Abstract
MicroRNAs are small noncoding RNA molecules that regulate gene expression posttranscriptionally through complementary base pairing with thousands of messenger RNAs. They regulate diverse physiological, developmental, and pathophysiological processes. Recent studies have uncovered the contribution of microRNAs to the pathogenesis of many human diseases, including liver diseases. Moreover, microRNAs have been identified as biomarkers that can often be detected in the systemic circulation. We review the role of microRNAs in liver physiology and pathophysiology, focusing on viral hepatitis, liver fibrosis, and cancer. We also discuss microRNAs as diagnostic and prognostic markers and microRNA-based therapeutic approaches for liver disease.
Keywords: Chronic Liver Disease, Liver Fibrosis, Cirrhosis, Hepatocellular Carcinoma
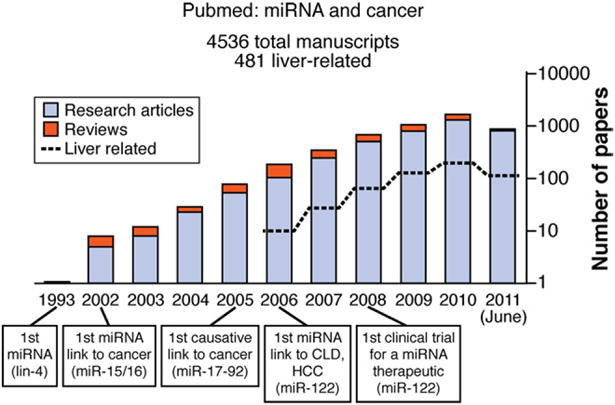
MicroRNAs (miRNAs) were first described in 1993, in developmental timing experiments in the nematode Caenorhabditis elegans.1 Since then, these small noncoding RNA molecules, about 22 nucleotides long, were found to be posttranscriptional regulators of gene expression in metazoans and plants.2,3 The human miRNA family comprises 1733 mature miRNAs, encoded by 1424 precursors (some have miRs annotated on both sides of the hairpin) (data from miRBase 17; www.mirbase.org/). A 2002 report that miRNAs were involved in tumorigenesis led to the identification of many other miRNAs and increased our understanding of their biogenesis and roles in oncogenesis.4 A PubMed search from June 2011 using the keywords “microRNA” or “miRNA” found these terms in the titles or abstracts of 7691 research articles and 1360 reviews or commentaries; 4536 were related to cancer and 482 were specific to liver diseases, including liver cancers (Figure 1). There has been exponential growth in the number of miRNA articles related to cancer since 2002.
Figure 1.
Timeline for studies of miRNAs in cancer. A PubMed search was conducted in June 2011 using the keywords “microRNA” or “miRNA” in titles or abstracts. A subsequent search was then restricted to liver or cancer.
The discovery of miRNAs has increased our understanding of the posttranscriptional regulation of genes and how this process contributes to development of cancer. More than 50% of genes that encode miRNAs are located at fragile sites or in cancer-associated regions of the genome, indicating that miRNAs are cancer related and could serve as diagnostic markers or therapeutic targets.5 Almost every type of human cancer analyzed has been associated with altered activities of miRNAs. These discoveries have accelerated our understanding of the pathogenesis of human cancer and provide tools for diagnosis and treatment of cancer.
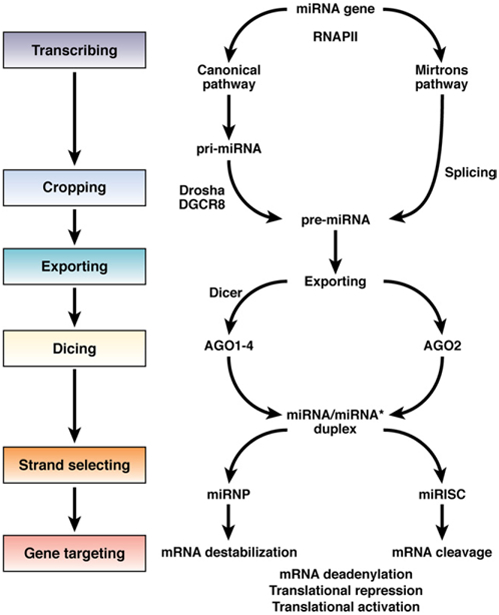
miRNA biogenesis has been well characterized and basically consists of 6 steps: transcription, cleavage, export, further cleavage, strand selection, and interaction with target messenger RNAs (mRNAs). Modification of any of these steps could contribute to development of liver or other diseases, including cancer (Figure 2). Briefly, miRNAs are transcribed from genes by RNA polymerase II into initial transcripts that are processed either via a canonical pathway (cleaved by the Drosha-DGCR8 complex) or the mirtrons pathway (processed by the spliceo-some) to form hairpin-like miRNA precursors called pre-miRNAs. These precursors are exported from the nucleus to the cytoplasm, which requires Exportin-5 and Ran-GTP. In the cytoplasm, they are processed by Dicer into imperfect duplexes that consist of mature miRNA and complementary fragments called miRNAs.
Figure 2.
Steps in miRNA biogenesis. miRNAs are mainly transcribed by RNA polymerase II (RNAPII) into initial transcripts know as pre-miRNA, which are processed either via a canonical pathway, in which they are cropped by the Drosha-DGCR8 complex, or via the Mirtrons pathway, in which they are spliced to form hairpin-like miRNA precursors called pre-miRNAs. These precursors are exported from the nucleus to the cytoplasm in an Exportin-5-RanGTP—dependent manner. In the cytoplasm, they are processed by Dicer into an imperfect duplex comprising mature miRNAs and a complementary fragment called miRNA. The processed miRNAs are then loaded onto the RISC and are guided to their mRNA targets through interacting with various members of the Argonaute family such as Ago1–4 or Ago2 to form RISCs that are known as miRNP or miRISC. miRNA-mediated gene silencing or activation could be achieved through several identified mechanisms such as mRNA deadenylation, translational repression or translational activation, and possibly many more mechanisms yet to be discovered. Different base-paring combinations of miRNA-mRNA through their unique complementarities could result in gene silencing or gene activation.
The processed miRNAs are loaded onto the RNA-induced silencing complex (RISC) and guided to their mRNA targets through interactions with members of the Argonaute family, such as Ago1–4 or Ago2, to form RISCs that are also called miRNP or miRISC. Gene expression can be reduced or increased by miRNAs via several mechanisms, such as mRNA deadenylation, translational repression or activation, or other undiscovered processes. Different base-paring combinations of miRNA-mRNA, through their unique complementarities, could reduce or increase production of gene products. It is not clear how miRNAs incorporate into RISCs and form complexes with different argonautes. However, miRNAs and their biogenesis pathways are highly conserved evolutionally, from plants to mammals, indicating their importance in cellular processes and development. Recent results indicate that miRNA from plants used as foodstuffs (eg, rice) can regulate gene expression in mammals.6
miRNAs regulate diverse physiological and developmental processes by controlling levels of specific mRNAs, so their own expression and processing must be tightly regulated for normal cell function.7, 8 Each miRNA could be transcribed and regulated independently, at the transcriptional levels by activators and repressors, or at the epigenetic level through DNA methylation.9–11 The expression levels of processing components are also tightly controlled to regulate the abundance of mature miRNAs. Alterations in any of these processes could lead to tumorigenesis or development of other diseases.12 Single nucleotide polymorphisms in genes that encode miRNAs can affect their processing and target binding, along with cancer risk, response to treatment, and disease progression.13 The complexity of miRNA regulation and the changes that contribute to tumorigenesis make it difficult to correlate specific miRNAs or features of their processing with particular tumor types. In fact, levels of miRNAs and miRNA processing are likely to vary throughout a tumor; tumor heterogeneity is a barrier to effective diagnosis and treatment.
