Abstract
The DNA loop extrusion model is a provocative new concept explaining the formation of chromatin loops, which revolutionizes our understanding of genome organization. Central to this model is the Structural Maintenance of Chromosomes (SMC) protein family that is now being ascribed a new function as a DNA motor. In this Perspective we review and reinterpret the current knowledge of SMC structure and function and propose a novel mechanism for SMC motor activity.
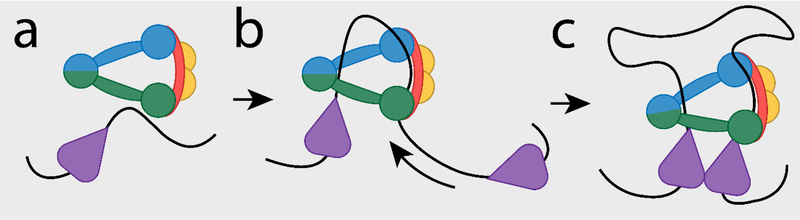
The spatial organization of DNA in the nucleus is critical to its function. A fundamental component of this organization involves DNA “loops”, physical point-to-point interactions between DNA sequences located far apart on the chromosome. These loops are key to chromatin condensation during mitosis and also regulate enhancer-promoter interactions during interphase. Several recent findings have led to the ‘DNA extrusion model’ of loop formation (Box 1). First theorized to explain how mitotic chromatin condensation might proceed without forming knots, the model also elegantly explains the observed CTCF motif orientation bias discussed in Box 1(ref. 1–6). The loop extrusion model posits that DNA loops begin as small pinches of the DNA molecule with each side held by one end of a proposed extrusion complex (Figure 1a). As the extrusion complex reels in DNA, the loop is progressively enlarged (Figure 1b). A stable loop is formed when the complex stops extruding (Figure 1c). This relatively straightforward model is a radical departure from previous thinking, and while it explains several puzzles it poses perhaps more.
Box 1. DNA loop extrusion model.
Loop extrusion has been independently proposed to explain the formation of numerous types of DNA loops, but the recent surge in interest is due to the ability of this model to explain the curious phenomenon of motif-oriented CTCF looping. CTCF loops are thought to be formed by two CTCF proteins bound to separate motifs on a chromosome. These loops are a clear and prominent feature of how the genome is organized. The asymmetric CTCF binding motif has an orientation that plays a fundamental role in the formation of these loops. As revealed by chromatin conformation capture assays, CTCF sites interact with each other significantly more when arranged in a convergent orientation. In agreement with this, CTCF loops form predominantly between CTCF sites oriented towards each other47. Conversely, CTCF sites oriented away from each other only rarely form loops. This finding has fundamental implications for the mechanism of loop formation. A simplistic model of loop formation via stabilization of stochastic collisions taking place in the three-dimensional space cannot account for this orientation bias. Rather, the loop formation mechanism must account for the orientation context of CTCF sites up to millions of base pairs apart. Loop extrusion solves this conundrum by having loops begin as small bends in the DNA that are progressively expanded (Figure 1). A loop extruder is theorized to expand the loop by translocating along the DNA, reeling the chromatin into the loop. The orientation bias of CTCF sites can then be explained by orientation-dependent interactions of CTCF with the extrusion machinery.
Figure 1: The Loop Extrusion Model.
a, Cohesin complexes load onto the DNA, either randomly or at specific sites, such as CTCF binding sites (purple). b, The cohesin complex reels in DNA, translocating over the DNA and expanding the loop. c, Cohesin complexes stop extruding when they meet a properly oriented CTCF site, leading to a loop between convergently-oriented CTCF anchors.
An important participant in loop extrusion is the highly conserved SMC family of proteins. SMC complexes assemble into large rings thought to encircle DNA strands . Entrapping DNA entirely within a protein complex leads to topological binding that will only be released by an opening of the protein complex. This renders the binding immune to disruption of the protein-DNA contacts and leads to exceptionally long residency times, while also permitting the free sliding of the SMC ring along the DNA.
