Abstract
Introduction:
The most cases of cancer death, which are in the first rank among cancers suffered by women is breast cancer. The breast cancer therapy for patients has been done, but still not optimal, so it is necessary to understand the mechanism of therapy in model cell of breast cancer.
Aim:
This study aim to develop an isolation technique of breast cancer cell from patients as a cancer cell model.
Material and methods:
Breast cancer cell isolation is performed by enzymatic methods using the collagen I and hyaluronidase. Then, breast cancer cells were characterized using flow cytometry based on the CD44/CD24 expression where MDA-MB468 and MCF-7 breast cancer cell lines were used as positive controls. Estrogen receptor (ER), progesterone receptor (PR), p53, HER2, and Ki67 expression were assessed using an immunohistochemistry assay.
Result and Discussion:
The morphology of cancer cells was fibroblast like cells on the day 7th after isolation. Isolated breast cancer cells expressed 95.33±0.47% of CD44+/CD24+ and human epidermal growth factor receptor 2 (HER2) low expressions. Isolation of breast cancer cells can use In-house enzymatic protocol. Isolated breast cancer showed the same expression as MDA-MB468 (CD44+/CD24+) and HER2- compared to MCF-7 cell lines (CD44-/CD24+).
Conclusion:
These cells belonged to a basal type of breast carcinoma and expressed CD44+/CD24+, then isolated BCCs can be used as model cancer cells for further research.
Keywords: breast cancer, cell lines, CD44 antigen, CD24 antigen, enzymatic assay
1. INTRODUCTION
Breast cancer is the first rank among cancers suffered by woman causing death. It often occurs recurrence after treatment (1). In the Asia-Pacific, nearly a quarter of the population of women were diagnosed with breast cancer (24%), with the largest percentage occurring in China (46%), Japan (14%), and Indonesia (12%). The type of cancer in Indonesia is dominated by breast cancer (30%) and cervical cancer (24%) (2).
Breast cancer classification was based on different parameter, i.e.: tumor grade, lymph node status, histological type, the presence of estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2). Molecular profiling proved that breast cancer could be classified into: normal, basal, luminal A, luminal B, and HER2 based on ERα, progesterone receptor (PR) and HER2 expression in immunohistochemical assay (3). MDA-MB453 is HER2+ type which expresses high Ki67 (4), while MDA-MB-468 is a basal type with ER-, PR- and HER2- expression and MCF7 is luminal A type with of ER+, PR+ and HER2- expression (5).
Breast cancer treatment depends on the stadium of breast cancer. Therefore, the selection of therapy at each stadium of the cancer is different (6). Surgery and chemotherapy are types of cancer therapy, which is effective to treat breast cancer (7). But, surgical therapy is costly and high risk of infection during surgery, as well as patient complaints regarding cosmetics (8). Chemotherapy can destroy normal cells and cause side effects, including: neutropenia, anemia, thrombocytopenia, alopecia, mucositis (8). The cell line is the most important factor in studying therapeutics target, carcinogenesis, and signal pathways for breast cancer (9).
2. AIM
The cell line of study is based on its biological aspects. Hence, research needs to be done on breast cancer isolation. Therefore, it is essential to have isolated breast cancer cells from breast cancer patient as a cancer cell model in order to determine the effective cancer therapy.
3. MATERIAL AND METHOD
Sample Preparation
The sample was the tissue from breast cancer patients 52 years old with ductal carcinoma mammae sinistra grade III and invasive to lymphovascular. The informed consent used the guidelines approved by the Institutional Ethics Committee collaboration between Maranatha Christian University, Bandung, Indonesia and Immanuel Hospital Bandung, Bandung, Indonesia. Tissue was then washed using phosphate buffer saline (PBS 1x, Gibco 1740576) and preserved in the transfer medium consist of PBS 1x + 1% Abam (Gibco, 1772653) + 1% Amphotericin B (Gibco, 15290026) + 0.1% Gentamicin (Gibco, 15750060) before being isolated (10).
Breast Cancer Cell Isolation
Medium preparation: Dulbecco’s Modified Eagle Medium (DMEM, Gibco 11995065) : F-12K (Sigma, I6634) (1:1), 5% Fetal Bovine Serum (FBS, Gibco, 10270106), 1% Antibiotic-antimycotic (Gibco, 1772653), 1% Amphotericin B (Gibco, 15290026), 0.1% Gentamicin (Gibco, 15750060) were dissolved then filtered using Syringe Filter PES 0.22µm (TPP, 99722). Enzyme preparation: Collagenase Type I (Gibco, 10114532) 4 mg/ml, hyaluronidase (Sigma, 515397) 1 mg/ml, trypsin 0.1% (Gibco, 25200072) were dissolved in basal medium that consists of DMEM: F-12K (1:1), then filtered using syringe filter 0,22 µm PES (TPP, 99722).
