Abstract
For several years, it has been known that histone deacetylase inhibitors have the potential to alter the immunogenicity of tumor cells exposed to checkpoint inhibitory immunotherapy antibodies. HDAC inhibitors can rapidly reduce expression of PD-L1 and increase expression of MHCA in various tumor types that subsequently facilitate the antitumor actions of checkpoint inhibitors. Recently, we have discovered that drug combinations which cause a rapid and intense autophagosome formation also can modulate the expression of HDAC proteins that control tumor cell immunogenicity via their regulation of PD-L1 and MHCA. These drug combinations, in particular those using the irreversible ERBB1/2/4 inhibitor neratinib, can result in parallel in the internalization of growth factor receptors as well as fellow-traveler proteins such as mutant K-RAS and mutant N-RAS into autophagosomes. The drug-induced autophagosomes contain HDAC proteins/signaling proteins whose expression is subsequently reduced by lysosomal degradation processes. These findings argue that cancer therapies which strongly promote autophagosome formation and autophagic flux may facilitate the subsequent use of additional antitumor modalities using checkpoint inhibitor antibodies.
1. TEXT ELEMENTS
Histone deacetylase inhibitors have been under investigation as anticancer agents for over 20 years (Zhan, Wang, Liu, & Suzuki, 2017). Simplistically, HDAC inhibitors regulate the acetylation status of histones, proteins that in turn regulate the condensation status of DNA, and the accessibility of promoter and suppressor elements to transcription factors, thereby regulating transcription. However, multiple other cytosolic and nuclear proteins are also regulated by reversible acetylation. Two of the most notable acetylated proteins whose functions are of prime importance in the survival of many tumor cell types are heat shock protein 90 (HSP90) and the p65 subunit of NFκB (Leus, Zwinderman, & Dekker, 2016; Rodrigues, Thota, & Fraga, 2016).
Acetylation of p65 NFκB plays a key role in activation of the transcription factor. For drugs that utilize NFκB signaling as a component of their “cell deathsignal,”e.g., byelevatingTNFαexpression,HDACinhibitorswillfacilitate p65 acetylation and tumor cell killing (Gang, Shaw, Dhingra, Davie, & Kirshenbaum, 2013). However, for drugs that use compensatory NFκB activation to protect themselves from a toxic stress, HDAC inhibitors have the potential via NFκB to suppress cell death (Karthik, Sankar, Varunkumar, Anusha, & Ravikumar, 2015). As single agents at clinically relevant concentrations, HDAC inhibitors often cause modest levels to tumor cell killing; the combination of HDAC inhibitors with agents that block NFκB activation, however, results in a synergy of tumor cell killing (Li, Li, et al., 2016; Li, Zhuang, et al., 2016). Multiple other transcription factors are regulated by reversible acetylation including p53, STAT3, GATA-1, and Sp3 (Formisano et al., 2015; Schäafer et al., 2017; Watamoto et al., 2003; Yuan, Guan, Chatterjee, & Chin, 2005). HSP90 acetylation is regulated by the enzyme HDAC6 and the acetyltransferase that also associates with HSP90, arrest defective-1 protein (ARD1) (DePaolo et al., 2016; Yang, Zhang, Zhang, Zhang, & Xu, 2013). Hyperacetylation of HSP90 has been proposed to cause the release of the cochaperone complex protein p23, and to inhibit the chaperone’s ATPase function, collectively reducing HSP90 chaperoning activity (Bali, Pranpat, Bradner, et al., 2005; Kekatpure, Dannenberg, & Subbaramaiah, 2009; Koga et al., 2006; Rao et al., 2008). Other chaperone proteins, e.g., HSP70 and GRP78 have also been found to be regulated by reversible acetylation (Chang et al., 2016; Li, Li, et al., 2016; Li, Zhuang, et al., 2016; Park, Seo, Park, Lee, & Kim, 2017; Seo et al., 2016). Acetylation of HSP90 has been proposed to regulate it and its client proteins ubiquitination and subsequent proteolytic breakdown (Mollapour & Neckers, 2012; Nanduri, Hao, Fitzpatrick, & Yao, 2015; Quadroni, Potts, & Waridel, 2015; Zhou, Agoston, Atadja, Nelson, & Davidson, 2008).
Immunotherapy, using checkpoint inhibitory antibodies, has become a first line therapeutic regimen in melanoma, NSCLC, bladder cancer, and H&N SCC. Antibodies that blockade the functions of PD-1, PD-L1, and CTLA-4 have all been approved as therapeutics within the last 5 years (Emens et al., 2017; Koller et al., 2016). Histone deacetylase inhibitors are known to increase MHC class I and II expression on the cell surface which would facilitate antitumor responses from both the innate and the adaptive immune systems (Nakajima et al., 2017). HDAC inhibitors have been shown to activate NK cells (Tiper & Webb, 2016). Other studies have linked HDAC inhibitors to both increased and decreased expression of PD-L1 and PD-L2 on tumor cells with the differential effects appearing to be dependent on HDAC inhibitor dose or the cell lines being tested, though all studies argue that HDAC inhibitors enhance the antitumor responses of the immune system using checkpoint inhibitory antibodies (Beg & Gray, 2016; Shen, Orillion, & Pili, 2016; Terranova-Barberio, Thomas, & Munster, 2016; Yang et al., 2015; Zheng etal.,2016). Thus, HDACinhibitors have the potential to enhance the efficacy of checkpoint inhibitory antibodies. And, several pan-HDAC inhibitors, such as sodium valproate, etinostat, panobinostat, and vorinostat, have all been shown in vivo to enhance the antitumor efficacy of checkpoint inhibitory antibodies, as well as promote activated T cell infiltration into the tumors (Booth, Roberts, Poklepovic, & Dent, 2017; Booth, Roberts, Poklepovic, Kirkwood, & Dent, 2017; Christiansen et al., 2011; Gameiro, Malamas, Tsang, Ferrone, & Hodge, 2016; Hornig, Heppt, Graf, Ruzicka, & Berking, 2016; Kroesen et al., 2016; Vo et al., 2009; West & Johnstone, 2014; West et al., 2013). Tumor types tested in these studies are diverse and include melanoma, breast, colorectal, glioblastoma, and hepatoma (Chae, Wang, Nimeiri, Kalyan, & Giles, 2017; Huang et al., 2017; Keller, Zhang, Li, Schaider, & Wells, 2017; Monnot & Romero, 2017; Rai et al., 2017).
