Abstract
Cholesterol is an essential component of cell membranes, and is required for mammalian pentameric ligand-gated ion channel (pLGIC) function. Computational studies suggest direct interactions between cholesterol and pLGIC’s but experimental evidence identifying specific binding sites is limited. In this study, we mapped cholesterol binding to Gloeobacter ligand-gated ion channel (GLIC), a model pLGIC chosen for its high level of expression, existing crystal structure, and previous use as a prototypic pLGIC. Using two cholesterol analogue photolabeling reagents with the photoreactive moiety on opposite ends of the sterol, we identified two cholesterol binding sites: an intersubunit site between TM3 and TM1 of adjacent subunits and an intrasubunit site between TM1 and TM4. In both the inter- and intrasubunit sites, cholesterol is oriented such that the 3-OH group points toward the center of the transmembrane domains rather than toward either the cytosolic or extracellular surfaces. We then compared this binding to that of the cholesterol metabolite, allopregnanolone, a neurosteroid that allosterically modulates pLGICs. The same binding pockets were identified for allopregnanolone and cholesterol, but the binding orientation of the two ligands was markedly different, with the 3-OH group of allopregnanolone pointing to the intra- and extracellular termini of the transmembrane domains rather than to their centers. We also found that cholesterol increases, whereas allopregnanolone decreases the thermal stability of GLIC. These data indicate that cholesterol and neurosteroids bind to common hydrophobic pockets in the model pLGIC, GLIC, but that their effects depend on the orientation and specific molecular interactions unique to each sterol.
1. Introduction
Mammalian pentameric ligand-gated ion channels (pLGICs), such as the nicotinic acetylcholine receptor (nAchR), gamma-aminobutyric acid receptor (GABAAR), and glycine receptor (GlyR) are important regulators of neuronal communication. Neurotransmitters bind to sites in the extracellular domain (ECD) and elicit structural changes in both the ECD and transmembrane domains (TMDs) that result in channel opening (1). The subsequent influx of ions regulates neuronal signaling. In addition to neurotransmitters, lipids, such as cholesterol and its derivatives, are important modulators of receptor function (2–6).
Cholesterol is an important determinant of receptor structure and function in both nAchRs (7–11) and GABAARs (12,13). Cholesterol or cholesterol mimetics such as cholesteryl hemisuccinate (CHS) are necessary for preserving the stability and function of purified nAchR and GABAAR (13,14). When nAchRs are reconstituted into lipid bilayers lacking cholesterol, agonist binding does not result in channel opening. Early studies suggested that cholesterol stabilizes the resting state of nAchRs, and permits transition from the resting to open state upon agonist binding (7). Cholesterol has also been shown to modulate GABAAR activity; enrichment and depletion of membrane cholesterol content reduces GABA potency (15).
Cholesterol can affect integral membrane protein (IMP) structure and function by two mutually non-exclusive mechanisms (6,16): 1) Cholesterol affects the physical properties of lipid bilayers such as membrane fluidity (6,17,18), membrane thickness (19), and the structure of lipid microdomains (20) and; 2) cholesterol can directly modulate protein function or stability by binding to specific sites on IMPs. Multiple computational studies suggest cholesterol binds to specific sites in the TMDs of pLGICs (21,22). Cholesterol docks to an intersubunit site in a homology model of GABAAR (21) and an intrasubunit site in nAchRs (22). Molecular dynamics (MD) simulations also identify an intrasubunit binding site for cholesterol in nAchR (22). While these studies suggest cholesterol directly interacts with pLGICs, experimental evidence for the location of cholesterol binding sites in pLGICs is limited (23).