The SMC complex condensin is known to organize DNA during mitosis. Processive expansion of initially small loops ensures that loop compaction occurs in order and only within a chromosome, precluding the formation of knots3. The SMC complex cohesin colocalizes with CTCF and is required for the formation of CTCF loops in interphase. Degradation of cohesin results in a complete loss of CTCF loops, while its stabilization via degradation of the cohesin release factor WAPL leads to additional loops7,8,9. This excessive looping condenses interphase chromatin into dense, mitotic-like “vermicelli” chromosomes. Importantly, this observation suggests that interphase loop formation by cohesin and mitotic condensation by condensin are fundamentally related processes. The SMC family also includes structurally similar members in Prokarya, where bacterial condensin juxtaposes the arms of replicating chromosomes in a manner reminiscent of loop extrusion10. It is thus likely that SMC complexes are part of an ancient mechanism of moving and organizing DNA via loop extrusion that has been repurposed to many ends over evolutionary time.
The Missing Motor
The loop extrusion model offers an attractive explanation for the reversible and orderly formation of DNA loops within chromosomes, but mechanistic details remain unknown, including how the proposed extrusion complex responsible for initiating and expanding DNA loops would work. The ability of the SMC family to bind DNA topologically recommends a model in which cohesin and condensin rings hold DNA loop ends, but the formation of loops up to millions of bases in size requires a motor: a mechanism by which DNA is pulled into the SMC loop. Numerous explanations have been proposed as to how loop extrusion is powered, including hitching rides with known DNA motors, such as RNA polymerase, pushing by DNA supercoiling, and passive diffusion along gradients of SMC complexes11,12,13,14. While each of these processes may be playing some role, there is now direct experimental evidence that the SMC complex condensin is capable of ATP-dependent unidirectional movement on a DNA substrate in vitro. In addition, strong circumstantial evidence suggests that loop extrusion is ATP dependent in vivo15,16. Condensin attached to DNA curtains was detected moving unidirectionally over a DNA molecule at ~60 base pairs per second17. Interestingly, on a relaxed single-te-thered DNA curtain, condensin compacts DNA through loop formation, but on the taut DNA of a double-tethered curtain, condensin translocates. This demonstrates that condensin can move along the DNA without forming an intramolecular loop. A subsequent experiment using Sytox Orange staining observed the extrusion of a loop on relaxed DNA at speeds up to ~1500 base pairs per second18. Importantly this study revealed that the SMC complex can extrude loops as a single complex, and that this extrusion is unidirectional in nature, with DNA being reeled into the loop from only one direction.
It is now clear that condensin is an ATP-powered DNA motor, but similar experiments performed with the cohesin complex have not detected motor activity19,20,21. Cohesin and condensin have remarkably similar architectures, and both have been independently hypothesized to form loops in DNA via loop extrusion. While it is possible that the intrinsic motor activity of cohesin has been replaced with an external process, it is also possible that in vitro assays are missing some critical component or post-translation modification. The extensive literature on the structure and function of the SMC complexes does not offer an immediate explanation for how these machines function as motors and a novel mechanism of active DNA translocation is required.
The Head, the Hinge, and the HAWKs
The core components of cohesin and condensin complexes are the SMC proteins (Figure 2a). These proteins have a complex structure with two globular domains, the head and the hinge, separated by long ~45 nm antiparallel coiled-coils. Pairs of SMC proteins heterodimerize at their hinges. SMC1 and SMC3 form the core of cohesin and SMC2 and SMC4 form condensin. The globular head domains contain ABC-type nucleotide binding domains that are thought to mediate dimerization between the two head domains of a complex. Each complex thereby cooperatively binds 2 ATP molecules. A third SMC component, kleisin, interacts with both head domains of the complex, linking them and forming a tripartite ring (Figure 2a). Kleisins are largely disordered peptide chains, much longer than is required to bind the two head domains. Kleisins are further bound by various members of a family of proteins that have come to be known as HEAT-repeat proteins associated with kleisins or HAWKs (Figure 2a). This family is rich in HEAT-repeat domains consisting of pairs of antiparallel alpha-helices linked together by just a few amino acids. Found in many proteins throughout the cell, HEAT-repeats are remarkable for their conformational flexibility. These structures adopt a horseshoe-like configuration capable of stretching and scrunching22. The kleisins of cohesin and condensin each interact with a number of these HAWKs, which regulate loading, unloading, and likely the motor activity of these complexes.