There are several FDA-approved HDAC inhibitors and also FDAapproved anti-PD-1 antibodies (pembrolizumab and nivolumab). And, at present there are a number of clinical trials are evaluating the HDAC inhibitor+anti-PD-1 antibody combination in a variety of tumor types. One phase II trial will use the anti-PD-1 antibody pembrolizumab in combination with pan-HDAC inhibitor vorinostat (NCT02638090). Pembrolizumab, nivolumab (anti-PD-1), and atezolizumab (anti-PD-L1) antibodies are being combined with the class I specific HDAC inhibitor entinostat in several clinical trials, respectively (NCT02437136, NCT02697630, NCT01928576, NCT02453620, and NCT02708680). At present, there is an open phase I clinical trial in which the stated HDAC6-specific inhibitor rocilinostat (ACY-241) is being combined with anti-PD-1 and anti-CTLA4 antibodies in malignant melanoma (NCT02935790). The trial NCT01928576 will determine the safety and efficacy of entinostat and azacytidine, a DNA methyl-transferase inhibitor, on the response to nivolumab.
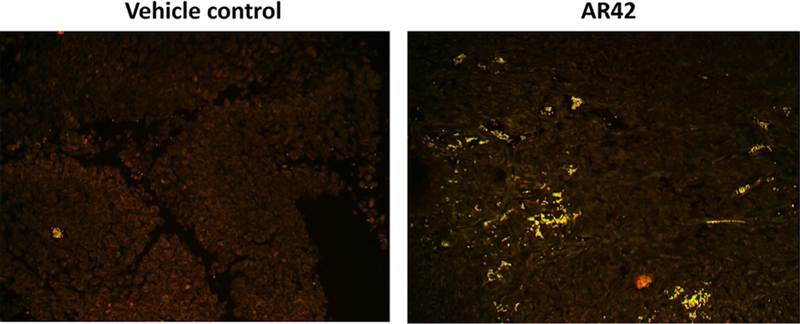
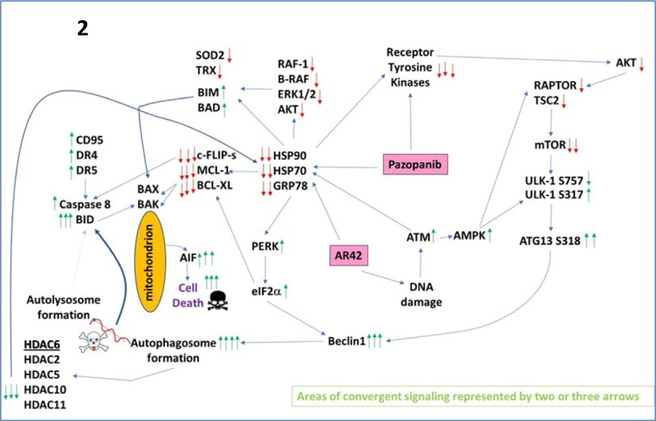
Studies by our laboratory in the field of immunotherapy were initiated in the autumn of 2016 using the pan-HDACi drugs sodium valproate and AR42, and using B16 melanoma cells as an in vivo model system (Booth, Roberts, Poklepovic, Kirkwood, & Dent, 2017; Booth, Roberts, Sander, et al., 2017). Our in vivo studies were predicated on in vitro findings which demonstrated valproate and AR42 both rapidly reduced the expression of PD-L1 and ornithine decarboxylase (ODC) in multiple tumor cell types, and enhanced the expression of class I MHCA. These effects on protein expression were enhanced when the HDAC inhibitor was combined with the multikinase and chaperone inhibitor pazopanib, which together also facilitated the release into the extracellular environment of the immunogenic nuclear protein HMGB1. As presented in Fig. 1, a 35-day prior treatment of B16 melanoma tumors with the pan-HDAC inhibitor AR42 results in the infiltration of tumors with M1-polarized macrophages, which predicts for a strong antitumor immune response. See Fig. 2 for the mechanisms by which pazopanib and HDAC inhibitors interact to kill melanoma cells. Both valproate and AR42 were shown to enhance the anti-B16 tumor efficacy of an anti-PD-1 antibody and of an anti-CTLA4 antibody.
Fig. 1.