Cells Isolation: Prepare a surgical device, PBS 1x and sterile petri dish. The tumor tissue was washed with PBS 1x on a petri dish and then cut it into small pieces using surgical scalpel. The sample fragments were washed again with PBS 1x (Gibco, 1740576), inserted into a 50 ml falcon tube (TPP, 91050) containing the mixed enzyme solution and then incubated 16 hours, 300 rpm, 37°C, 5% CO2. After incubation sample was filtered using 100 μm cell strainer (Corning, 431752) then centrifuged at 1600 rpm for 10 min. Supernatant was discarded and the pellet was washed with PBS 1x and centrifuged at 1600 rpm for 5 min. The pellet was then resuspended with the complete growth medium. The cell suspension was next incubated at 37°C, 5% CO2 (10).
Breast Cancer Cells Characterization
Breast Cancer Cells (BCCs) at P3 were characterized using flow cytometry (Macsquant, Analyzer 10) for positive and negative markers of breast cancer cells at density 1x105 cells/tube. The cells were then stained with CD44 and CD24 according to manufacturer protocol (BioLegend kit, 338804/ 311118). The experiments and measurements of surface marker were performed in triplicate (11).
Proliferation Assay
BCC was cultured in medium consisting of DMEM: F-12K (1:1), 5% FBS, 1% Antibiotic-antimycotic, 1% Amphotericin B, 0.1% Gentamicin and the medium was replaced every 3 days. Briefly 3 x 105 cells were seeded on T25 flask and cultured in complete medium until reaching confluence 80% around 3 days for proliferation assay. Briefly, cultured cells was detached using trypsin, then incubated for 1-3 min at 37oC, complete medium was added to neutralize trypsin and the detached cells were centrifuged (MPW-2000) at 1600 rpm for 5 min at room temperature. The cell pellet was resuspended with trypan blue solution (Sigma, 25200072) and diluted in 1:1 dilution. Then, cells were counted using a haemocytometer (Neubauer, 17849). Population Doubling (PD) was counted at every passage with the formula:
PD = [log10(NH) − log10(NI)]/log10
NI is the inoculum cell number and NH is the cell harvest number. Then, to determine cumulative PD data by added the PD at the previous passage.
The PD time (PDT) was determined by the formula (11).
PD time = t (time)/PD (in days)
Immunohistochemistry (IHC) Analysis
Breast cancer cells were processed using anti-S727p-STAT3 (BD Biosciences), anti-CA-IX and anti-EGFR (clone 31G7, Zymed Technologies, Invitrogen, Burlington, ON) protocols. The Idetect Ultra HRP Detection System (ID Labs Inc, London, Ontario) was used to visualize. Counterstaining was performed with Gill modified hematoxylin (Harleco®) (12).
Statistical Analysis
Statistical analysis was conducted using SPSS software (version 20.0). Data were presented as Mean ± Standard Deviation. Significant differences among treatments were determined using the one-way Analysis of variance (ANOVA) and p < 0.05 were considered as statistically significant, along with Turkey honestly significant difference post hoc test and 95% confidence interval. Z score values were used for plotting the protein expression based on immunohistochemistry signal. Clustering methods based UPGMA statistical analysis were performed by R software ver.3.4.3.
4. RESULTS
Isolation of BCCs with enzymatic method
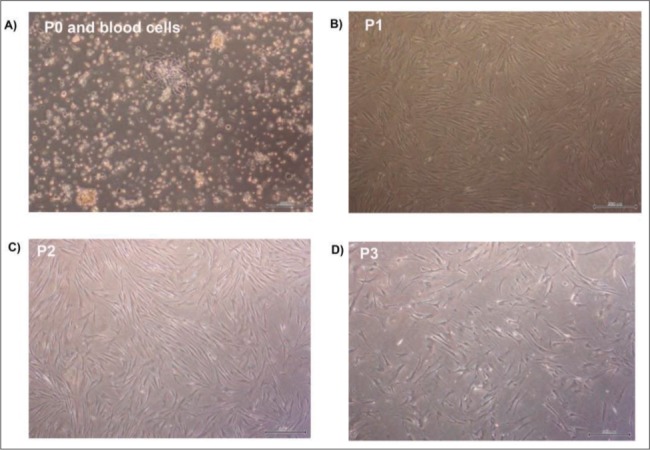
BCCs were successfully isolated using an enzymatic method and cultured until passage 9. The cells attached to the surface of the plastic disc a day after seeding (P0) and were maintained up to two weeks. The cells showed short fibroblastic-like morphology in growth medium and there were still blood cells at the beginning of P0 (Figure 1A). From the first passage onwards, particularly at P1, P2, P3, the cells in growth medium changed their shape into long fibroblast cells (Figure 1B-1D).