Neurosteroids, which are synthesized from cholesterol in the brain, bind in the transmembrane domains of pLGICs and affect channel gating by either modulating the effect of neurotransmitters or directly activating channel opening. (2,3,24). Photolabeling studies have identified neurosteroid binding to an intersubunit site in β3 GABAAR homopentamers (25). Crystal structures of chimeric proteins GLIC-α1GABAAR (26) and β3GABAAR-α5GABAAR (27) provide further evidence of neurosteroids binding to a canonical intersubunit site. Neurosteroid binding to pLGICs has also been explored by photolabeling Gloeobacter ligand-gated ion channel (GLIC), a bacterial homologue and important structural model of mammalian pLGICs. Two neurosteroid binding sites were identified in each subunit of GLIC: the canonical intersubunit site as well as a novel intrasubunit site (28). Computational studies of GLIC suggest that bacterial hopanoids, which resemble eukaryotic sterols, may also bind to this intrasubunit site (29).
In order to investigate the sites and orientation of cholesterol binding in pLGICs, we used GLIC as a model protein. We performed photoaffinity labeling-mass spectrometry (PAL-MS) with two cholesterol analogues in which a photolabeling group is placed either on the 7position of the steroid ring structure or on the aliphatic tail. We have previously found that MS analysis of steroid-photolabeled sites in the TMDs of IMPs is limited by the poor solubility and recovery of steroid-labeled hydrophobic peptides (30,31). Here, we employ a middle-down MS approach (28) coupled with FLI-tag for enhanced identification of sites of photolabel incorporation. FLI-tag is coupled via click chemistry to the photolabeling reagent after it has covalently modified its binding site(s). It is a hydrophilic, isotopically-labeled tag that adds both charge and a heavy/light stable isotope pair that simplifies analysis by creating doublets in the MS1 spectra (31). Using these methods, we show that cholesterol binds to the same pockets in GLIC as do neurosteroids. Surprisingly, our results show that cholesterol and neurosteroids occupy these sites with opposite orientations.
2. Methods
2.1. Expression and purification of GLIC from E.coli
GLIC was expressed and purified as previously described (32). Briefly, pET26-MBP-GLIC, a gift from Raimund Dutzler, was expressed in OverExpress™ C43(D3) Escherichia coli, grown in Terrific broth supplemented with 0.2% Glucose and 30 mM potassium phosphate buffer, pH 7.5, and induced with 0.2 mM isopropyl-1-thio-β-D-galactopyranoside. Cells were pelleted, reconstituted in 20 mM Tris pH 7.5, 100 mM NaCl (with protease inhibitor and DNase), lysed, and solubilized in 1% n-dodecyl-β-D-maltoside (DDM). GLIC was then purified with amylose resin and eluted with 40 mM maltose in buffer containing 0.02% DDM. His-MBP-GLIC was digested with HRV 3c to remove the affinity tag and further purified with reverse purification on a Ni-NTA column to remove the His-MBP and size exclusion chromatography (SEC) on a Sephadex 200 Increase 10/300 column. Purified protein was eluted from SEC in 50 mM Tris pH 7.5, 150 mM NaCl, and 0.02% DDM.
2.2. Click chemistry of photolabeled GLIC and refolding into DDM
10–45 μg purified GLIC in 0.02% DDM, 50mM Tris, 150mM NaCl was photolabeled with 100 μM LKM38 or KK174. The syntheses of LKM38 and KK174 were reported previously (30,33). The photolabeled protein samples were processed in two ways: 1) Samples were buffer exchanged into 50 mM triethylammonium bicarbonate buffer (TEABC) (using BioRad© buffer exchange columns) followed by endoproteolytic digestion of photolabeled GLIC or; 2) 1% Sodium dodecyl sulfate (SDS) was added to the sample to a concentration of 1% and FLI-tag was attached to the photolabeled protein via click chemistry using 2 mM C6H7NaO6, 0.1 mM Tris(benzyltriazolylmethyl)amine, and 1 mM CuSO4. The click chemistry reaction mixture was run overnight at room temperature. 30 kDa cut off filters were pre-equilibrated with 8 M urea, 100 mM Tris-pH 8.0. The clicked sample was added to 200 μL 8 M urea, 100 mM Tris-pH 8.0 and allowed to sit at RT for approximately 5 min before centrifuging at 12,000 × g. 200 μL of 8 M urea was added three more times and the sample was centrifuged at 12,000 × g after each addition. This allowed the protein to be retained in the 30 kDa cut off filter and the SDS to be removed. The urea was removed by two sequential Bio-Rad© buffer exchange columns pre-equilibrated with 50 mM TEABC in 0.02% DDM.