Chronic liver diseases such as viral hepatitis, which can be caused by infection with hepatitis B or C viruses (HBV or HCV), alcohol consumption, or obesity, are major global health burdens that can increase the risk of hepatocellular carcinoma (HCC). Although vaccination can prevent HBV infection,14 strategies to eliminate HBV from chronic carriers are ineffective and there are no vaccines for HCV. With 2 recently approved drugs and dozens more in the pipeline, HCV treatment strategies are likely to improve.15 HCC is a common form of primary liver cancer; it is the third most deadly and fifth most common cancer in the world.16–18 Despite many years of research into treatment and causes of HCC, it remains one of the most difficult-to-treat malignancies, with a 5-year survival rate of less than 12% in the United States.18
miRNAs in Liver Disease
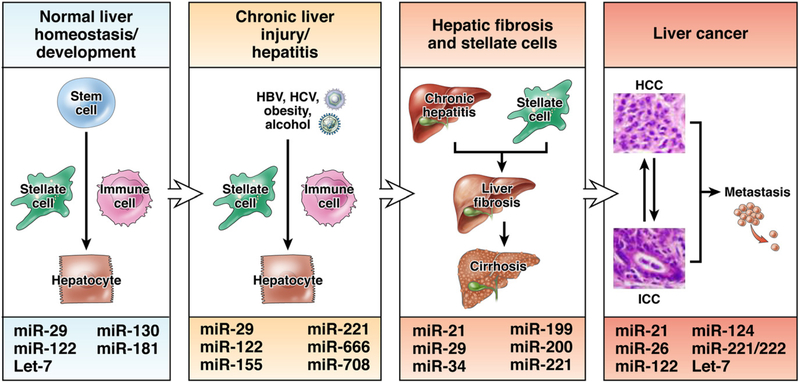
The roles of miRNAs in regulation of gene transcription in animal development have been well documented.19 Organogenesis and development of the liver have also been well studied.20 Although we have limited understanding of the role of miRNAs in liver development, these molecules are likely to regulate cell lineages and differentiation. Expression of miR-122 is liver specific and inhibition of miR-122 expression in mice leads to a down-regulation of cholesterol and lipid-metabolizing enzymes.21 Members of the miR-181 family are highly expressed in embryonic liver and can target the mRNA that encodes GATA6,22 a transcription factor that regulates liver organogenesis.20 Increasing our knowledge about the roles of miRNAs in normal liver development will help us understand liver pathophysiology and cancer pathogenesis (Figure 3). Because of a close link between miRNAs and development, it is not surprising that so many different types of miRNAs are involved in different stages of liver diseases (Figure 3). Most of the recent studies have focused on the roles of miRNAs in initiation and progression of liver cancer, although we are learning more about the roles of miRNAs in chronic liver disease.
Figure 3.
Roles of miRNAs in the pathogenesis of chronic liver disease and cancer. Cells in the liver, including hepatic stem and progenitor cells, hepatocytes, hepatic stellate cells, and immune cells, contribute to the progression of chronic liver disease to liver cancer. Factors such as HBV and HCV infection, obesity, and alcohol contribute to development of chronic liver disease, which can lead to fibrosis and cirrhosis and increase risk for HCC and intrahepatic cholangiocarcinoma (ICC). The figure shows miRNAs that are involved in different stages of liver disease.
Viral Hepatitis
Patients with viral hepatitis or chronic liver disease are at increased risk for developing cirrhosis and primary liver cancer. The liver-specific miR-122 could contribute to the liver tropism of HCV by accelerating the binding of ribosomes to the viral RNA and thereby stimulating HCV translation.23 This exciting finding indicates that inhibition of miR-122 could block HCV replication. The finding resulted in a successful preclinical study24 and the first miRNA-based clinical trial (Figure 1).
Alcoholic and Nonalcoholic Fatty Liver Disease
Alcoholic liver disease (ALD) is a major cause of chronic liver disease worldwide and can lead to fibrosis and cirrhosis.25 Nonalcoholic fatty liver disease (NAFLD) has been recognized as the most common chronic liver disease in Western industrialized countries. NAFLD affects 20%−30% of adults in developed countries25 and is associated with insulin resistance and metabolic syndrome, as in obesity and type 2 diabetes mellitus. Like viral hepatitis, ALD involves activation of intracellular signaling pathways in various cell types, including hepatocytes, hepatic stem cells, stromal cells, and inflammatory cells, although different molecular events could be involved. miRNAs seem to be involved in the pathogenesis of ALD. miR-125b, miR-146a, and miR-155 can regulate inflammatory responses to lipopolysaccharide-induced tumor necrosis factor (TNF)-α in Kupffer cells.26 Recent studies indicate that Kupffer cell–specific miR-155 contributes to alcohol-induced activation of TNF-α in macrophages from patients with ALD.27 Levels of circulating miRNAs such as miR-34a and miR-122 were found to be increased in patients with NAFLD.28 These miRNAs might be used as markers for diagnosis or therapeutic targets for ALD and NAFLD.
Fibrosis
Liver fibrosis is a consequence of chronic damage to the liver. It develops through a complex network of signaling pathways that regulate the deposition of extracellular matrix proteins and fibrogenesis, a characteristic of most types of chronic liver disease. Hepatic stellate cells and inflammatory cells such as Kupffer cells could contribute to development of chronic liver disease. Recent studies indicate that miR-29 regulates liver fibrosis and is part of a signaling nexus that involves transforming growth factor β and nuclear factor κB in hepatic stellate cells.29 Members of the miR-29 family were significantly down-regulated in livers of mice following induction of fibrosis with CCl4 and in livers of patients with advanced fibrosis. In addition, miR-29b suppresses activation of hepatic stellate cells and might slow or prevent liver fibrosis.30 Other studies have associated overexpression of the miR-199 and −200 families with progression of liver fibrosis.31 Interestingly, overall down-regulation of miRNAs was observed in a feedback mechanism that occurs during the early phases of liver regeneration.32 These studies reiterated the functional importance of miRNA during liver regeneration and consequent liver fibrosis.
HCC
Numerous reports have linked deregulation of miRNA expression to liver cancers, but studies related to viral hepatitis-related HCC are limited. Gene expression profiling studies have shown different expression patterns of miRNA in HBV- and HCV-related HCC samples at various stages of tumor progression and in patients from different ethnic backgrounds.33–37 Through the examination of differentially expressed miRNAs that are associated with chromosome amplification or deletion, miR-151 was identified as an oncomir, an miRNA with an oncogenic potential.38 Abnormal expression patterns of miRNA have been associated with stem cell-like HCC cells.22,39,40 Another recent study indicated that miR-199a/b-3p was down-regulated in HCC cells and can suppress growth of HCCs by inhibiting the PAK4-Raf-MEK-ERK pathway.41 The roles of miRNA in hepatic carcinogenesis are complex; different studies have reported unique profiles, with only a few miRNAs in common, indicating the heterogeneity of HCC. The miRNAs that have been identified in several studies, such as let-7, miR-122, miR-26, and miR-101, which are all down-regulated in HCC, and miR-221, miR-181, and miR-17-92, which are all up-regulated in HCC, could serve as biomarkers or therapeutic targets for HCC.