Figure 1. The morphology of breast cancer cells in the different passages. (A) BCCs at P0 and blood cells. (B) BCCs at P1. (C) BCCs at P2. (D) BCCs at P3. Magnification 40x. Scale Bars: 500uM.
Characteristic of Breast Cancer Cells
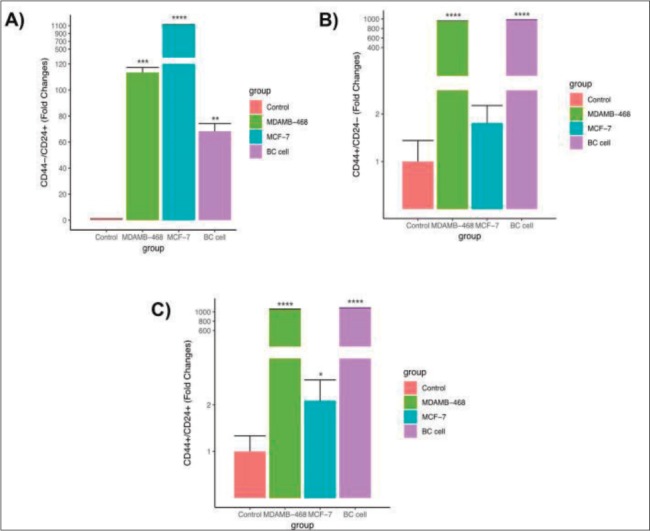
The flow cytometry analysis displayed the biological properties of BCCs, which was compared to MDA-MB468 and MCF-7 as cells control. BCCs showed a highest CD44+ cell intensity expression compared to the MCF-7 cell line, but had no significant difference with MDA-MB468. The intensity of CD44+ in most cells around 101 (log1) (Figure 2). However, MDA-MB468 and BCCs had a lower number of CD44-/CD24+ cells than MCF-7. On the other hand, CD44+/CD24- and CD44+/CD24+ of BCCs and MDA-MB468 were higher than MCF-7 (Figure 3).
Figure 2. Density plot CD44 expressions in the unstained cell as a control (A), MDAMB-468 cell (B), MCF-7 cell (C), and breast cancer cell/BC cell (D).
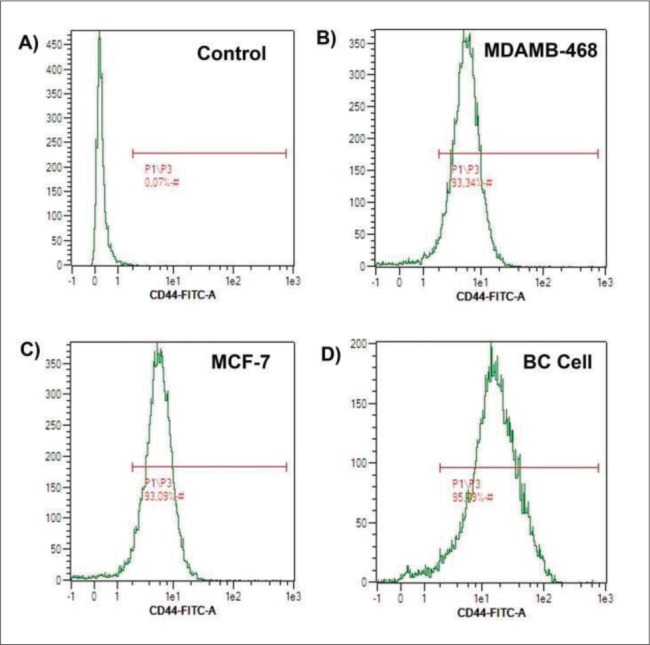
Figure 3. Expression of CD44-/CD24+ cell (A), CD44+/CD24- cell (B) and CD44+/CD24+ cell (C) characterization in breast cancer cell (BC cell) compared to MCF-7 cell lines and MDAMB-468 cell lines. Significance difference with p value p<0.05.