2.2. Cross Competition assay
60 μg GLIC in 0.02%DDM, 50 mM Tris, 150 mM NaCl was incubated with 10 μM LKM38 or 10 μM 5α−6AziP (28) on ice, in the dark, for ~1 h. Samples were split into 3 groups and 100 μM Allopregnanolone, 100 μM Cholesterol, or vehicle (ethanol) was added to each group, respectively. Samples were incubated on ice, in the dark, for ~1 h. Each sample was photolabeled for 5 min, buffer exchanged using Bio-Rad© buffer exchange columns pre-equilibrated with 50 mM TEABC in 0.02% DDM, and digested with trypsin for one day at 37 °C. Formic acid was added to 1% and samples were analyzed by LCMS on a Thermo™ Orbitrap Elite mass spectrometer. Labeling efficiencies (TM3=Intersubunit site, TM4=Intrasubunit site) were calculated by determining the area under the curve of extracted ion chromatograms (XICs) of the photolabeled peptide/(the unlabeled peptide + labeled peptide). Reduction in labeling efficiency (competition) in the presence of 100 μM sterol was assessed by calculating the relative labeling efficiencies of 5α−6AziP or LKM38 with adding 100 μM sterol prior to photolableing vs adding vehicle only. Analysis of statistical significance was performed using a one-way ANOVA with a post hoc Dunnett’s test.
2.4. Endoproteolytic digestion in DDM for middle-down MS of photolabeled GLIC
Protein was digested with trypsin (3–6 μg) in 50 mM TEABC, 0.02% DDM for one day at 37 °C or for one week at 4 °C. The digestion was stopped by adding formic acid to 1% and the peptides were analyzed by liquid chromatography tandem mass spectrometry as previously described (28).
2.5. Intact MS of photolabeled GLIC
100 μg GLIC was sequentially photolabeled three times with either 100 μM LKM38 or KK174 and photolabeled protein was processed and analyzed as previously described (30). Briefly, photolabeled protein was precipitated with CH3Cl:MeOH:H2O in a 4:4:1 ratio and resuspended in formic acid. Formic acid was diluted to less than 5% with 4:4:1 CH3Cl:MeOH:H2O and directly injected into a Thermo™ Orbitrap Elite mass spectrometer. A spray voltage of 4 kV, capillary temperature of 320 °C, and insource dissociation of 30 V was used. Full spectra were acquired in the linear trap quadrupole. Spectra were deconvoluted using MagTran (34).
2.6. MS1-based pair search and MS1-based extracted ion chromatograms of tryptic peptides
Photolabeled peptides were identified either by a search of MS1 spectra for pairs of features differing by 10 mass units (31) or by a manual search of MS1 data using Xcalibur™ (Thermo Fisher Scientific) as previously described (28). For the MS1 extracted ion chromatograms the mass-to-charge ratio of photolabeled and unlabeled tryptic peptides were calculated using ProteinProspector (© UCSF) and programmed into Xcalibur™ layouts. MS1 features were manually confirmed for accurate mass error less than 30 ppm, accurate charge state, and chromatographic elution of labeled peptides after unlabeled peptides (supplementary table 1). PEAKS™ database searches aided in identification of labeled peptides in β–loops at the TMD/ECD interface. Any peptide identified by a PEAKS search was confirmed or rejected manually at the MS1 level using the above criteria. Sites of ligand incorporation were identified by manual assignment of features in the product ion spectra based on the expected m/z of fragment ions with and without a photolabel modification. The mass tolerance for fragment ion identification was 30 ppm.