Combinations of genomic analyses and functional studies have identified miRNAs that function as oncogenes (oncomirs) or tumor suppressors. For example, up-regulation of miR-21 can activate PTEN, which activates phosphatidylinositol 3-kinase signaling to AKT and contributes to progression of HCC.42 miR-21 is frequently up-regulated in other human solid malignancies, such as tumors of breast, colon, lung, pancreas, prostate, and stomach.43 This miRNA is druggable and could be a good target for common human cancers, including HCC.
In a comparison of samples of primary metastatic and nonmetastatic HCCs from patients, altered levels of 20 miRNAs (including let-7g and miR-122a) were associated with venous metastasis of HCC.34 Sixteen of these were down-regulated in metastatic tumors, indicating that reducing their expression contributes to tumor progression. Down-regulation of one of these, the hepatocyte-specific miRNA miR-122, promoted growth of HCCs in mice, regulated expression of cell cycle components, and increased migration of HCC cells and their invasive activities. miR-122 might therefore be a suppressor of HCC metastasis.44,45 The miRNA let-7g could suppress metastasis of HCC, in part, by targeting soluble collagens.46 Moreover, a feedback loop that includes the transcription factor hepatocyte nuclear factor (HNF)-4α and miRNA regulates inflammation and hepatocellular oncogenesis.47
The incidence of HCC is 2- to 6-fold higher in men than in women.48 Moreover, women with this disease tend to survive longer than men, indicating a sex-related mechanism that prevents HCC formation or slows progression. Researchers identified 5 miRNAs (miR-26a, miR-10b, miR-125b, miR-99b, and miR-325) that were expressed at higher levels in livers of women than men; 3 were down-regulated in HCCs, indicating that they might act as tumor suppressors.49 miR-26 has been consistently found to be silenced in metastatic HCC and its levels are associated with patient survival time.49,50 miR-26 targets mRNAs that encode cell cycle components and systemic administration of this miRNA in a mouse model of HCC inhibited cancer cell proliferation, induced tumor-specific apoptosis, and protected mice from disease progression.50 Similarly, expression of let-7 can reduce growth of lung tumors in mice.51
Similar to HCC, miRNAs are involved in the pathogenesis of cholangiocarcinoma, the second most common type of primary liver cancer. miR-21 and many other miRNAs are expressed at high levels in cholangiocarcinoma and regulate programmed cell death 4 and tissue inhibitor of metalloproteinase 3; they also prevent gemcitabine-induced apoptosis by PTEN-dependent activation of phosphatidylinositol 3-kinase signaling.52,53 Interleukin-6 epigenetically controls expression of miR-370 in malignant cholangiocytes.54 miR-494 is down-regulated in cholangiocarcinoma; up-regulation of this miRNA can reduce cancer cell proliferation by affecting multiple targets involved in the G1–S transition.55 These results indicate that miRNAs are useful markers and possible therapeutic targets for cholangiocarcinoma.
Liver Cell Types and Associated miRNAs
Hepatocytes
Hepatocytes make up 70%–80% of the liver mass and mediate detoxification, modification, and excretion of exogenous and endogenous substances. The function of miRNAs in normal liver physiology was assessed in mice that lack functional Dicer1 in hepatocytes.56 Hepatic function was maintained in the absence of mature miRNAs. However, chronic miRNA deficiency led to hepatocyte apoptosis, hepatocyte regeneration, and portal inflammation, which are processes associated with chronic liver disease. HNF-4α regulates hepatocyte differentiation20; miR-24 and miR34a have each been found to target HNF4α mRNA.57 miR-122 comprises more than 70% of the miRNA in the liver58 and can be regulated by HNF-4α.59 Therefore, HNF-4α might regulate hepatocyte differentiation through miR-122. miRNAs might regulate hepatocyte differentiation and metabolism during cellular stress, triggered by various etiologic factors.
Kupffer Cells
Kupffer cells are macrophages in the liver. There have been few studies of the functions of miRNAs in Kupffer cells, although they appear to be the major source of TNF-α production in patients with ALD. miR-155 has been associated with ALD and might perpetuate production of TNF-α production by stabilizing its mRNA.27
Hepatic Stellate Cells
Hepatic stellate cells mediate hepatic fibrosis and have been found to express miR-29, the miR-199 and −200 families, and miR-221/222.29–31,60,61 Expression of the miR-199 and −200 families has been correlated with progression of liver fibrosis.31 Hepatocyte growth factor could up-regulate miR-29, which regulates collagen synthesis to repress stellate cell activity. The transcription factor nuclear factor κB can activate expression of miR-221 and miR-222, which can promote proliferation of stellate cells. These miRNAs might be therapeutic targets for liver fibrosis.
Liver Sinusoidal Endothelial Cells
Liver sinusoidal endothelial cells comprise approximately 50% of nonparenchymal hepatic cells and serve as a scavenger system in the exchange of material between the blood and liver parenchyma. Long-term exposure to alcohol can induce endothelin 1 and hypoxia-inducible factor 1α and inhibit expression of miR-199 in liver sinusoidal endothelial cells.62 Overexpression of miR-199 can reduce expression of hypoxia-inducible factor 1α and endothelin 1. miR-199 might therefore function as a negative regulator of the level of endothelin 1 and the function of liver sinusoidal endothelial cells.
miRNA Regulatory Networks
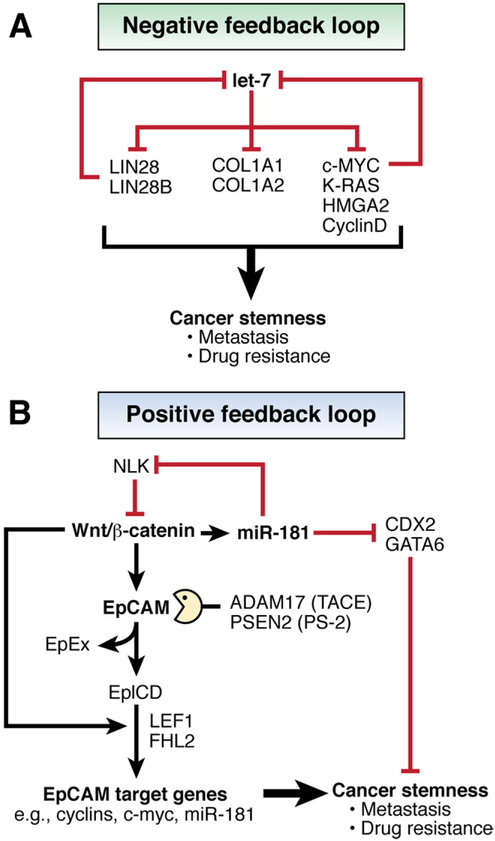
Cell homeostasis is maintained by signaling networks that, when disrupted, cause diseases such as cancer.63 miRNAs participate in many negative and positive feedback loops that control cell processes, including stem cell properties (Figure 4). The let-7 miRNA precursor, which binds to the mRNA Lin28 (a marker of human embryonic stem cells), is regulated by the product of the oncogene c-Myc, which contains let-7 binding sites, indicating the presence of a feedback loop. Let-7 regulates other oncogenes as well as cell proliferation and apoptosis. The let-7 family consists of 13 members, conserved in sequence and located on 9 different chromosomes; its functions are conserved from C elegans to humans. Levels of let-7 family members are down-regulated in malignancies, including HCC, and associated with cancer stem cells.