Population Doubling Time of Breast Cancer Cells
PDT and PD rate were used to evaluate the breast cancer cell proliferation capacity. PDT is the time needed by cell to divide into two cells. According to the results shown in Table 1, BCCs suggested high PDT at early P2 and P3 around 21.38±1.34 and 18.10±1.69 respectively compared to late passages (P7 and P8). It could be seen that BCCs reduced their PDT after every passaging (Table 1).
Immunohistochemistry (IHC) and Gene Correlation Analysis
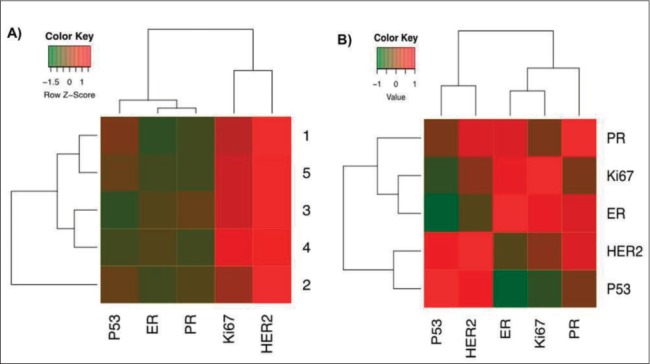
IHC analysis was identified for immunoprofile of breast tissue from breast cancer patient (Figure 4). This procedure was performed to double check flow cytometry analysis of isolated BCCs in order to determine the molecular classification of cancer cell subtype. Five gene analysis (p53, ER, PR, Ki67, HER2) using an IHC staining method showed that HER2 was the strongest upregulated gene expression followed by Ki67 and other genes were down regulated (Figure 5A). In contrast, DeFazio-Eli et al. (2011), Holliday and Speirs (2011) reported that MDA-MB468 suggested very low level of HER2. In a gene correlation analysis using Pearson method, HER2 had a high positive correlation with p53, PR, ER and low positive correlation with Ki67 (Figure 5B).
Figure 4. Immunoprofile of breast tissue from breast cancer patient. Immunohistochemistry result of HER2 expression in breast cancer.
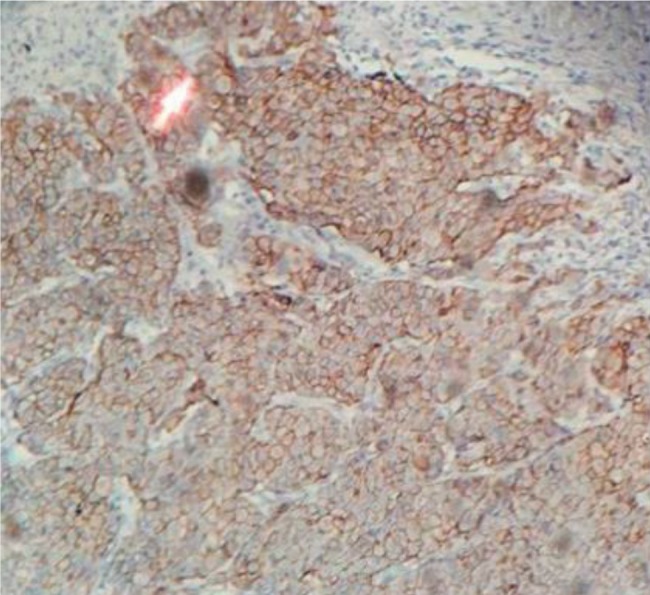
Figure 5. Gene Correlation Analysis (A) Heatmap protein expressions in breast cancer were analyzed by immunohistochemistry. (B) heatmap correlation graph by “Pearson” analysis in five surface protein expressions.
5. DISCUSSION
There are two kinds of breast cancer, namely ductal and lobular carcinoma that attack the milk duct and the milk producing gland respectively (13). Breast cancer therapy, which has existed is still not optimal. Surgery therapy was costly and has a risk of infection during surgery (8). It also often occurs recurrence after treatment (1). Chemotherapy also has not shown optimal results, because the chemicals also target normal cells in the body, so that they can harm cells in the body. This causes new issues are alternative therapy is needed to determine the mechanism and causes of breast cancer. Therefore, to understand the mechanism in breast cancer cell, model cell is needed. Even though utilization of cell lines models cell for most the basic research, they are already engineered for some reasons which may not clinically related to the population of breast cancer.