The pan-HDAC inhibitor AR42 enhances M1 macrophage infiltration into the tumor 5 weeks after drug exposure. Trametinib/dabrafenib-resistant MEL28 melanoma cells were implanted into the rear flanks of athymic mice. Tumors were treated with vehicle control or with AR42 (15mg/kg) for 2 days. Thirty-five days later, at the time of animal nadir, tumors were fixed, embedded in paraffin, and 4-μm sections obtained. Sections were deparaffinized, renatured, and stained to examine (10 × mag.) the colocalization of F4/80 staining and iNOS staining (that stains for M1 macrophages).
Fig. 2.
A simplified model of the molecular pathways by which pazopanib and AR42 combine to kill cancer cells. As an HDAC inhibitor, AR42 causes DNA damage and causes inhibitory chaperone acetylation. Pazopanib, as a multikinase and chaperone inhibitor, also inhibits chaperone activities as well as many class III receptor tyrosine kinases. DNA damage causes activation of ATM. ATM signals to activate the AMPK. AMPK signaling inactivates RAPTOR and TSC2 resulting in the inactivation of mTORC1 and mTORC2. Downstream of mTOR is the kinase ULK-1; the drug combination via AMPK promotes ULK-1S317 phosphorylation which activates the kinase; the drug combination via mTOR inactivation reduces ULK-1S757 phosphorylation which also activates the kinase. Activated ULK-1 phosphorylates ATG13 which is the key gate-keeper step in permitting autophagosome formation. AR42-induced ATM signaling also acts to reduce the activities of multiple chaperone proteins. Reduced HSP90 and HSP70 function lowers the expression of all receptor tyrosine kinases and the activities of STAT3, STAT5, ERK1/2, and AKT that results in lower expression of ROS/RNS detoxifying enzymes such as TRX and SOD2. Reduced GRP78 function causes activation of PERK and subsequently eIF2α. Enhanced eIF2α signaling reduces the transcription of proteins with short halflives such as c-FLIP-s, MCL-1, and BCL-XL, and enhances expression of Beclin1, DR4, and DR5. Thus, the convergent actions of reduced HSP90 and HSP70 chaperone activity and eIF2α signaling lead to a profound reduction in the protein levels of c-FLIP-s, MCL-1, and BCL-XL which facilitates death receptor signaling through CD95, DR4, and DR5 to activate the extrinsic apoptosis pathway. Enhanced Beclin1 expression converges with elevated ATG13 phosphorylation to produce high levels of autophagosome formation that acts to reduce HDAC2/5/6/10/11 expression but also to stall autophagosome fusion with lysosomes and to stall autolysosome maturation, which likely through cytosolic cathepsin proteases converges with the extrinsic apoptosis pathway to cleave BID and cause mitochondrial dysfunction. Tumor cell killing downstream of the mitochondrion was mediated by AIF and not caspases 3/7. The tumoricidal actions of AIF were facilitated by reduced HSP70 functionality as this chaperone can sequester AIF in the cytosol and prevent its translocation to the nucleus.
In contemporaneous studies, we had discovered that the drug combination of [pemetrexed+sildenafil] reduced the expression of HDAC6 which resulted in elevated HSP90 acetylation (Booth, Roberts, Poklepovic, Gordon, & Dent, 2017). Acetylated HSP90 exhibited lower ATPase activity and had reduced association with client proteins. Although HDAC6 is known to associate with components of the ubiquitin/proteasome system and be ubiquitinated itself, the rapid reduction in HDAC6 expression was prevented by knock down of Beclin1 or ATG5, essential proteins in the regulation of autophagosome formation. The proteasome inhibitor bortezomib did not prevent the degradation of HDAC6. These findings raised the possibility that autophagosome formation could regulate the acetylation of proteins, indirectly, through modulation of HDAC protein levels.
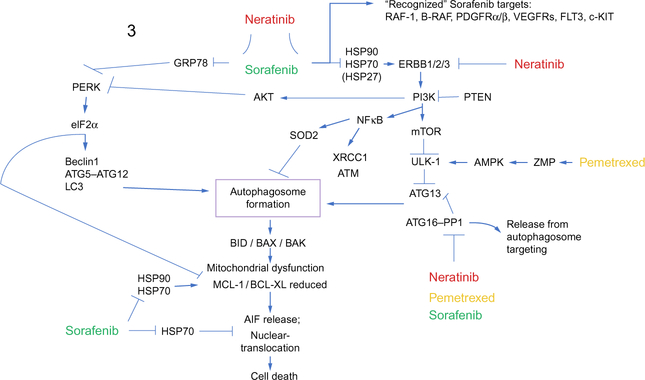
Thus, additional studies were performed to determine whether the [pazopanib+HDAC inhibitor] combination or the previously established drug combinations of [pemetrexed+sorafenib] or [pemetrexed+sildenafil], that we have shown to also kill through autophagy, regulated the expression of not only HDAC6 but also of HDACs1–11 (Booth, Roberts, Poklepovic, Gordon, et al., 2017; Booth, Roberts, Poklepovic, Kirkwood, & Dent, 2017; Booth et al., 2016). It was discovered that [pemetrexed+sildenafil], [pazopanib+HDAC inhibitor], and [pemetrexed+sorafenib] all acted to rapidly, within 6h, reduce the expression of multiple HDAC proteins, and effect blocked by knock down of Beclin1 or of ATG5. See Fig. 3 for the mechanisms by which pemetrexed, sorafenib, and neratinib interact to kill tumor cells. Again, use of bortezomib to block proteasome function did not prevent HDAC degradation. In tumor types tested, including melanoma, sarcoma, lung and ovarian, [pemetrexed+sildenafil], and [pemetrexed +sorafenib] routinely reduced the expression of HDAC2, HDAC4, HDAC6, HDAC9, and HDAC11, respectively. Treatment of tumor cells with[pazopanib+HDACinhibitor]routinelyreducedthelevelsofHDAC1, HDAC2, HDAC3, HDAC5, HDAC6, HDAC7, HDAC9, HDAC10, and HDAC11. Based on the tumor cell, the HDACs responsible for the regulation/inhibition of promoter elements for PD-L1, MHCA, and ODC are reported to be HDAC1, HDAC2, HDAC3, and HDAC10. Molecular studies, knocking down the expression of individual HDAC proteins or combinations of HDAC proteins revealed that PD-L1 expression was coordinately regulated by HDACs1/2/3/10. The expression of MHCA was most often regulated by HDACs1/3/10 and the expression of ODC regulated by HDACs2/3/10. Of note was that the expression of PD-L1, MHCA, and ODC was more weakly modulated by knock down of HDAC6, compared to HDACs1/2/3/10, arguing that the changes in PD-L1, MHCA, and ODC expression were primarily due to altered transcription and not protein stability.