Figure 4.
miRNAs participate in negative and positive feedback loops that regulate stem cell properties of cancer cells. (A) During normal development, the RNA-binding protein Lin28 is highly expressed in stem and progenitor cells. Lin28 blocks processing of let-7 miRNA precursor molecules into mature miRNAs to maintain expression of genes that promote self-renewal and proliferation. As progenitor cells differentiate, Lin28 expression decreases, which allows let-7 processing and increased production of mature let-7 miRNAs. These further inhibit Lin28, c-Myc, and several other cancer-associated genes. Similarly, c-Myc can promote self-renewal and proliferation in stem and progenitor cells, in part by inhibiting let-7 expression. Let-7 then represses the expression of genes involved in self-renewal, resulting in lineage commitment and terminal differentiation. In liver cancer cells, especially those with stem cell features, Lin28 and c-myc are highly activated, which results in an inactivation of let-7 and the maintenance of stem cell–like features of cancer cells, in a negative feedback loop. Let-7 can directly target Lin28, c-myc, K-Ras, HMGA2, Cyclin D1, and soluble collagens such as COL1A1 and COL1A2. (B) Signaling pathways such as Wnt–β-catenin, EpCAM, and miR-181 are activated in hepatic cancer stem cells. miR-181 promotes stem cell features of HCC cells by targeting CDX2 and GATA6 (hepatic transcriptional regulators of differentiation) and NLK (an inhibitor of Wnt–β-catenin signaling); this inhibition of NLK up-regulates transcription of EpCAM. Upon cleavage by TACE/PS-2, an intracellular ectodomain of EpCAM, referred as EpICD, translocates to the nucleus in a multiprotein complex that contains FHL2, β-catenin, and Lef-1 to induce transcription of genes that encode cyclin D1, c-Myc, and miR-181. This positive feedback loop promotes the stem cell-like characteristics of HCC cells.
Given the functional redundancy and corepression of let-7 family members in tumor cells, there might be a common mechanism that coordinately inhibits multiple members of this family. The lin28 family members lin28 and lin28B each target and inhibit let-7.64,65 Let-7 suppresses expression of c-Myc, which inhibits transcription of let-7. Loss of such a negative feedback loop appears to be a common event in cancer cells from advanced-stage tumors such as HCC.66
In contrast, miR-181 regulates the Wnt-β-catenin signaling pathway in a positive feedback loop in stem cells. Members of the miR-181 family are highly expressed in HCC cells that have stem cell properties.22 miR-181 promotes stem-cell-like features of HCC cells by targeting mRNAs that encode CDX2 and GATA6, which are hepatic transcriptional regulators of differentiation. miR-181 also inhibits the mRNA that encodes NLK, an inhibitor of Wnt-β-catenin signaling. Activation of this pathway induces transcription of epithelial cell adhesion molecule (EpCAM),67 which is cleaved by the metallopeptidase ADAM17, in conjunction with PSEN2, to produce an intracellular ectodomain called EpICD. EpICD complexes with FHL2, β-catenin, and Lef1 to induce transcription of cyclin D1, c-myc, and miR-181.68,69 This type of positive feedback loop could be used by cancer cells to continuously self-propagate and contribute to metastasis and drug resistance.
During hepatocarcinogenesis, inactivation of HNF-4α leads to an activation of miR-24 and miR-629 via interleukin-6 signaling to Stat3. Activation of these miRNAs keeps HNF-4α inactivated, leads to production of more miR-124, and promotes oncogenesis.47 Gaining better understanding of miRNA-regulated feedback loops and finding ways to manipulate them might lead to strategies to inhibit liver cancer stem cells.70
Biomarkers
Diagnostics
MiRNAs are good biomarkers because they are well defined, chemically uniform, restricted to a manageable number of species, and stable in cells and in the circulation.71–73 Challenges to their development as markers include their small size, the presence of target sequences in precursor miRNAs, and absence of specific features such as caps or tails. Analyzing patterns of miRNAs requires polymerase chain reaction (PCR)-based techniques, hybridization assays, or next-generation sequencing methods.74,75
Standard procedures for complementary DNA synthesis and amplification are modified to function adequately with small RNAs.76,77 Quantitative reverse transcriptase (qRT)-PCR– based methods are generally difficult to reconcile with large sample numbers and probe sets. If samples from a homogeneous patient population are available, one viable approach for exploratory studies could be to perform multiplex PCR78 with pooled samples, followed by analyses of individual samples.79 Conventional qRT-PCR and microfluidic qRT-PCR platforms each have better limits of detection and dynamic ranges than expression microarrays80 but cannot compete with the ability of these assays to analyze thousands of targets. Direct hybridization with RNA in solution, using color-coded probes, offers a more highly multiplexed analysis without a need for amplification.81
Different studies of miRNA expression profiles report different results despite analyses under similar conditions. These differences could be caused by variations in patient populations (differences in diagnostic classification, sex and race composition,82–84 lifestyle factors such as alcohol or narcotic consumption,85–87 nutrition, or disorders such as obesity88,89 or fibrosis29,90,91), sampling differences, or use of primary tissues versus cell lines. Furthermore, different methods are used for miRNA isolation92 or analysis (microarray and qRT-PCR data from the same samples do not always correlate well).93 Multicenter studies are needed to settle at least some of these issues and to cross-validate findings.94
Another challenge to the use of miRNAs as markers is that their mRNA targets are not easily identified by computational methods.95 Targets should be validated biochemically, such as via immunoprecipitation of argonaute-miRNA-mRNA complexes.96 Also, it is important to characterize miRNA targets not only at the mRNA level but also at the protein level, such as by using proteome methods.97–99 Network analyses might show association of a set of down-regulated miRNAs with common cellular pathways or even convergence on a common protein or set of related proteins.100–106 It is also important to consider epigenetic factors, copy number variations, and polymorphisms in the regulatory elements of genes that encode miRNAs.107,108
Circulating miRNA
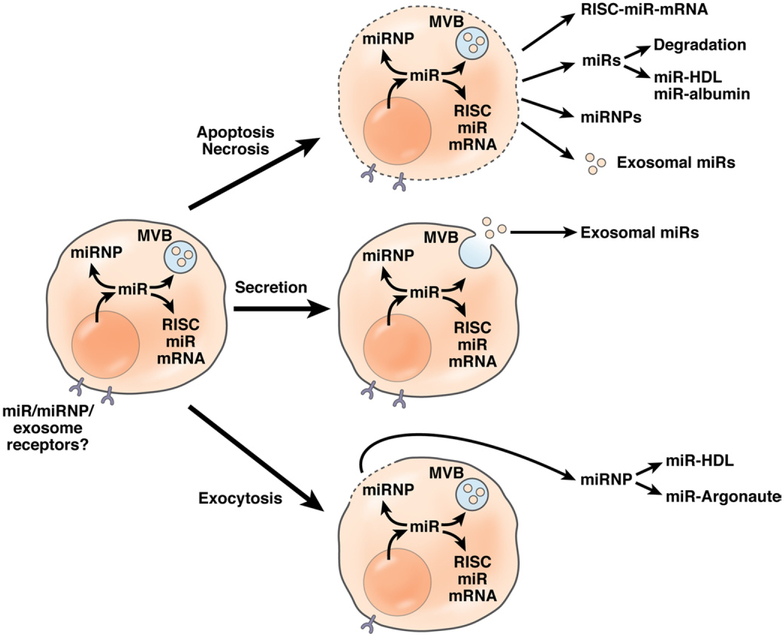
Extracellular miRNAs (cell-free nucleic acids in the circulation)109 are stable and can be recovered from serum, plasma, and other biologic fluids.72,110 Extracellular miRNA is found in exosomes and in exosome-independent forms; the latter are presumably protected from degradation by forming complexes with proteins, such as argonaute proteins (mainly Ago2) or high-density lipoproteins.111,112 Some miRNAs (probably the minority)111,113 are packaged into exosomes and microvesicles114 in multivesicular bodies; others exist as protein-miRNA complexes (miRNPs) (Figure 5). Some miRNAs might even originate from external sources (eg, derived from ingested food).6 The capacity of miRNAs for export might depend on their specific sequence or specific export mechanisms of cell types. Liver-specific miRNA-122 is released as a microvesicle-independent species.113,115 The average half-life of intracellular miRNA appears to be about 5 days, although some species are more stable than others.71 The half-life of extracellular miRNA is unknown but probably depends on the type of carrier (exosomes or free-protein complexes).
