI] [PubMed] [Google Scholar]
- 37).Tian B; Ma J; Qiu Z; Zardán Gómez de la Torre T; Donolato M; Hansen MF; Svedlindh R; Strömberg M Optomagnetic detection of microRNA based on duplex-specific nuclease-assisted target recycling and multilayer core-satellite magnetic superstructures. ACS nano 2017, 11, 1798–1806. [DOI] [PubMed] [Google Scholar]
- 38).Chen S; Wan Q; Badu-Tawiah AK Mass spectrometry for paper-based immunoassays: toward on-demand diagnosis. J. Am. Chem. Soc 2016, 138, 6356–6359. [DOI] [PubMed] [Google Scholar]
- 39).Nakayama H; Yamauchi Y; Taoka M; Isobe T Direct Identification of Human Cellular MicroRNAs by Nanoflow Liquid Chromatography-High-Resolution Tandem Mass Spectrometry and Database Searching. Anal. Chem 2015, 87, 2884–2891. [DOI] [PubMed] [Google Scholar]
- 40).Kullolli M; Knouf E; Arampatzidou M; Tewari M; Pitteri SJ Intact microRNA analysis using high resolution mass spectrometry. J. Am. Soc. Mass. Spectrom 2014, 25, 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Takebayashi K; Hirose K; Izumi Y; Bamba T; Fukusaki E Application of ion mobility-mass spectrometry to microRNA analysis. J. Biosci. Bioeng 2013, 115, 332–338. [DOI] [PubMed] [Google Scholar]
- 42).Konishi K; Kojima K; Watanabe M; Shimizu K; Tanaka K; Sato TA In C71. MAJOR IMPACT OF MICRO-RNA FUNCTION, Am Thoracic Soc: 2012; pp A4976–A4976. [Google Scholar]
- 43).Kim S; Park J; Na J; Jung GY; Hwang J Simultaneous Determination of Multiple microRNA Levels Utilizing Biotinylated Dideoxynucleotides and Mass Spectrometry. PloS one 2016, 11, e0153201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Grimson A A targeted approach to miRNA target identification. Nat. Methods 2010, 7, 795–797. [DOI] [PubMed] [Google Scholar]
- 45).Liu L; Xu Q; Hao S; Chen YA Quasi-direct LC-MS/MS-based Targeted Proteomics Approach for miRNA Quantification via a Covalently Immobilized DNA-peptide Probe. Scientific reports 2017, 7. doi: 10.1038/s41598-017-05495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Zhang S; Liu R; Xing Z; Zhang S; Zhang X Multiplex miRNA assay using lanthanide-tagged probes and the duplex-specific nuclease amplification strategy. Chem. Commun 2016, 52 (99), 14310–14313. [DOI] [PubMed] [Google Scholar]
- 47).Li X; Xu R; Wei X; Hu H; Zhao S; Liu Y-M Direct Analysis of Biofluids by Mass Spectrometry with Microfluidic Voltage-Assisted Liquid Desorption Electrospray Ionization. Anal. Chem 2017, 89, 12014–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Ma Y; Chiao YA; Clark R; Flynn ER; Yabluchanskiy A; Ghasemi O; Zouein E; Lindsey ML; Jin Y-F Deriving a cardiac ageing signature to reveal MMP-9-dependent inflammatory signalling in senescence. Cardiovascular Res 2015, 106 (3), 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Ma Y; Halade GV; Zhang J; Ramirez TA; Levin D; Voorhees AP; Jin Y-F; Han H-C; Manicone AM; Lindsey M Matrix Metalloproteinase-28 Deletion Exacerbates Cardiac Dysfunction and Rupture Following Myocardial Infarction in Mice. Circul. Res 2012, CIRCRESAHA. 111.300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Théry C; Amigorena S; Raposo G; Clayton A Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology 2006, 30(1), 1–22. [DOI] [PubMed] [Google Scholar]
- 51).Tauro BJ; Greening DW; Mathias RA; Ji H; Mathivanan S; Scott AM; Simpson RJ Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [DOI] [PubMed] [Google Scholar]
- 52).Xu R; Yang T; Chen Y Quantification of microRNA by DNA-Peptide probe and liquid chromatography-tandem mass spectrometry-based quasi-targeted proteomics. Anal. Chem 2015, 88 (1), 754–763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