Fig. 3.
A simplified model of the molecular pathways by which pemetrexed, sorafenib, and neratinib combine to kill cancer cells. As a thymidylate synthase inhibitor, pemetrexed causes DNA damage and elevates ZMP levels. ATM phosphorylates the AMPK and ZMP allosterically activates the AMPK which leads to inactivation of mTOR, activation of ULK-1 and ATG13 phosphorylation concomitant with formation of toxic autophagosomes. Sorafenib, in addition to inhibiting RAF kinases and class III RTKs, is an HSP90 family and HSP70 family chaperone inhibitor. Reduced GRP78 function facilitates and endoplasmic reticulum stress response which acts to enhance the levels of Beclin1 and ATG5, facilitating autophagosome formation. The induction of autophagy results in mitochondrial dysfunction which is facilitated by ER stress acting to downregulate MCL-1 and BCL-XL expression. Sorafenib, via inhibition of HSP90 and HSP70, permits AIF, released from the mitochondria, to dissociate from inhibitory chaperones and where it then translocates to the nucleus.
In vivo studies using HDACinhibitors alone or in combination with antiPD-1 or anti-CTLA4 antibodies demonstrated that a transient exposure of established B16 melanoma tumors to HDAC inhibitors resulted in a permanent (~20 days) upregulation of MHCA expression and downregulation of PD-L1 levels within the tumor cells (Booth, Roberts, Poklepovic, Kirkwood, Dent, 2017). These observations in drug-treated tumors correlated with increased infiltration into the tumors of M1 macrophages, NK cells, and activated T cells (see Fig. 1). The presence of M1 macrophages, NK cells, and activated T cells would all be predicted to act in an antitumor fashion to suppress tumor growth. The HDAC inhibitors sodium valproate and AR42 each enhanced the antitumor efficacy of an anti-PD-1 antibody and of an anti-CTLA4 antibody. Other in vivo studies treated Lewis lung carcinoma tumors with [pemetrexed+sildenafil], and determined that this drug combination also enhanced the antitumor efficacy of an anti-PD-1 antibody and of an anti-CTLA4 antibody (Booth, Roberts, Poklepovic, & Dent, 2017). Collectively, these findings all confirm that HDAC inhibitors, and combinations of agents that act to reduce HDAC levels, can enhance the efficacy of checkpoint immunotherapeutic antibodies in vivo.
More recent studies from our laboratory have taken the concept of autophagic degradation of HDAC proteins and other signaling mediators to a new level of complexity. Treatment of tumor cells with clinically relevant concentrations of the irreversible ERBB1/2/4 inhibitor neratinib (100nM), and to a greater extent [neratinib+sodium valproate] also can reduce the expression of HDACs via autophagic degradation (Booth, Roberts, Poklepovic, Avogadri-Connors, et al., 2017). In turn, this results in reduced PD-L1, PD-L2, and ODC expression, and increased MHCA levels. We discovered that [neratinib+sodium valproate] enhanced the antitumor activity of an anti-PD-1 antibody using the 4T1 mouse mammary carcinoma model system. Thus, we predict that many drug combination therapies that promote autophagosome formation and autophagic flux could act to reduce HDAC expression, and thus regulate the protein levels of PD-L1, MHCA, and ODC, and tumor cell immunogenicity.
During our neratinib studies we became aware that neratinib, acting solely as an irreversible ERBB1/2/4 inhibitor, was only one component of a complicated biology for this drug (Booth, Roberts, Poklepovic, AvogadriConnors, et al., 2017; Booth, Roberts, Poklepovic, Kirkwood, Sander, et al., 2017). Initial studies with neratinib were designed to examine the effects ofneratinib on ERBB1phosphorylation in NSCLC cells expressing a mutated active ERBB1 protein. Although neratinib significantly reduced ERBB1 phosphorylation, it also rapidly, within 6h, reduced total ERBB1 expression. In mammary and ovarian cancer cells expressing ERBB2, a similar phenomenon occurred when cells were treated with neratinib. Combined incubation of neratinib-treated cells with either sodium valproate or the novel HDAC inhibitor AR42 enhanced the downregulation of ERBB1 and ERBB2 expression. Knock down of either Beclin1 or ATG5 prevented neratinib or [neratinib+HDACinhibitor] from reducing ERBB1 and ERBB2 expression.