Figure 5.
Origins of extracellular, circulating miRNA. Schematic summary of miRNA translocation from intracellular forms (free, complexed, and located in exosomes in multivesicular bodies) to extracellular forms. Three main mechanisms are believed to contribute to this as indicated. One mechanism is cell death (apoptosis or necrosis) with release of intracellular species such as protein-associated miRs (in RISC together with mRNA), in argonaute-miR complexes (miRNA-miRNPs), in exosomes, and as free miRNA that is quickly degraded or stabilized by association with plasma proteins such as high-density lipoproteins and albumin. Another mechanism is through secretion of exosomes by fusion of multivesicular bodies with the plasma membrane. Finally, exocytosis of miRNPs is believed to occur. Different miRNAs appear to participate in different complexes and to different degrees in exosomes and exosome-independent forms, indicating specific roles in intercellular signaling, such as through putative miRNA, miRNP, and exosome receptors.
Use of circulating molecules as disease markers requires that enough material can be detected and analyzed. miRNAs could be used as markers for cancer if cancer-specific miRNAs existed, but such species have not been found, and only those normally present in low concentrations could be used. However, systemic effects, such as altered levels or profiles of miRNAs, could be associated with disease risk and identified. Levels of miRNA in whole blood are much higher than miRNA levels in plasma or serum.116 Serum and plasma levels of specific miRNAs are not readily comparable,82 and the preparation of plasma and sera (especially centrifugation) affects the amount of cellular miRNAs in the presumed extracellular material.84,94 Despite a number of proposals116–118 and good candidates for endogenous normalizers for tissue miRNA measurements,119 no circulating miRNA has been identified and validated for use as a standard. Variations in miRNA levels with tissue type, donor sex and race, and to some degree technical features of collection and assay methods have prevented their use as diagnostic or prognostic markers.94
Because of issues associated with purification of circulating miRNA, samples must be processed uniformly and RNA must be isolated using a consistent, robust, simple, and quick method to ensure valid comparisons of miRNA profiles in large studies. Direct qRT-PCR–based miRNA assays using samples of plasma or serum without RNA extraction could be feasible.120,121
Given the technical challenges, it is not surprising that there are conflicting data regarding up-regulation or down-regulation of circulating miRNA in various pathologies.122–124 The profiles of circulating miRNAs from patients with HCC and viral hepatitis in some respects reflect those of tissue miRNAs from these patients.49,125 The alterations in circulating miRNA most consistently reported in patients with liver disease are therefore increases in levels of miRNA-221 and −223 in patients with HCC.118,126 Circulating levels of miRNA-21 and −122 are also increased in patients with HCC, but more so among patients with chronic hepatitis or other types of toxic or viral liver injury.85,118
A mouse model of drug-induced liver injury was also reported to have increased circulating levels of liver-specific miRNAs (−122 and −192).127 Levels of miRNA-25, miRNA-375, and let-7f are increased in the circulation of patients with HBV infection or HBV with HCC.128 Increased levels of miR-885-5p were significantly associated with liver disorders (HCC, cirrhosis, and chronic HBV infection) but not with gastric malignancies.79 In a study of HCV infection and NAFLD, serum levels of miRNA-34a and −122 correlated with fibrosis, steatosis, and inflammatory activity.28 Encouragingly, a panel of plasma miRNAs (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) can be used to diagnose patients with HBV-related HCC.129 In patients with advanced liver fibrosis, circulating levels of miR-29a are significantly reduced.29 In patients with cirrhosis, miR 513-3p and miR-571 are increased whereas miR-652 is reduced.130 The combination of these miRNAs was highly predictive for the presence of liver disease or cirrhosis and superior to classic markers such as albumin, international normalized ratio, or platelet count.
Therapy
As expected from the broad regulatory roles of miRNAs, disrupting their expression has been associated with the onset and progression of several diseases,131–134 making them interesting candidates for therapeutic targeting. However, interest in the biological activities of miRNAs themselves, and their roles in posttranscriptional processing, is what has fueled research in this field. Compounds that disrupt or mimic miRNAs could simultaneously modify multiple pathways and networks, a different approach from inhibitors of single pathway constituents. There are 2 different strategies to miRNA therapeutics that have different strengths and limitations.
miRNA Replacement
miRNA replacement therapy utilizes short RNA duplexes that mimic miRNAs that are underexpressed in certain diseases. In mouse models in which down-regulation of a particular miRNA was associated with disease development, the miRNA mimic was able to reverse or reduce the disease phenotype.50,135,136 As gain-of-function reagents, miRNA mimics restore normal level of the target mRNAs to diseased cells but may also alter processes in healthy cells that normally do not express the miRNA, with unpredictable outcomes. Not surprisingly, therefore, therapeutic development has been focused on miRNAs that are underexpressed in diseased cells but highly expressed in normal tissues, where additional levels would not be likely to have an effect.
Delivery of miR-26 suppresses Myc-induced tumorigenesis in mice.50 Systemic administration of miR-124 can prevent and suppress hepatocarcinogenesis in mice by inducing tumor-specific apoptosis without toxic side effects.47 Mimics to the miRNAs Let-7 and miR-34 are in preclinical development for cancer therapy (see www.mirnatherapeutics.com).
The therapeutic objectives of miRNA mimics (supplementing a missing or underexpressed miRNA) and small inhibitory RNAs (siRNAs) (which inhibit specific mRNA targets) are essentially the same. However, it has been a challenge to develop the technology to safely and effectively deliver siRNAs to target cells. This obstacle has slowed the development of siRNAs as therapeutics and has been more difficult to overcome than initially believed; it is expected to also affect the progress of miRNA replacement therapy.137
miRNA Inhibitors
miRNA inhibitors are chemically modified, single-stranded oligonucleotides that antagonize overexpressed miRNAs. The most widely used miRNA inhibitors, which have been reported to work by either degradation or sequestration of the mature miRNA, are termed antagomirs or antimiRs, respectively. Antagomirs have a complementary sequence to the entire, mature miRNA target, are based on the medium affinity chemistry termed 2′-O-methyl (2′-OMe), are partially phosphorylated and are conjugated to cholesterol.21 Antagomirs have been reported to inhibit target miRNAs, in a dose-dependent manner, in different tissues in mice when administered intravenously as naked molecules.21,138 An antagomir of miR-221 blocked growth of HCC xenograft tumors in mice and prolonged their survival.139
AntimiRs are usually complementary to only part of the mature miRNA target, but bind it with high affinity, and were developed using locked nucleic acid (LNA) chemistry. These small anti-miRNAs have potent activity in a range of tissues in mice, rats, monkeys, and chimpanzees following systemic administration as naked molecules, at doses considerably lower than those of other inhibitors.24,140–145 The antimiR SPC3649 was tested in a phase 2a clinical trial of patients with chronic HCV infection (ClinicalTrials.gov number NCT01200420).