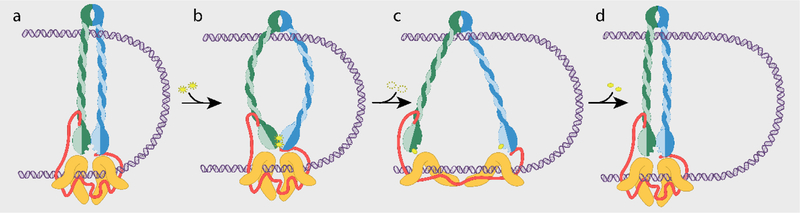
Figure 2: The Tethered Inchworm Model.
a, The SMC complex is composed of two SMC proteins (green and blue) which dimerize at the hinge (top) and at the head domain (bottom). Tethering the two heads together is a kleisin (red) further bound by HAWK proteins (orange). The SMC complex forms a small loop in the DNA (purple) by binding with both the hinge dimer and the kleisin-HAWK subcomplexes. b, Binding of 2 ATP molecules (yellow) by the ATPase head domains (green and blue) induces a conformational rotation of each head. This movement forces the coiled-coil arms apart, bending them, and propagating the strain to the hinge domains. c, ATP hydrolysis causes dissociation of the head domains and opening of the SMC arms. In this model, the leading HAWK would slide forward along the DNA due to its weaker affinity for DNA. The kleisin would straighten and unfurl to accommodate this movement and in doing so pull on the HAWKs, stretching these spring-like proteins. d, In the extended configuration the DNA binding affinities of the HAWKs then reverse causing the lagging HAWK to catch up as the head domains reunite, completing a mechanochemical cycle that has enlarged the DNA loop.
For a single SMC ring to achieve unidirectional movement, it must possess two means of interacting with the DNA simultaneously: one that will act as a stationary anchor and another that will produce movement along the DNA. The SMC complex has two reported mechanisms of binding DNA, the hinge domains on one end of the molecule and the kleisin and HAWK subcomplex on the other. The hinge domains of cohesin and condensin have high affinity for single-stranded DNA and some affinity for double-stranded DNA23. How the hinge interacts with DNA is still uncertain, but some evidence points to a positively charged groove formed by the inner-side of the hinge and the nearby coiled-coils24. DNA binding by the hinge has been shown to catalyze ATP hydrolysis by the head domains, and disruption of the hinge can disrupt the function of the entire complex18. The SMC hinge is a critical component of the complex that is likely key to the mechanochemical cycle driving SMC movement. In condensin, the kleisin and HAWK subcomplex forms a positively-charged pocket that wraps around the DNA fiber in what is described as a “safety belt” binding mechanism25. This creates a topological engagement that holds DNA in a sequence-independent manner. While this specific DNA-binding conformation has only been directly observed in the Brn1-Ycg1 kleisin-HAWK complex of S. cerevisiae, many HAWKs have DNA binding affinity. Structural similarities between the kleisin-HAWK subcomplexes that form part of cohesin suggest this may be a conserved mechanism of DNA binding. The kleisins of cohesin and condensin bind to at least two HAWK components simultaneously potentially forming multiple DNA binding pockets in each complex. These subcomplexes could bind to the same molecule of DNA or possibly hold two separate molecules of DNA together.