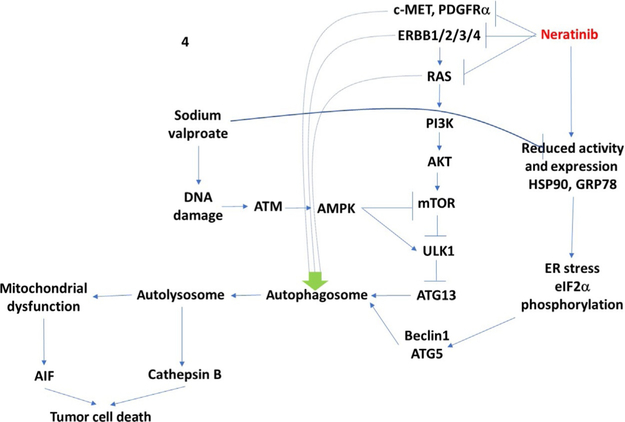
As negative controls in our neratinib studies, we included assessments of changes in ERBB3 and c-MET expression. ERBB3 lacks an active catalytic site and in theory should not bind neratinib, and c-MET, which is not part of the ERBB receptor family, should not bind neratinib. To our surprise, in addition to ERBB1 and ERBB2, neratinib could also reduce both ERBB3 and c-MET expression. The time course of ERBB3 and c-MET downregulation appeared to be slightly slower than that observed for ERBB1 and ERBB2 suggesting their degradation were secondary processes. Knock down of Beclin1 or ATG5 prevented the downregulation of c-MET. Follow up findings also demonstrated that the levels of PDGFRα could be reduced by neratinib in GBM cells (Booth, Roberts, Poklepovic, Kirkwood, Sander, et al., 2017). See Fig. 4 for the mechanisms by which neratinib and sodium valproate interact to kill tumor cells.
Fig. 4.
A simplified model of the molecular pathways by which neratinib and valproate combine to kill cancer cells. As an HDAC inhibitor, sodium valproate causes DNA damage. This activates an ATM–AMPK pathway that inactivates mTOR, activated ULK1, and results in ATG13 phosphorylation and autophagosome formation. Neratinib both inhibits and downregulates the expression of ERBB family receptors as well as other fellow-traveler RTKs and mutant RAS proteins. Sodium valproate enhances HSP90 and GRP78 acetylation which reduces chaperone activity and neratinib reduces the protein levels of HSP90 and GRP78; this results in an endoplasmic reticulum stress response, phosphorylation of eIF2α, and increased expression of Beclin1 and ATG5, that in turn facilitate autophagosome formation. Cathepsin B, from autolysosomes both cleaves BID to cause mitochondrial dysfunction and acts as a direct cell killing protease. Elevated ER stress signaling reduces MCL-1 and BCL-XL levels which facilitates AIF release and AIF translocation to the nucleus where it executes the tumor cell.
The logical progression from these discoveries was to examine the impact of neratinib on the levels of other plasma membrane-associated proteins which can play a tumor-promoting role. Mutated RAS proteins have been considered for several decades to be one of the most important cancerdriving proteins to therapeutically target (McCormick, 2015). RAS proteins, to be active and plasma membrane localized, are prenylated, and this essential modification can be targeted using specific farnesyl transferase inhibitors or inhibitors of HMG CoA reductase (i.e., statins) (Hamed et al., 2008). However, K-RAS and N-RAS proteins, in addition to farnesylation, can also be alternatively geranyl geranylated. As a result, farnesyl transferase inhibitors have largely been unsuccessful in the clinic at controlling RAS-dependent tumors. In our most recent studies, we demonstrated that clinically relevant concentrations of neratinib and [neratinib + valproate] activate the AMPK which in turn phosphorylates and inactivates HMG Co-A reductase (Booth, Roberts, Poklepovic, Kirkwood, Sander, et al., 2017). Reduced HMG Co-A reductase activity will lower the levels of mevalonate that ultimately results in reduced levels of both the farnesyl and geranyl substrates. Thus, the use of the [neratinib + valproate] combination attacks the activity of mutant RAS proteins by direct and indirect mechanisms. The drug combination directly reduces RAS protein levels through a process requiring autophagy and lysosomal degradation, and the combination indirectly reduces RAS function by impeding the ability of the cell to prenylate and facilitate plasma membrane localization of RAS proteins. Thus, our data argue that the degradation of other receptors and RAS proteins by neratinib is a secondary on-target effect of the drug and occurs because of the primary inhibition, internalization, and degradation of ERBB family receptors.
This review began by discussing how the modulation of HDAC function, via HDAC inhibitors or via HDAC protein degradation, could regulate the expression of PD-L1 and MHCA, and tumor cell immunogenicity. As our studies led us toward a more complicated understanding of the mechanisms of action for neratinib, we realized that neratinib alone, or in combination with HDAC inhibitors, could suppress signaling from multiple growth factor receptors and oncogenic mutant RAS proteins. Thus, does receptor-RAS signaling play a role in the protein levels of PD-L1 and MHCA? In NSCLC cells, and other tumor types, e.g., HNSCC, breast cancer, signaling by activated ERBB1 or mutant K-RAS, via the ERK1/2 pathway, acts to increase the expression of PD-L1 and MHCA (El-Jawhari et al., 2014; Ji et al., 2016; Loi et al., 2016; Sers et al., 2009; Sumimoto, Takano, Teramoto, & Daigo, 2016; Yang et al., 2017; Zhang et al., 2017). Inhibitors of ERBB1 or of MEK1/2 suppress the abilities of mutant ERBB1 or mutant RAS from maintaining PD-L1 and MHCA expression. Thus, neratinib and valproate exposure act to enhance tumor cell immunogenicity both via reducing HDAC expression and derepressing ERK1/2 signaling that prevents upregulation of immunotherapy biomarkers.