2.7. AutoDock analysis of cholesterol binding sites
Docking was performed using AutoDock 4.2 as previously described (28), using the GLIC structure PDB 4FHI. The cholesterol ligand was prepared by converting an sdf file from pubchem into a pdb file using Open Babel (35), and partial charges and torsion angles were calculated using PRODRG set for reduced charges and energy minimization (36). A 30 Å × 30 Å × 30 Å grid box was centered in an intrasubunit or intersubunit site and 1000 docking poses were obtained for each site.
2.8. Thermal stability of GLIC
Samples containing 5 μg GLIC in 100 μL of 50 mM Tris pH 8.0, 150 mM NaCl, and 0.02% DDM were either left on ice or heated to 40 °C, 50 °C, 60 °C, or 70 °C for one hour. After heating, samples were placed on ice until injection onto a Sephadex 200 Increase 10/300 column. Running buffer was 50 mM Tris pH 8.0, 150 mM NaCl, and 0.02% DDM. Absorbance was measured at 280 nm, and the maximal intensity of the monodisperse pentamer peak was recorded for each sample. Thermal destabilization of the native protein structure leads to oligomer dissociation and/or aggregation. Reduction in the intensity of the monodisperse pentamer peak observed on SEC reports on the loss of pentamer without the complication of discriminating between oligomer dissociation and aggregation (26,27,37).
The effects of allopregnanolone and cholesterol on GLIC thermal stability were tested by adding 100 μM sterol to 10 μg GLIC in 200 μL. The samples were split in half and either left on ice at 4 °C, or heated to 55 °C for one hour. Control samples without sterol were prepared and analyzed in the same way. Maximum peak height for the monodisperse pentamer peak from the absorbance profiles were determined in Microsoft Excel. For each sample, the ratio of maximum peak heights at 4 °C vs 55 °C was calculated. The 4 °C / 55 °C ratios of each sterol (allopregnanolone, cholesterol) were normalized to the 4 °C / 55 °C ratios of their respective controls. Experiments were performed in triplicate and statistical significance was determined using one-sample, two-tailed, t-tests.
3. Results
3.1. Cholesterol analogue labels GLIC with a stoichiometry of two
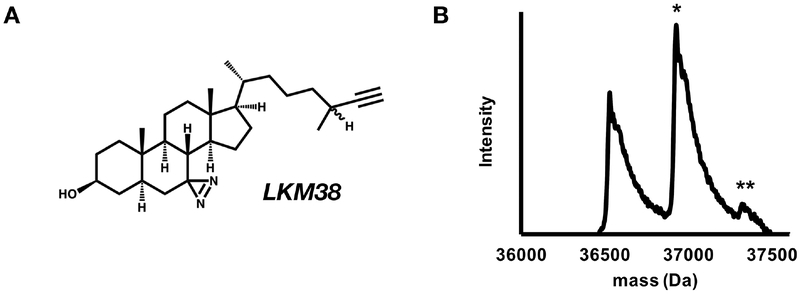
To determine the stoichiometry of labeling, we analyzed the intact mass of GLIC photolabeled with LKM38, a cholesterol analogue with a diazirine in the 7-position of the steroid backbone and an alkyne for click chemistry attachment of tags (Fig. 1A). A single cholesterol pocket per GLIC subunit (i.e. stoichiometry of one) would give a feature equal to the mass of a GLIC monomer + one LKM38 ligand, or 36,928 Da. If labeling efficiency is less than 100%, an unlabeled feature at 36,532 Da, the mass of an unlabeled GLIC monomer, will also be observed. If each GLIC monomer contains multiple cholesterol pockets, we expect to observe features at the mass of GLIC monomers + multiple LKM38 ligands. The expected mass of GLIC + 2 LKM38 ligands is 37,324 Da. Assuming the labeling at both sites is independent of each other, the efficiency of labeling of the doubly labeled monomer is the product of the labeling efficiencies at each of the two sites. If more than two sites are labeled the doubly labeled feature would be the sum of products at each combination of two sites. If labeling efficiencies are very small, a multiply labeled feature may not be observed. As such, a single labeled feature represents one or more labeling sites, a doubly labeled feature represents two or more labeling sites, etc. In addition to affinity, efficiency of labeling is influenced by photochemical specificity, and, at high concentrations, the maximal solubility of the ligand. Therefore, binding affinity is only roughly approximated by labeling efficiency.