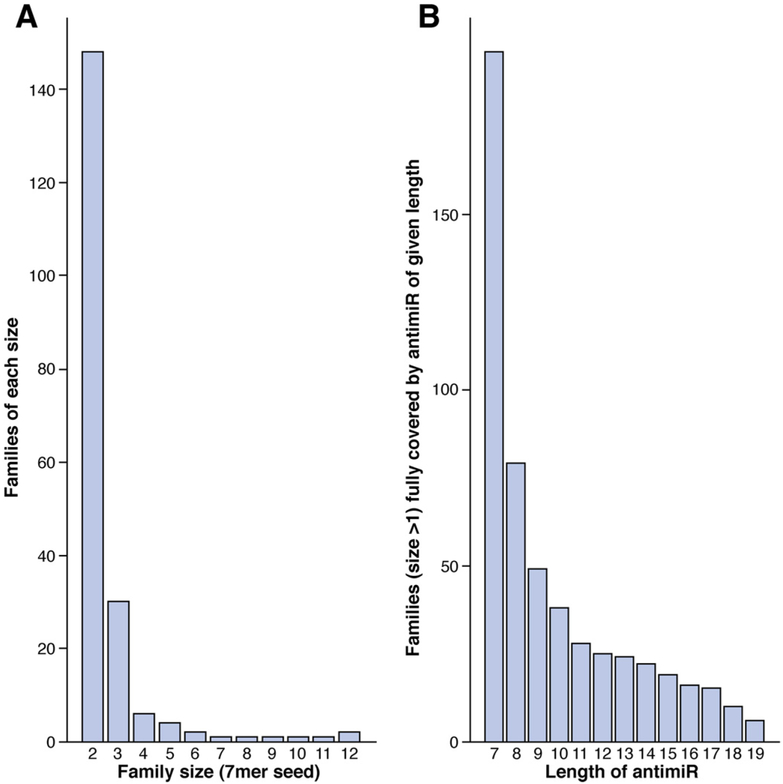
A particular challenge to developing miRNA antagonist therapies is that many human miRNAs are members of functionally redundant families146,147 yet have insufficient sequence similarity to allow them to be simultaneously bound by a typical-size oligonucleotide. According to the miRBase 17, more than 25% of all mature human miRNAs (511 of 1733) are members of a family (based on a 7-seed sequence identity) comprising 2 or more members; some families have more than 10 members (Figure 6A). Oligonucleotides with strong binding at very short sizes are required to target any of these families. Pharmacologically active oligonucleotides (as short as 8-mers) are required to target a significant part of these miRNA families and as short as 7-mers to target all families (see Figure 6B).
Figure 6.
Size of human miRNA families and potential interactions with antimiRs. (A) Number of human miRNA families with 2 or more family members. (B) Number of all human miRNA families that can be targeted by antimiRs with lengths from 7 to 19 nucleotides.
LNA chemistry allows creation of very short oligonucleotides with sufficient affinities to disrupt miRNA targets and such oligonucleotides have strong pharmacologic effects in animals.24 Obad et al recently showed that fully modified, seed-targeted LNA oligos as short as 8-mers, termed tiny-LNAs, concomitantly and potently suppressed entire families of miRNAs in cultured cells.148 Moreover, systemically administered, naked molecules of tiny-LNAs distributed broadly throughout tissues of mice and had potent, dose-dependent, long-lasting activity in liver, kidney, lung, and lung xenograft tumors.
Because they are only 8 nucleotides long, tiny-LNAs encounter many fully complementary sequences in nontarget RNAs and therefore could cause undesired, off-target effects. However, microarray analysis of cultured cells and livers of mice given tiny-LNAs against miR-21, let-7, and miR-122 had the expected up-regulation of the mRNAs targeted by the miRNAs but no detectable effects on mRNAs with target sites complementary to the tiny-LNA. These findings indicate that off-target effects could be limited. This finding was supported at the proteomic level; analyses of protein samples from livers of mice given tiny-LNAs showed the expected effects on protein products of mRNAs with canonical miR-122 target sites but no significant changes in protein products of mRNAs with binding sites for the anti-miR-122 tiny-LNA.
Clinical Development
miRNA-based therapeutics are still in early its stages. More work needs to be done to understand the complex biological function of miRNAs, their roles in pathogenesis, and the safety of compounds that alter their levels. Despite significant advances in recent years, the miRNA mimic and antagonist technologies have also not yet been validated as platforms for the development of safe and effective human therapeutics. However, a fair number of miRNAs have been characterized to the point that they can be considered as attractive therapeutic targets.
The most advanced program is for an antimiR, Miravirsen, that targets miR-122 and which is being developed as a therapeutic for chronic infection with all subtypes of HCV. MiR-122 is a highly abundant, liver-specific miRNA involved in lipid and cholesterol metabolism and in HCV replication.23,149 In preclinical studies in mice140 and green African monkeys,141 Miravirsen, given intravenously as a naked molecule, potently inhibited miR-122 in a dose-dependent manner, resulting in expected increases in predicted mRNA targets and reductions in plasma cholesterol levels. In chimpanzees chronically infected with HCV subtype 1a or 1b, intravenous dosing with Miravirsen once weekly for 12 weeks reduced the viral load in a dose-dependent manner, with a maximum decrease of 2.6 orders of magnitude in serum levels of HCV RNA in the high-dose group.24 Notably, viral suppression lasted up to 3 months after the last dose, with no evidence of viral resistance or adverse effects in any of the treated animals.
In May 2008, Miravirsen became the first miRNA inhibitor to enter clinical trials. Since then, the compound has been tested in 2 phase 1 safety studies in healthy volunteers, a single ascending dose and a 4-week multiple ascending dose study, and a phase 2a study in treatment-naive patients with chronic HCV genotype 1 infection (data not published, November 2011). In all of these studies, the compound was found to be safe, with no dose-limiting toxicities and no discontinuations, but further confirmation is warranted. The 2 phase 1 studies showed some signs of efficacy, which included significant and dose-dependent reductions in plasma levels of cholesterol—a surrogate marker of miRNA-122 disruption by the inhibitor. Preliminary data show a significant decrease of HCV RNA in patients treated with Miravirsen,150 providing the first piece of clinical evidence for miRNAs as therapeutic targets (unpublished observations, March 2012).
Future Directions
Recent findings from integrative and mechanism-based profiling studies have provided important information about the roles of miRNAs in normal cell functions and disease. These studies could improve our understanding of the molecular mechanisms of chronic liver diseases and liver cancer. Viral load affects risk for HCC, so development of antiviral agents will aid chemopreventive measures to reduce the incidence of HCC. Promising agents such as synthetic anti-miR-122 molecules could control HCV infection and reduce the prevalence of HCC. The ability of miR-122 to reduce metastasis of HCC and its down-regulation in metastatic HCC are interesting areas for future studies. Further analyses are needed to determine whether inhibiting miR-122 promotes metastasis.
Liver diseases, including cancer, are complex. Tracking molecular mechanisms of their progression would be better than using phenotypic surrogate parameters, such as patients’ age, sex, tumor size, and liver function, to define outcomes of diseases such as HCC. It is therefore important to increase our understanding of the molecular mechanisms of tumor formation and development. miRNAs appear to be stable and can be readily recovered from formalin-fixed, paraffin-embedded tissues and body fluids such as serum and plasma. These molecules could therefore serve as biomarkers for use in diagnosis of tumor type or predicting response to therapy. Furthermore, miRNAs frequently target mRNAs that encode multiple proteins, such as tumor suppressors and products of oncogenes. HCC cell activities might depend on deregulation of miRNAs, and strategies designed to restore miRNA function might be effective therapies.