In conclusion, we have demonstrated that multiple drug combinations that promote autophagosome formation all have the theoretical potential to downregulate the expression of HDAC proteins thereby opsonizing tumor cells to checkpoint immunotherapies. By also reducing signaling through the ERK1/2 pathway, many of these drug combinations also derepress gene expression elements facilitating increases in MHCA and decreases in PD-L1.
ACKNOWLEDGMENTS
Support for the present study was funded from philanthropic funding from Massey Cancer Center, the Universal Inc. Chair in Signal Transduction Research and PHS R01-CA192613.
REFERENCES
- Bali P, Pranpat M, Bradner J, et al. (2005). Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90—A novel basis for antileukemia activity of histone deacetylase inhibitors. The Journal of Biological Chemistry, 280, 26729–26734. [DOI] [PubMed] [Google Scholar]
- Beg AA, & Gray JE (2016). HDAC inhibitors with PD-1 blockade: A promising strategy for treatment of multiple cancer types? Epigenomics, 8, 1015–1017. [DOI] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Poklepovic A, Avogadri-Connors F, Cutler RE, Lalani AS, et al. (2017). HDAC inhibitors enhance neratinib activity and when combined enhance the actions of an anti-PD-1 immunomodulatory antibody in vivo. Oncotarget, 8(52), 90262–90277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Poklepovic A, & Dent P (2017). [Pemetrexed + sildenafil] regulates the immunotherapy response of tumor cells. Cancer Biology and Therapy, 18(9), 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Poklepovic A, Gordon S, & Dent P (2017). PDE5 inhibitors enhance the lethality of pemetrexed through inhibition of multiple chaperone proteins and via the actions of cyclic GMP and nitric oxide. Oncotarget, 8, 1449–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Poklepovic A, Kirkwood J, & Dent P (2017). HDAC inhibitors enhance the immunotherapy response of melanoma cells. Oncotarget, 8, 83155–83170. 10.18632/oncotarget.17950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Poklepovic A, Kirkwood J, Sander C, Avogandri-Connors F, et al. (2017). The levels of mutant K-RAS and mutant N-RAS are rapidly reduced in a Beclin1/ATG5-dependent fashion by the irreversible ERBB1/2/4 inhibitor neratinib. Cancer Biology & Therapy, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Sander C, Lee J, Kirkwood JM, Poklepovic A, et al. (2017). The HDAC inhibitor AR42 interacts with pazopanib to kill trametinib/dabrafenibresistant melanoma cells in vitro and in vivo. Oncotarget, 8, 16367–16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Tavallai M, Chuckalovcak J, Stringer DK, Koromilas AE, et al. (2016). [Pemetrexed + Sorafenib] lethality is increased by inhibition of ERBB1/2/3-PI3K-NFκB compensatory survival signaling. Oncotarget, 7, 23608–23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YK, Wang S, Nimeiri H, Kalyan A, & Giles FJ (2017). Pseudoprogression in microsatellite instability-high colorectal cancer during treatment with combination T cell mediated immunotherapy: A case report and literature review. Oncotarget, 8, 57889–57897. 10.18632/oncotarget.18361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW,Tseng CF,Wang MY,Chang WC,Lee CC,Chen LT,et al. (2016). Deacetylation of HSPA5 by HDAC6 leads to GP78-mediated HSPA5 ubiquitination at K447 and suppresses metastasis of breast cancer. Oncogene, 35, 1517–1528. [DOI] [PubMed] [Google Scholar]
- Christiansen AJ, West A, Banks KM, Haynes NM, Teng MW, Smyth MJ, et al. (2011). Eradication of solid tumors using histone deacetylase inhibitors combined with immune-stimulating antibodies. Proceedings of the National Academy of Sciences of the United States of America, 108, 4141–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaolo JS, Wang Z, Guo J, Zhang G, Qian C, Zhang H, et al. (2016). Acetylation of androgen receptor by ARD1 promotes dissociation from HSP90 complex and prostate tumorigenesis. Oncotarget, 7, 71417–71428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Jawhari JJ, El-Sherbiny YM, Scott GB, Morgan RS, Prestwich R, Bowles PA, et al. (2014). Blocking oncogenic RAS enhances tumour cell surface MHC class I expression but does not alter susceptibility to cytotoxic lymphocytes. Molecular Immunology, 58, 160–168. [DOI] [PubMed] [Google Scholar]
- Emens LA, Ascierto PA, Darcy PK, Demaria S, Eggermont AMM, Redmond WL, et al. (2017). Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. European Journal of Cancer, 81, 116–129. [DOI] [PubMed] [Google Scholar]
- Formisano L, Guida N, Valsecchi V, Cantile M, Cuomo O, Vinciguerra A, et al. (2015). Sp3/REST/HDAC1/HDAC2 complex represses and Sp1/HIF-1/p300 complex activates ncx1 gene transcription, in brain ischemia and in ischemic brain preconditioning, by epigenetic mechanism. The Journal of Neuroscience, 35, 7332–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro SR, Malamas AS, Tsang KY, Ferrone S, & Hodge JW (2016). Inhibitors of histone deacetylase 1 reverse the immune evasion phenotype to enhance T-cell mediated lysis of prostate and breast carcinoma cells. Oncotarget, 7, 7390–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang H, Shaw J, Dhingra R, Davie JR, & Kirshenbaum LA (2013). Epigenetic regulation of canonical TNFα pathway by HDAC1 determines survival of cardiac myocytes. American Journal of Physiology. Heart and Circulatory Physiology, 304, H1662–9. [DOI] [PubMed] [Google Scholar]