The morphology of isolated BCCs at P0 show different shape compared to BCCs at P1 onwards. This is caused by the adaptation ability of isolated BCCs in a new environment. Since isolated BCCs have not only similar biological properties to MDA-MB468 but also its morphology like fibroblast cells (14). Isolated BCCs seem to keep with a good state in fibroblast like morphology at each passage.
This study uses MCF-7 and MDA-MB468 as a comparison because both cell lines are breast cancer cell lines, MCF-7 is the luminal type with ER+ and PR+ and MDA-MB468 is the basal type with ER-, PR- and HER2- breast cancer cell line (5). IHC analysis suggests that isolated BCCs are the basal type of breast cancer cell. This finding highlights the importance of classifying isolated BCCs based on molecular classification using IHC assay and surface marker expression using flow cytometry analysis into consideration of clinical cancer therapy.
The isolated BCCs express low HER2 expression and high Ki67 expression based on IHC analysis. It is in line with research from DeFazio-Eli et al. that MDA-MB468 cell line has also very low levels of HER2. Also, Holliday and Speirs (3) mention that MDA-MB468 is a basal type because this cell line has ER-, PR- and HER2- and high Ki67 characteristic around 20%. Ki67 expression ranging under 50%, while cancer cell lines ranging from 20%-100% is one explanation of striking differences between cell lines and isolated primary tumor cell (9). The similarities between basal and triple-negative phenotypes cannot guarantee the type of breast cancer. In line with recent studies, there is still no convincing that triple negative is a feature of basal type cancer (15). Therefore, our finding can be assumed that isolated BCCs have the same characteristics as MDA-MB468 according to IHC assay and flow cytometry analysis.
According to the results shown in Table 1, breast cancer cell proliferation in the second passage need 21 days to doubling their population. While in the third and fourth passage need 18 day and 13 day to doubling their population respectively. In earlier passages, the cell takes time to proliferate because it adapts to the new environment, however, in the next passage the cell has adapted well and high proliferate. Furthermore, doubling time in cancer cells is faster than other cells isolated from human. It is because cell cancer is a malignant cell that has a fast proliferation time than normal cells. It is supported by Liu et al. (16) she states that malignant cells have faster doubling times compared to other normal cells. The doubling time for MDA-MB468 (ATCC) should be 30-40 hours (17), while in this study PDT is 5,73 days in passage 8. This indicates that the isolated cells had a lower proliferation rate compared to MDA-MB468.
6. CONCLUSION
This study concludes that BCCs could express CD44+/CD24+, then expected act as model breast cancer cells effectively.
Acknowledgment:
This research was financially supported by the Ministry of Research, Technology and Higher Education of the Republic of Indonesia and for research grant 2018 (Penelitian Dasar Unggulan Perguruan Tinggi). This research was also supported by Al Ihsan Hospital, Bandung also Aretha Medika Utama Biomolecular and Biomedical Research Center, Bandung, Indonesia for the research methodology and laboratory facilities. We are thankful to Ni Luh Wisma Eka Yanti, Rismawati Laila Qodariah, Hanna Sari W Kusuma, Satrio Haryo Wibowo, Ubaydillah Zedd Munshy from Aretha Medika Utama Biomolecular and Biomedical Research Center, Bandung, Indonesia for their valuable assistance.
Author’s contribution:
This work was carried out in collaboration between all authors. W.W., supervised the laboratory work, contributed to the experimental design and the protocol (writing of the manuscript). Y.H., D. R. L., D. K. J. and T. L. W. carried out laboratory work, contributed to the experimental design and the protocol. R. R., F. S. P., A. A., A. A., and Z. K. carried out laboratory work, the experimental design and contributed to the protocol. A. F. and M. S. performed the statistical analysis and contributed to the protocol. All authors read and approved the final manuscript.
Financial support and sponsorship:
None.
Conflict of interest:
There are no conflict of interest.