Acknowledgments
Funding Supported in part by the Intramural Research Program of the Center for Cancer Research, the US National Cancer Institute (Z01 BC 010876). Dr Heegaard was supported by research grants from the Lundbeck Foundation, the Danish Rheumatism Association, Fonden til Lægevidenskabens Fremme, and Aage Bangs Foundation. Work on SPC3649 at Santaris has been supported by the Danish National Advanced Technology Foundation.
Abbreviations used in this paper:
- ALD
alcoholic liver disease
- HNF
hepatocyte nuclear factor
- LNA
locked nucleic acid
- miRNA
microRNA
- miRNP
protein-microRNA complex
- NAFLD
nonalcoholic fatty liver disease
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- RISC
RNA-induced silencing complex
- siRNA
small inhibitory RNA
- TNF
tumor necrosis factor
Footnotes
Conflicts of Interest
The authors disclose the following: H.Ø. is an employee of Santaris, which develops miR-based therapy. The remaining authors disclose no conflicts.
The authors regret being unable to cite many relevant primary references due to space limitations and thank members of their research teams for their invaluable contribution.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 2006;57:19–53. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002; 99:15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012;22:107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 8.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011;12:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 2006;38:1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008;105:13556–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji J, Wang XW. New kids on the block: diagnostic and prognostic microRNAs in hepatocellular carcinoma. Cancer Biol Ther 2009; 8:1686–1693. [DOI] [PubMed] [Google Scholar]
- 13.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang MH, Chen CJ, Lai MS, et al. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan Childhood Hepatoma Study Group. N Engl J Med 1997;336:1855–1859. [DOI] [PubMed] [Google Scholar]
- 15.Schlutter J Therapeutics: new drugs hit the target. Nature 2011; 474:S5–S7. [DOI] [PubMed] [Google Scholar]
- 16.Carr BI, Flickinger JC, Lotze MT. Hepatobiliary cancers: cancer of the liver In: DeVita VT Jr, Hellman S, Rosenberg SA, eds. Cancer principles & practice of oncology. 5th ed. Philadelphia, PA: Lippincott-Raven, 1997:1087–1114. [Google Scholar]
- 17.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 18.El-Serag HB. Hepatocellular carcinoma. New Engl J Med 2011; 365:1118–1127. [DOI] [PubMed] [Google Scholar]
- 19.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008;9:219–230. [DOI] [PubMed] [Google Scholar]
- 20.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet 2002;3:499–512. [DOI] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–689. [DOI] [PubMed] [Google Scholar]
- 22.Ji J, Yamashita T, Budhu A, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 2009;50:472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005;309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 24.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 2010;327:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011;141:1572–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baltimore D, Boldin MP, O’Connell RM, et al. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 2008;9:839–845. [DOI] [PubMed] [Google Scholar]
- 27.Bala S, Marcos M, Kodys K, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem 2011;286:1436–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cermelli S, Ruggieri A, Marrero JA, et al. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One 2011;6:e23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roderburg C, Urban GW, Bettermann K, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011;53:209–218. [DOI] [PubMed] [Google Scholar]
- 30.Sekiya Y, Ogawa T, Yoshizato K, et al. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem Biophys Res Commun 2011;412:74–79. [DOI] [PubMed] [Google Scholar]
- 31.Murakami Y, Toyoda H, Tanaka M, et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One 2011;6:e16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu J, Kren BT, Xia Z, et al. Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology 2011;54:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006;25:2537–2545. [DOI] [PubMed] [Google Scholar]
- 34.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008;47:897–907. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res 2008;14:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 2008; 47:1955–1963. [DOI] [PubMed] [Google Scholar]
- 37.Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008;47:1223–1232. [DOI] [PubMed] [Google Scholar]
- 38.Ding J, Huang S, Wu S, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol 2010;12:390–399. [DOI] [PubMed] [Google Scholar]
- 39.Cairo S, Wang Y, de Reynies A, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A 2010;107:20471–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma S, Tang KH, Chan YP, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010;7:694–707. [DOI] [PubMed] [Google Scholar]
- 41.Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011;19:232–243. [DOI] [PubMed] [Google Scholar]
- 42.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007;67:6092–6099. [DOI] [PubMed] [Google Scholar]
- 45.Coulouarn C, Factor VM, Andersen JB, et al. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009;28:3526–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji J, Zhao L, Budhu A, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol 2010;52:690–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011;147:1233–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132: 2557–2576. [DOI] [PubMed] [Google Scholar]
- 49.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med 2009;361: 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kota J, Chivukula RR, O’donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008;7:759–764. [DOI] [PubMed] [Google Scholar]
- 52.Meng F, Henson R, Lang M, et al. Involvement of human microRNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006;130:2113–2129. [DOI] [PubMed] [Google Scholar]
- 53.Selaru FM, Olaru AV, Kan T, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 2009;49:1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng F, Wehbe-Janek H, Henson R, et al. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 2008;27:378–386. [DOI] [PubMed] [Google Scholar]
- 55.Olaru AV, Ghiaur G, Yamanaka S, et al. MicroRNA down-regulated in human cholangiocarcinoma control cell cycle through multiple targets involved in the G1/S checkpoint. Hepatology 2011;54: 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hand NJ, Master ZR, Le Lay J, et al. Hepatic function is preserved in the absence of mature microRNAs. Hepatology 2009;49:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takagi S, Nakajima M, Kida K, et al. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 2010;285:4415–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002;12:735–739. [DOI] [PubMed] [Google Scholar]
- 59.Li ZY, Xi Y, Zhu WN, et al. Positive regulation of hepatic miR-122 expression by HNF4alpha. J Hepatol 2011;55:602–611. [DOI] [PubMed] [Google Scholar]
- 60.Kwiecinski M, Noetel A, Elfimova N, et al. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS One 2011;6:e24568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogawa T, Enomoto M, Fujii H, et al. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut 2012. January 20 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 62.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J Immunol 2009;183:5232–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 64.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science 2008;320:97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heo I, Joo C, Cho J, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 2008;32:276–284. [DOI] [PubMed] [Google Scholar]
- 66.Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009;41:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamashita T, Budhu A, Forgues M, et al. Activation of hepatic stem cell marker EpCAM by Wnt-β-catenin signaling in hepatocellular carcinoma. Cancer Res 2007;67:10831–10839. [DOI] [PubMed] [Google Scholar]
- 68.Maetzel D, Denzel S, Mack B, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol 2009;11:162–171. [DOI] [PubMed] [Google Scholar]
- 69.Ji J, Yamashita T, Wang XW. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci 2011;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oishi N, Wang XW. Novel therapeutic strategies for targeting liver cancer stem cells. Int J Biol Sci 2011;7:517–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gantier MP, McCoy CE, Rusinova I, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res 2011;39:5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu A, Tetzlaff MT, Vanbelle P, et al. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol 2009;2:519–527. [PMC free article] [PubMed] [Google Scholar]
- 74.de Planell-Saguer M, Rodicio MC. Analytical aspects of microRNA in diagnostics: A review. Anal Chim Acta 2011;699:134–152. [DOI] [PubMed] [Google Scholar]
- 75.Cissell KA, Deo SK. Trends in microRNA detection. Anal Bioanal Chem 2009;394:1109–1116. [DOI] [PubMed] [Google Scholar]