- Hamed H, Mitchell C, Park MA, Hanna D, Martin AP, Harrison B, et al. (2008). Human chorionic gonadotropin (hCG) interacts with lovastatin and ionizing radiation to modulate prostate cancer cell viability in vivo. Cancer Biology & Therapy, 7, 587–593. [DOI] [PubMed] [Google Scholar]
- Hornig E, Heppt MV, Graf SA, Ruzicka T, & Berking C (2016). Inhibition of histone deacetylases in melanoma—A perspective from bench to bedside. Experimental Dermatology, 25, 831–838. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu F, Liu Z, Tang H, Wu H, Gong Q, et al. (2017). Immune checkpoint in glioblastoma: Promising and challenging. Frontiers in Pharmacology, 8, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji M, Liu Y, Li Q, Li X, Ning Z, Zhao W, et al. (2016). PD-1/PD-L1 expression in non-small-cell lung cancer and its correlation with EGFR/KRAS mutations. Cancer Biology & Therapy, 17, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik S, Sankar R, Varunkumar K, Anusha C, & Ravikumar V (2015). Blocking NF-κB sensitizes non-small cell lung cancer cells to histone deacetylase inhibitor induced extrinsic apoptosis through generation of reactive oxygen species. Biomedicine & Pharmacotherapy, 69, 337–344. [DOI] [PubMed] [Google Scholar]
- Kekatpure VD, Dannenberg AJ, & Subbaramaiah K (2009). HDAC6 modulates Hsp90 chaperone activity and regulates activation of aryl hydrocarbon receptor signaling. The Journal of Biological Chemistry, 284, 7436–7445. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Keller HR, Zhang X, Li L, Schaider H, & Wells JW (2017). Overcoming resistance to targeted therapy with immunotherapy and combination therapy for metastatic melanoma. Oncotarget, 8, 75675–75686. 10.18632/oncotarget.18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga F, Xu W, Karpova TS, McNally JG, Baron R, & Neckers L (2006). Hsp90 inhibition transiently activates Src kinase and promotes Src-dependent Akt and Erk activation. Proceedings of the National Academy of Sciences of the United States of America, 103, 11318–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller KM, Wang W, Schell TD, Cozza EM, Kokolus KM, Neves RI, et al. (2016). Malignant melanoma—The cradle of anti-neoplastic immunotherapy. Critical Reviews in Oncology/Hematology, 106, 25–54. [DOI] [PubMed] [Google Scholar]
- Kroesen M, Büll C, Gielen PR, Brok IC, Armandari I, Wassink M, et al. (2016). Anti-GD2 mAb and Vorinostat synergize in the treatment of neuroblastoma. Oncolmmunology, 5, e1164919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leus NG, Zwinderman MR, & Dekker FJ (2016). Histone deacetylase 3 (HDAC 3) as emerging drug target in NF-κB-mediated inflammation. Current Opinion in Chemical Biology, 33, 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li QZ, Chen L, Chen BD, Wang B, Zhang XJ, et al. (2016). Histone deacetylase inhibitor RGFP109 overcomes temozolomide resistance by blocking NF-κB-dependent transcription in glioblastoma cell lines. Neurochemical Research, 41, 3192–3205. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhuang M, Zhang L, Zheng X, Yang P, & Li Z (2016). Acetylation modification regulates GRP78 secretion in colon cancer cells. Scientific Reports, 6, 30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, et al. (2016). RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: Therapeutic cooperation between MEK and PD-1/PDL1 immune checkpoint inhibitors. Clinical Cancer Research, 22, 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F (2015). KRAS as a therapeutic target. Clinical Cancer Research, 21, 1797–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, & Neckers L (2012). Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochimica et Biophysica Acta, 1823, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnot GC, & Romero P (2017). Rationale for immunological approaches to breast cancer therapy. Breast. 10.1016/j.breast.2017.06.009. pii: S0960–9776 (17)30477–0. [DOI] [PubMed] [Google Scholar]
- Nakajima NI, Niimi A, Isono M, Oike T, Sato H, Nakano T, et al. (2017). Inhibition of the HDAC/Suv39/G9a pathway restores the expression of DNA damagedependent major histocompatibility complex class I-related chain A and B in cancer cells. Oncology Reports, 38, 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri P, Hao R, Fitzpatrick T, & Yao TP (2015). Chaperone-mediated 26S proteasome remodeling facilitates free K63 ubiquitin chain production and aggresome clearance. The Journal of Biological Chemistry, 290, 9455–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, Seo JH, Park JH, Lee HS, & Kim KW (2017). Hsp70 acetylation prevents caspase-dependent/independent apoptosis and autophagic cell death in cancer cells. International Journal of Oncology, 51, 573–578. 10.3892/ijo.2017.4039. [DOI] [PubMed] [Google Scholar]
- Quadroni M, Potts A, & Waridel P (2015). Hsp90 inhibition induces both proteinspecific and global changes in the ubiquitinome. Journal of Proteomics, 120, 215–229. [DOI] [PubMed] [Google Scholar]