The Anchor and the Motor
Even with an understanding of how SMC complexes might engage DNA, it is not immediately obvious which end of the SMC complex would remain stationary and which end would move along the DNA. It has been proposed that the kleisin-HAWK topological binding pocket may serve as the anchor26, which would leave the comparatively simple hinge domain to serve as a motor. In one proposed model the SMC arms and hinge act as a DNA pump26,27. In this model, DNA loops are loaded into the ring formed by the SMC arms and ATP driven conformational changes close the ring, driving the loop into a smaller chamber formed by the kleisin and SMC heads where it combines with a larger loop. This model posits a topological binding of the DNA by the SMC-kleisin ring. However, a recent study of cohesin suggests that SMC rings incapable of topologically binding the DNA are still capable of extrusion28. Another potential model for hinge-mediated motor activity might be ATP-driven dissociation of the hinge leading to a walking mechanism. However, studies of the DNA binding capabilities of the hinge monomers have seen little to no independent DNA binding ability23.
Alternatively, the hinge domain could serve as the anchor, while the kleisin and HAWK subcomplexes move along the DNA. Several features of the structure support this model. The topological engagement of the kleisin-HAWK binding domain would allow movement of the DNA through the groove without release. Indeed, the loose nature of the DNA binding pocket results in low binding affinity for short DNA fragments, suggesting they could slide out of the groove25. Additionally, the kleisin and HAWK components appear uniquely suited for large conformational changes. The kleisin-HAWK DNA binding domain is not conformationally frozen, with different configurations of the HAWK and DNA observed in different crystals25. Between the terminal domains of kleisins, the protein is mostly unstructured, and much longer than would seem necessary to connect the two head domains together, suggesting there may exist some “slack” in this tether. The HEAT-repeats of the HAWKs are found in many other proteins, where they are known to stretch and compress in response to mechanical force22. Indeed, HEAT-repeats can be thought of as springs capable of stretching and contracting while storing and releasing potential energy29. Cryo-EM analysis of the HAWK protein Scc2 revealed a high degree of conformational flexibility with an estimated capacity to stretch lengthwise up to ~11 nm30. Taken together, the kleisin-HAWK DNA binding domain would appear to be capable of undergoing large conformational changes and sliding along the DNA. We therefore propose that the kleisin-HAWK subcomplexes represent the mobile DNA binding domain.
A model for SMC complex translocation on DNA must be compatible with both eukaryotic and prokaryotic SMC members. Prokaryotic SMC complexes lack HAWK proteins. Instead their kleisins are bound by much smaller Kite proteins that nevertheless appear to have functional similarities to the HAWKs31. Kite proteins are composed of two Winged-Helix Domains (WHD) connected by an intrinsically disordered linker. Each WHD binds to the kleisin creating the potential for two topological DNA binding grooves. Indeed, the eukaryotic Kites of the SMC5/6 complex have recently been found to bind DNA32. The disordered linker would permit the orientations of the WHDs to change dramatically allowing for folding and opening that could mimic the conformational flexibility of the HAWK proteins33. That the unrelated Kite and HAWK families share distinctive functional characteristics suggests that they might play a conserved role as flexible DNA binding components of the SMC complexes.
Kinetics
The step rate and step size of the extrusion process are important criteria for evaluating potential models of SMC motors. Unfortunately, the existing estimates of SMC motor kinetics are rough and ambiguous. The speed at which the SMC complex moves depends on the rate at which it steps and the size of its steps. If the SMC heads function similarly to related ABC-type domains, then each ATPase cycle most likely corresponds to the hydrolysis of 1 or 2 molecules of ATP. In the presence of DNA, condensin hydrolyzes ATP at a rate of ~2 ATP per second17. However, this bulk rate represents a mixture of condensin molecules in various states: actively extruding complexes, DNA-bound but stationary, non-extruding complexes, and non-DNA bound complexes. Therefore, the rate of hydrolysis of an actively extruding complex could be significantly higher than this average rate. The most unambiguous observation of the extrusion speed of condensin shows a single condensin extruding up to ~1,500 bp or ~500 nm per second18. However, the rate of extrusion displays a strong dependence on the tension on the DNA fiber and slows to a more modest ~600 bp per second rate at physiological tensions of ~0.4 pN. Whether this reduction in speed is a result of changes in step sizes, step rates, or the proportion of productive steps, will have important implications for the mechanism of the SMC motor. Importantly, the experiments discussed above were performed on naked DNA lacking nucleosomes. ATP-independent diffusion of cohesin on DNA is significantly impeded by the presence of nucleosomes19. Additionally, the force generated by condensin extrusion, estimated at ~1 pN, would be insufficient to evict the histone octamer34. This suggests that SMC complexes likely possess the ability to actively translocate past nucleosomes on chromatin.