Figure 1: Cholesterol analogues efficiently photolabel GLIC with a stoichiometry of two.
(A) Structure of LKM38. (B) Deconvoluted spectra of intact GLIC exhaustively photolabeled with 3 × 100 μM LKM38 showing an unlabeled GLIC, GLIC with one LKM38 (*) bound and GLIC with 2 LKM38 molecules bound (**).
GLIC photolabeled with LKM38 was analyzed by intact (denatured) mass spectrometry and deconvolution of the mass spectra showed an unlabeled feature of 36,531 Da, a feature labeled with one LKM38 ligand at 36,929 Da, and a feature labeled with two LKM38 ligands at 37,322 Da (Fig. 1B). This indicates that LKM38 photolabels GLIC with a stoichiometry of at least two (i.e. two cholesterol analogues per GLIC subunit).
3.2. Sites of cholesterol labeling suggest two shared sterol pockets
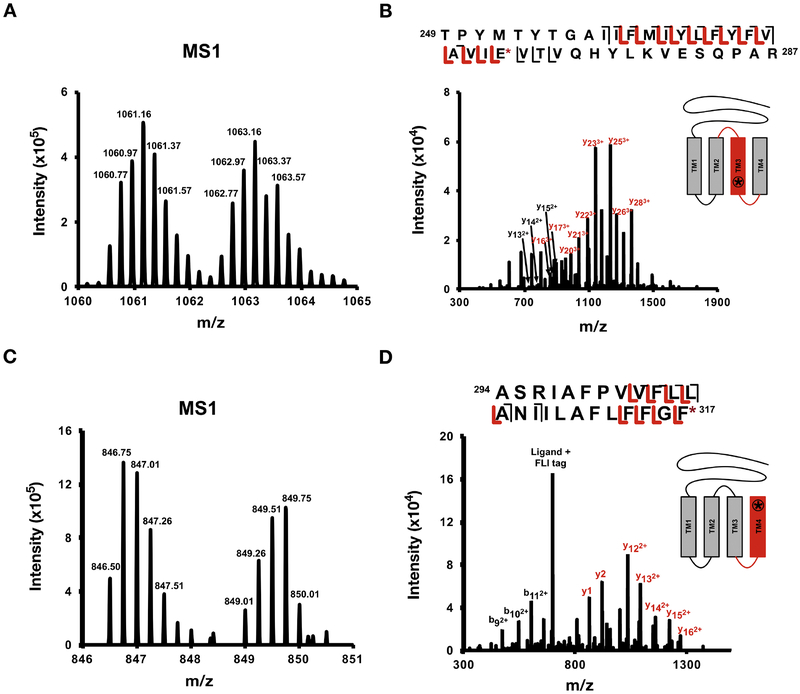
To determine the sites of photolabeling, GLIC was photolabeled with 100 μM LKM38. The photolabeled protein was clicked to FLI-tag in 1% SDS, refolded in DDM, digested with trypsin, and analyzed by middle-down MS. Attempts to perform the click reaction in DDM were unsuccessful, likely because the hydrophilic FLI-tag is unable to partition into the DDM micelles and reach labeled sites within the transmembrane domains. Denaturing in 1% SDS enabled successful azide-alkyne cycloaddition of FLI-tag. To avoid loss of hydrophobic peptides associated with conventional urea-based digests and post-digestion desalting steps, the photolabeled, clicked protein was refolded into DDM in the absence of salt prior to trypsin digestion. The peptides were analyzed by tandem MS and the spectra were searched at the MS1-level for pairs separated by 10 Daltons. This was performed by an automated MS1-level pair search and by manual analysis of XICs of labeled and unlabeled tryptic peptides. Searches revealed multiple pairs, all representing labeling of transmembrane domain 3 (TM3) or TM4 peptides (Fig. 2A and 2C, supplementary table 1). Analysis of fragmentation (product-ion) spectra revealed labeling at Glu272 in TM3 (Fig. 2B) and Phe317 in TM4 (Fig. 2D). Both of these residues are sites previously shown to be labeled by 5α−6-AziP, a neurosteroid analogue photolabeling reagent (28).