- Rai V, Abdo J, Alsuwaidan AN, Agrawal S, Sharma P, & Agrawal DK (2017). Cellular and molecular targets for the immunotherapy of hepatocellular carcinoma. Molecular and Cellular Biochemistry. 10.1007/s11010-017-3092-z. [DOI] [PubMed] [Google Scholar]
- Rao R, Fiskus W, Yang Y, Lee P, Joshi R, Fernandez P, et al. (2008). HDAC6 inhibition enhances 17-AAG-mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood, 112, 1886–1893. [DOI] [PubMed] [Google Scholar]
- Rodrigues DA, Thota S, & Fraga CA (2016). Beyond the selective inhibition of histone deacetylase 6. Mini Reviews in Medicinal Chemistry, 16, 1175–1184. [DOI] [PubMed] [Google Scholar]
- Schäfer C, Göder A, Beyer M, Kiweler N, Mahendrarajah N, Rauch A, et al. (2017). Class I histone deacetylases regulate p53/NF-κB crosstalk in cancer cells. Cellular Signalling, 29, 218–225. [DOI] [PubMed] [Google Scholar]
- Seo JH, Park JH, Lee EJ, Vo TT, Choi H, Kim JY, et al. (2016). ARD1mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nature Communications, 7, 12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sers C, Kuner R, Falk CS, Lund P, Sueltmann H, Braun M, et al. (2009). Downregulation of HLA Class I and NKG2D ligands through a concerted action of MAPK and DNA methyltransferases in colorectal cancer cells. International Journal of Cancer, 125, 1626–1639. [DOI] [PubMed] [Google Scholar]
- Shen L, Orillion A, & Pili R (2016). Histone deacetylase inhibitors as immunomodulators in cancer therapeutics. Epigenomics, 8, 415–428. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Takano A, Teramoto K, & Daigo Y (2016). RAS-mitogen-activated protein kinase signal is required for enhanced PD-L1 expression in human lung cancers. PLoS One, 11, e0166626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova-Barberio M, Thomas S, & Munster PN (2016). Epigenetic modifiers in immunotherapy: A focus on checkpoint inhibitors. Immunotherapy, 8, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiper IV, & Webb TJ (2016). Histone deacetylase inhibitors enhance CD1d-dependent NKT cell responses to lymphoma. Cancer Immunology, Immunotherapy, 65, 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo DD, Prins RM, Begley JL, Donahue TR, Morris LF, Bruhn KW, et al. (2009). Enhanced antitumor activity induced by adoptive T-cell transfer and adjunctive use of the histone deacetylase inhibitor LAQ824. Cancer Research, 69, 8693–8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watamoto K, Towatari M, Ozawa Y, Miyata Y, Okamoto M, Abe A, et al. (2003). Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation. Oncogene, 22, 9176–9184. [DOI] [PubMed] [Google Scholar]
- West AC, & Johnstone RW (2014). New and emerging HDAC inhibitors for cancer treatment. The Journal of Clinical Investigation, 124, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, Mattarollo SR, Shortt J, Cluse LA, Christiansen AJ, Smyth MJ, et al. (2013). An intact immune system is required for the anticancer activities of histone deacetylase inhibitors. Cancer Research, 73, 7265–7276. [DOI] [PubMed] [Google Scholar]
- Yang H, Chen H, Luo S, Li L, Zhou S, Shen R, et al. (2017). The correlation between programmed death-ligand 1 expression and driver gene mutations in NSCLC. Oncotarget, 8, 23517–23528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Lan P, Hou Z, Guan Y, Zhang J, Xu W, et al. (2015). Histone deacetylase inhibitor SAHA epigenetically regulates miR-17–92 cluster and MCM7 to upregulate MICA expression in hepatoma. British Journal of Cancer, 112, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PH, Zhang L, Zhang YJ, Zhang J, & Xu WF (2013). HDAC6: Physiological function and its selective inhibitors for cancer treatment. Drug Discoveries & Therapeutics, 7, 233–242. [DOI] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, & Chin YE (2005). Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science, 307, 269–273. [DOI] [PubMed] [Google Scholar]
- Zhan P, Wang X, Liu X, & Suzuki T (2017). Medicinal chemistry insights into novel HDAC inhibitors: An updated patent review (2012–2016). Recent Patents on Anti-Cancer Drug Discovery, 12, 16–34. [DOI] [PubMed] [Google Scholar]
- Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu S, et al. (2017). Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. OncoTargets and Therapy, 10, 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhao W, Yan C, Watson CC, Massengill M, Xie M, et al. (2016). Inhibitors enhance T-cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clinical Cancer Research, 22, 4119–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Agoston AT, Atadja P, Nelson WG, & Davidson NE (2008). Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Molecular Cancer Research, 6, 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
FURTHER READING
- Mazzone R, Zwergel C, Mai A, & Valente S (2017). Epi-drugs in combination with immunotherapy: A new avenue to improve anticancer efficacy. Clinical Epigenetics 9, 59 10.1186/s13148-017-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann I, Greve G, Jung M, & Lübbert M (2016). Epigenetic therapy approaches in non-small cell lung cancer: Update and perspectives. Epigenetics, 11, 858–870. [DOI] [PMC free article] [PubMed] [Google Scholar]