- 76.Raymond CK, Roberts BS, Garrett-Engele P, et al. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 2005;11:1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005;33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mestdagh P, Feys T, Bernard N, et al. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res 2008;36:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gui J, Tian Y, Wen X, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jang JS, Simon VA, Feddersen RM, et al. Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays. BMC Genomics 2011;12:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317–325. [DOI] [PubMed] [Google Scholar]
- 82.Heegaard NH, Schetter AJ, Welsh JA, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012;130:1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Law PT, Wong N. Emerging roles of microRNA in the intracellular signaling networks of hepatocellular carcinoma. J Gastroenterol Hepatol 2011;26:437–449. [DOI] [PubMed] [Google Scholar]
- 84.Duttagupta R, Jiang R, Gollub J, et al. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One 2011;6: e20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y, Jia Y, Zheng R, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem 2010;56:1830–1838. [DOI] [PubMed] [Google Scholar]
- 86.Miranda RC, Pietrzykowski AZ, Tang Y, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res 2010;34:575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eipper-Mains JE, Kiraly DD, Palakodeti D, et al. microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA 2011;17:1529–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng L, Lv GC, Sheng J, et al. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol 2010;25:156–163. [DOI] [PubMed] [Google Scholar]
- 89.Jin X, Ye YF, Chen SH, et al. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis 2009;41:289–297. [DOI] [PubMed] [Google Scholar]
- 90.Jiang X, Tsitsiou E, Herrick SE, et al. MicroRNAs and the regulation of fibrosis. FEBS J 2010;277:2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marquez RT, Bandyopadhyay S, Wendlandt EB, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest 2010;90:1727–1736. [DOI] [PubMed] [Google Scholar]
- 92.Ach RA, Wang H, Curry B. Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol 2008;8:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Camarillo C, Swerdel M, Hart RP. Comparison of microarray and quantitative real-time PCR methods for measuring MicroRNA levels in MSC cultures. Methods Mol Biol 2011;698:419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 2011;57:833–840. [DOI] [PubMed] [Google Scholar]
- 95.Dweep H, Sticht C, Pandey P, et al. miRWalk - Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 2011;44:839–847. [DOI] [PubMed] [Google Scholar]
- 96.Beitzinger M, Meister G. Experimental identification of microRNA targets by immunoprecipitation of Argonaute protein complexes. Methods Mol Biol 2011;732:153–167. [DOI] [PubMed] [Google Scholar]
- 97.Vinther J, Hedegaard MM, Gardner PP, et al. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res 2006;34:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang Y, Chaerkady R, Kandasamy K, et al. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol Biosyst 2010;6:1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ebner OA, Selbach M. Whole cell proteome regulation by microRNAs captured in a pulsed SILAC mass spectrometry approach. Methods Mol Biol 2011;725:315–331. [DOI] [PubMed] [Google Scholar]
- 100.Seliger B, Jasinski S, Dressler SP, et al. Linkage of microRNA and proteome-based profiling data sets: a perspective for the priorization of candidate biomarkers in renal cell carcinoma? J Proteome Res 2011;10:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seliger B, Dressler SP, Wang E, et al. Combined analysis of transcriptome and proteome data as a tool for the identification of candidate biomarkers in renal cell carcinoma. Proteomics 2009;9:1567–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tseng CW, Lin CC, Chen CN, et al. Integrative network analysis reveals active microRNAs and their functions in gastric cancer. BMC Syst Biol 2011;5:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Satoh J, Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Min 2011;4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaller M, Liffers ST, Oeljeklaus S, et al. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and micro-array analysis. Mol Cell Proteomics 2011;10:M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boutz DR, Collins PJ, Suresh U, et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem 2011;286:18066–18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Le BA, Portales-Casamar E, Vetter G, et al. MIR@NT@N: a framework integrating transcription factors, microRNAs and their targets to identify sub-network motifs in a meta-regulation network model. BMC Bioinformatics 2011;12:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol 2009;41:87–95. [DOI] [PubMed] [Google Scholar]
- 108.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426–437. [DOI] [PubMed] [Google Scholar]
- 110.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39: 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010;285:17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- 116.Heneghan HM, Miller N, Lowery AJ, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 2010;251:499–505. [DOI] [PubMed] [Google Scholar]
- 117.Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu J, Wu C, Che X, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 2011;50:136–142. [DOI] [PubMed] [Google Scholar]
- 119.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008;14:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo HQ, Huang GL, Guo CC, et al. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers 2010;29:251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Asaga S, Kuo C, Nguyen T, et al. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem 2011;57:84–91. [DOI] [PubMed] [Google Scholar]
- 122.Albulescu R, Neagu M, Albulescu L, et al. Tissular and soluble miRNAs for diagnostic and therapy improvement in digestive tract cancers. Expert Rev Mol Diagn 2011;11:101–120. [DOI] [PubMed] [Google Scholar]
- 123.Gao W, Liu L, Lu X, et al. Circulating microRNAs: possible prediction biomarkers for personalized therapy of non-small-cell lung carcinoma. Clin Lung Cancer 2011;12:14–17. [DOI] [PubMed] [Google Scholar]
- 124.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Panarelli NC, Yantiss RK. MicroRNA expression in selected carcinomas of the gastrointestinal tract. Patholog Res Int 2011; 2011:124608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li J, Wang Y, Yu W, et al. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun 2011;406:70–73. [DOI] [PubMed] [Google Scholar]
- 127.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 2009;106:4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li LM, Hu ZB, Zhou ZX, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 2010;70:9798–9807. [DOI] [PubMed] [Google Scholar]
- 129.Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 2011;29:4781–4788. [DOI] [PubMed] [Google Scholar]
- 130.Roderburg C, Mollnow T, Bongaerts B, et al. Micro-RNA profiling in human serum reveals compartment-specific roles of miR-571 and miR-652 in liver cirrhosis. PLoS One 2012;7:e32999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis 2007;28:2–12. [DOI] [PubMed] [Google Scholar]
- 132.Montgomery RL, van Rooij E. Therapeutic advances in MicroRNA targeting. J Cardiovasc Pharmacol 2011;57:1–7. [DOI] [PubMed] [Google Scholar]
- 133.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res 2010;70:7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stenvang J, Lindow M, Kauppinen S. Targeting of microRNAs for therapeutics. Biochem Soc Trans 2008;36:1197–1200. [DOI] [PubMed] [Google Scholar]
- 135.Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 2011;19:1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Takeshita F, Patrawala L, Osaki M, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther 2010;18:181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ledford H Drug giants turn their backs on RNA interference. Nature 2010;468:487. [DOI] [PubMed] [Google Scholar]
- 138.Krutzfeldt J, Kuwajima S, Braich R, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res 2007;35:2885–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Park JK, Kogure T, Nuovo GJ, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res 2011;71:7608–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 2008;36:1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452:896–899. [DOI] [PubMed] [Google Scholar]
- 142.Worm J, Stenvang J, Petri A, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res 2009;37:5784–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kocerha J, Faghihi MA, Lopez-Toledano MA, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A 2009;106:3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rayner KJ, Suarez Y, Davalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010;328: 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010;328:1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 2009;23:2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007;26:3699–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Obad S, dos Santos CO, Petri A, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 2011;43:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87–98. [DOI] [PubMed] [Google Scholar]
- 150.Janssen HL, Reesink HW, Zeuzem S, et al. A randomized, double-blind, placebo (Plb) controlled safety and anti-viral proof of concept study of Miravirsen (Mir), an oligonucleotide targeting miR-122, in treatment naïive patients with genotype 1 (Gt1) chronic HCV infection. Hepatology 2011;54:1430A. [Google Scholar]