Step size can be directly measured by experiments using magnetic tweezers, which precisely detect the compaction of DNA with high temporal resolution. Several magnetic tweezer experiments using condensin and cohesin from S. cerevisiae as well as condensin I from X. laevis have demonstrated DNA compaction on naked DNA occurring in highly variable steps larger than 100 nm in size35,36,37. Such large step sizes are incompatible with models that limit themselves to the ~50 nm length of SMC complexes. However, there is evidence to suggest these steps represent a mechanism of compaction distinct from extrusion. Similar large DNA compaction steps are observed for budding yeast condensin in the absence of ATP; these have been demonstrated to be distinct from smaller, co-occurring steps34. Two separate DNA compaction mechanisms have been reported for bacterial SMC complexes as well38. Most likely these large steps represent some form of loop capture distinct from extrusion. Both cohesin and condensin have demonstrated some capability to form inter-complex interactions that could explain these large compaction steps. Further studies will be needed to distinguish between these processes and to establish the kinetics of the SMC motors.
The Tethered Inchworm Model
While the kleisin-HAWK DNA bound subcomplex is in principle capable of accommodating large conformational changes, it must be the ATP-hydrolyzing head domains that provide the motive force. The ABC-type ATPase domains located in the SMC head domains form 2 ATP binding sites when engaged. ABC-type domains are thought to have a conserved mechanism of action where ATP binding and hydrolysis correspond to head engagement and disengagemen39. ATP-mediated head engagement is accompanied by a conformational shift, often a rotation, of the interface between the two domains to accommodate the nucleotides. Commonly this rotation is propagated into adjacent domains to perform mechanical work. Crystal structures of SMC heads reveal that ATP-bound forms are rotated ~30 degrees in relation to their unbound form40. This rotation dramatically increases the angle between the coiled-coil arms as they exit the head domains. Driving the coiled-coil arms apart likely forces them to bend, widening the ring and propagating this steric strain all the way to the hinge domain (Figure 2b). It has been proposed that this tension is relieved by ATP hydrolysis followed by disengagement and separation of the head domains40. In this way, ATP binding and hydrolysis could force the head domains apart, using the arms as force-amplifying levers (Figure 2c). No structural data for this open conformation exists, however AFM images of SMC dimers often show large, >50 nm distances between the head domains41.
Taking into consideration the conformational flexibility present in the kleisin-HAWK subcomplexes linking the two head domains, it is possible the kleisin might remain bound to both heads as they are pulled apart42. The disordered structure of kleisin could straighten and unfurl to accommodate this motion. In doing so, this could stretch the HAWK subunits bound at multiple points to the kleisins. If the kleisin-HAWK subcomplexes are topologically engaged with the DNA molecule, then this movement could be permitted by sliding the proteins along the DNA. Together these conformational changes would spread the SMC complex along the DNA. These motions could generate productive unidirectional movement if they were coordinated with changes in DNA binding affinity in the kleisin-HAWK subcomplexes. If in the closed configuration two kleisin-HAWK binding domains had differing affinities for DNA, then the side more weakly bound would preferentially move upon head separation. This would cause the less tightly bound HAWK to slide forward along the DNA (Figure 2c). A subsequent closing motion would pull the lagging end of the complex forward, assuming that the stretching of the kleisin-HAWK subcomplexes reversed the DNA binding affinities of the proteins (Figure 2d). The HEAT-repeats of the HAWKs act as springs, storing potential energy in their conformational changes. This energy might help drive the lagging step by pulling the head domains back together. Dimerization of the reunited SMC heads would complete a mechanochemical cycle in which ATP binding and hydrolysis powers net unidirectional movement along the DNA. This model, in which opening of the SMC ring pushes the leading end forward along the DNA and subsequent closing pulls the lagging end up, is akin to an inchworm motor. The interesting topology of the DNA-bound complex leads us to suggest the more descriptive term “tethered inchworm” for this model of SMC locomotion.