REFERENCES
- 1.Huang J, Wu L, Tashiro S, Onodera S, Ikejima T. The augmentation of TNFα induced cell death in murine L929 fibrosarcoma by pan-caspase inhibitor Z-VAD-fmk through premiochondrial and MAPK-dependent pathways. Acta Med Okayama. 2016;59:253–260. doi: 10.18926/AMO/31959. [DOI] [PubMed] [Google Scholar]
- 2.Wahidin M, Noviani R, Hermawan S, Andriani V, Ardian A, Djarir H. Population-based cancer registration in Indonesia. Asian Pac J Cancer Prev. 2012;13(4):1709–1710. doi: 10.7314/apjcp.2012.13.4.1709. [DOI] [PubMed] [Google Scholar]
- 3.Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13(4)(215):1–7. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeFazio-Eli L, Strommen K, Dao-Pick T, Parry G1, Goodman L, Winslow J. Quantitative assays for the measurement of HER1-HER2 heterodimerization and phosphorylation in cell lines and breast tumors: applications for diagnostics and targeted drug mechanism of action 2011. Breast Cancer Res. 2011;13(R44):1–18. doi: 10.1186/bcr2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padua MB, Bhat-Nakshatri P, Anjanappa M, Hao Y, Liu Y, McElyea K, Sandusky G, Althouse S, Perkins S, Nakshatri H. Disruption of the estradiol-regulated NTN1-UNC5A dependence receptor signaling axis causes a hybrid basal/luminal molecular phenotype in estrogen receptor-positive breast cancer cells. Cancer Res. 2017 doi: 10.1158/1538-7445.SABCS16-P6-08-02. [DOI] [Google Scholar]
- 6.Ruslim SK, Purwoto G, Widyahening IS, Ramli I. Stadium IB – IIA cervical cancer patient’s survival rate after receiving definitive radiation and radical operation therapy followed by adjuvant radiation therapy along with analysis of factors affecting the patient’s survival rate. J Physics: Conf. Series. 2017;884(1):012121. [Google Scholar]
- 7.Fulda S. Targeting apoptosis signalling pathway for anticancer therapy. Frontiers Oncol. 2011;1(23):1–7. doi: 10.3389/fonc.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray J, Donahue D, Head H, Mansell M, Margraf L. Low-Grade glioma combination chemotherapy may be effective in braf V600e codon-mutated optic pathway gangliogliomas. Neuro Oncol. 2017;19(Suppl 4):iv–34. [Google Scholar]
- 9.Subik K, Lee J-F, Baxter L, Strzepek T, Cstell D, Crowley P. The expression of patterns of ER, PR, HER2, CK5/6, EGFR, Ki67, and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer (Auckl) 2010:35–41. [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao X, Cristofanilli M, Rizvanov A, Pestel R. Breast cancer stem cell isolation. Breast Cancer Methods Protocols. 2016:121–135. doi: 10.1007/978-1-4939-3444-7_10. [DOI] [PubMed] [Google Scholar]
- 11.Widowati W, Wijaya L, Bachtiar I, Gunanegara R, Sugeng S U, Irawan YA, Sutiman BS, Widodo MA. Effect of oxygen tension on proliferation and characteristics of Wharton’s jelly-derived mesenchymal stem cells. Biomarkers Genomic Med. 2014;6:43–48. [Google Scholar]
- 12.Pham N, Morrison A, Schowk J, Aviel-Ronen S, Lakovlev V, Tsao M. Quantitative image analysis of immunohistochemical stains using a CMYK color model. Diagn Pathol. 2007;2(8):1–10. doi: 10.1186/1746-1596-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.American Cancer Society. 2018. [August 16 2018]. www.cancer.org.
- 14.Popolin C, Becceneri AB, Graminha AE, Batista AA, Cominetti MR. Evaluation of ruthenium complex [Ru(AmSal)(dppe)2] PF6 on the proliferation, morphology, and migration of triple-negative MDA-MB-231 breast tumor cells. Mol Cancer Res; AACR Special Conference: Advances in Breast Cancer Research. 2017 Oct;:7–10. [Google Scholar]
- 15.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Rakha EA, Richardson AL, Schmitt FC, Tan PH, Tse GM, Weigelt B, Ellis IO, Reis-Filho JS. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Krawczyk E, Suprynowicz FA, Palechor-Ceron N, Yuan H, Dakic A, Simic V, Zheng YL, Sripadhan P, Chen C, Lu J, Hou TW, Choudhury S, Kallakury B, Tang DG, Darling T, Thangapazham R, Timofeeva O, Dritschilo A, Randell SH, Albanese C, Agarwal S, Schlegel R. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat Protoc. 2017;12(2):439–451. doi: 10.1038/nprot.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leibniz Institute DSMZ-German Collection of Microorganism and Cell Culture. 2018. [September 11 2018]. https://www.dsmz.de/catalogues/details/culture/ACC-738.html.