The tethered inchworm model is a general framework lacking in specifics and leaves several important questions unanswered. A wide range of step sizes would be compatible with this model due to the extremely flexible nature of each component. Step sizes upwards of ~50 nm could be accommodated by eukaryotic kleisins but will ultimately depend on the separation driven by ATP-binding and hydrolysis, which is likely smaller. A related question is how SMC complexes navigate obstacles such as nucleosomes. While the DNA binding grooves of the kleisin-HAWK subcomplexes are not large enough to permit ~11 nm sized nucleosomes, it is conceivable that HAWKkleisin dissociation during the walking cycle would allow SMC complexes to step over nucleosomes. It is also unclear in which direction the SMC complex moves, which would be determined by the order of changes in binding affinity of the kleisin-HAWK subcomplexes. Nevertheless, this putative model may begin to explain the known regulatory roles of various HAWKs on SMC function. Chromatin-bound cohesin consists of both mobile and immobile fractions43. Depleting the cohesin HAWK PDS5, rather than stopping extrusion, results in enhanced extrusion and condensation of the genome, suggesting that PDS5 may function as a component of immobile cohesin complexes9. PDS5 competes for its kleisin binding site with the HAWK NIPBL, whose depletion results in a loss of loops and extrusion44,45. PDS5 and NIPBL may represent static and mobile HAWK components, respectively, which compete to turn the cohesin motor off and on46. The tethered inchworm model is highly speculative, but our new perception of SMC complexes as loop extruding motors requires a bold reimagining of previous knowledge. Our proposal that the HAWK proteins are conformationally flexible and dynamic DNA binding elements is conjecture that is required to create a functional model of motor activity. Further study of the enigmatic HAWK family will be needed to evaluate this proposition. Our relatively better understanding of the core SMC proteins is unable to account for the motor activity of the SMC complex. Thus, understanding the functions of kleisin and HAWK proteins, namely whether and how they bind to DNA, what conformational changes they undergo during the ATP-hydrolysis cycle, and what roles different subunits play in regulating the complexes, will likely prove key to elucidating the motor function of SMC complexes. Future work on the structures and kinetics of SMC complexes will refine our understanding of this fascinating protein family responsible for DNA organization across all domains of life.
Acknowledgments
Work in the authors’ lab is supported by U.S. Public Health Service Award R01 GM035463 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Riggs AD DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Trans. R. Soc. Lond., B, Boil. Sci 326, 285-297 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet 35, 673–745 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Alipour E & Marko JF Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 40, 11202–11212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols MH & Corces VG A CTCF Code for 3D Genome Architecture. Cell 162, 703–705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanborn AL et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. PNAS 112, E6456–E6465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fudenberg G et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao SSP et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haarhuis JHI et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wutz G et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. The EMBO Journal 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Brandão HB, Le TBK, Laub MT & Rudner DZ Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrington C, Finn R & Hadjur S Cohesin biology meets the loop extrusion model. Chromosome Res 25, 51–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Racko D, Benedetti F, Dorier J & Stasiak A Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Research 46, 1648–1660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto T & Schiessel H Osmotic mechanism of the loop extrusion process. Phys. Rev. E 96, 030402 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Brackley CA et al. Extrusion without a motor: a new take on the loop extrusion model of genome organization. Nucleus 9, 95–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vian L et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell 173, 1165–1178.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X et al. In vivo evidence for ATPase-dependent DNA translocation by the Bacillus subtilis SMC condensin complex. Molecular Cell In Press, [DOI] [PMC free article] [PubMed]
- 17.Terakawa T et al. The condensin complex is a mechanochemical motor that translocates along DNA 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganji M et al. Real-time imaging of DNA loop extrusion by condensin. Science eaar7831 (2018). doi: 10.1126/science.aar7831This study directly imaged real-time unidirectional loop extrusion by single condensin complexes. This represents the strongest evidence to date that SMC complexes are DNA motors and provides important insights into the mechanism by which extrusion occurs.
- 19.Stigler J, Çamdere G, Koshland DE & Greene EC Single-Molecule Imaging Reveals a Collapsed Conformational State for DNA-Bound Cohesin. Cell Rep 15, 988–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanke M, Tahara E, Veld PJH in’t & Nishiyama T Cohesin acetylation and WaplPds5 oppositely regulate translocation of cohesin along DNA. The EMBO Journal 35, 2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson IF et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J 35, 2671–2685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura SH & Hirano T HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments? J. Cell. Sci 129, 3963–3970 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Hirano M & Hirano T Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell 21, 175–86 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Chiu A, Revenkova E & Jessberger R DNA interaction and dimerization of eukaryotic SMC hinge domains. J Biol Chem 279, 26233–42 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Kschonsak M et al. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell 171, 588–600.e24 (2017).This study identified the ability and mechanism by which a kleisin-HAWK subcomplex binds DNA. The topological and labile nature of this interaction is, we believe, illustrative of all kleisin-HAWK-DNA interactions and is integral to the tethered inchworm model.
- 26.Diebold-Durand M-L et al. Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization. Molecular Cell 67, 334–347.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marko JF, Rios PDL, Barducci A & Gruber S DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. bioRxiv 325373 (2018). doi: 10.1101/325373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srinivasan M et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Nontopological as well as Topological Mechanisms. Cell 173, 1508–1519.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kappel C, Zachariae U, Dölker N & Grubmüller H An Unusual Hydrophobic Core Confers Extreme Flexibility to HEAT Repeat Proteins. Biophys J 99, 1596–1603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao WCH et al. Structure of the cohesin loader Scc2. Nature Communications 8, 13952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells JN, Gligoris TG, Nasmyth KA & Marsh JA Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr Biol 27, R17–R18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zabrady K et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res 44, 1064–1079 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamada K, Miyata M & Hirano T Molecular Basis of SMC ATPase Activation: Role of Internal Structural Changes of the Regulatory Subcomplex ScpAB. Structure 21, 581–594 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Keenholtz RA et al. Oligomerization and ATP stimulate condensin-mediated DNA compaction. Scientific Reports 7, 14279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eeftens JM et al. Real‐time detection of condensin‐driven DNA compaction reveals a multistep binding mechanism. EMBO J 36, 3448–3457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strick TR, Kawaguchi T & Hirano T Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol 14, 874–80 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Sun M, Nishino T & Marko JF The SMC1-SMC3 cohesin heterodimer structures DNA through supercoiling-dependent loop formation. Nucleic Acids Res 41, 6149–6160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H & Loparo JJ Multistep assembly of DNA condensation clusters by SMC. Nat Commun 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopfner KP & Tainer JA Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol 13, 249–55 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Kamada K, Su’etsugu M, Takada H, Miyata M & Hirano T Overall Shapes of the SMC-ScpAB Complex Are Determined by Balance between Constraint and Relaxation of Its Structural Parts. Structure 25, 603–616.e4 (2017).This study crystallized ATP-bound condensin heads and showed that they undergo a large conformational shift. The authors propose a model of ATP binding and hydrolysis driving apart the SMC heads that is key to the tethered inchworm model.
- 41.Eeftens JM et al. Condensin Smc2-Smc4 Dimers Are Flexible and Dynamic. Cell Rep 14, 1813–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasmyth K & Haering CH The Structure and Function of Smc and Kleisin Complexes. Annual Review of Biochemistry 74, 595–648 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Gerlich D, Koch B, Dupeux F, Peters JM & Ellenberg J Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16, 1571–8 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Kikuchi S, Borek DM, Otwinowski Z, Tomchick DR & Yu H Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc. Natl. Acad. Sci. U.S.A 113, 12444–12449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petela N et al. Multiple interactions between Scc1 and Scc2 activate cohesin’s DNA dependent ATPase and replace Pds5 during loading. bioRxiv 205914 (2017). doi: 10.1101/205914 [DOI] [Google Scholar]
- 47.Rao SSP et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]