Figure 2: Residues labeled by LKM38.
(A) Representative MS1 pair of LKM38-peptide adduct with five charges and (B) corresponding CID fragmentation spectra of TM3 + LKM38 + FLI-tag with site defining ions placing LKM38 on Glu272. (C) Representative MS1 pair of LKM38-peptide adduct with four charges and (D) corresponding CID fragmentation spectra of TM4 + LKM38 + FLI-tag with site defining ions placing LKM38 on Phe317.
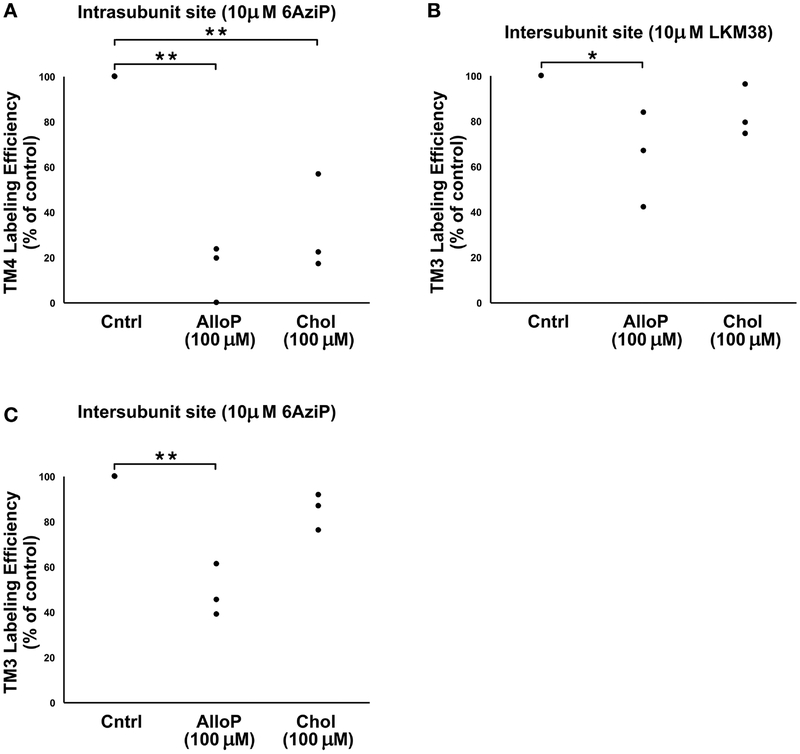
To confirm that the labeling was identifying specific binding sites and to further explore the possibility of overlapping sterol sites, we performed a cross-competition assay by testing the ability of excess cholesterol (100 μM) or allopregnanolone (100 μM) to prevent photolabeling by a neurosteroid (10 μM 5α−6-AziP) and of excess allopregnanolone (100 μM) or cholesterol (100 μM) to prevent labeling by LKM38 (10 μM). 10 μM LKM38 labeling of the intersubunit site was reduced in the presence of 100 μM allopregnanolone (p<0.05), and 10 μM 5α−6-AziP labeling of the intrasubunit site was reduced by the presence of 100 μM cholesterol (p<0.01) (Fig. 3). These results are consistent with cholesterol and neurosteroids binding to shared pockets in GLIC. Consistent with previous results, labeling of both the intra- and intersubunit sites by 5α−6-AziP were reduced by allopregnanolone. We could not assess competitive inhibition of LKM38 labeling at the intrasubunit site due to the low efficiency of LKM38 labeling of this site. We did not observe statistically significant reduction of LKM38 labeling in the presence of excess cholesterol. This may be due to technical limitation associated with the low aqueous solubility of cholesterol. Out-competing an irreversible photolabeling process (LKM38 labeling) with a reversible ligand (cholesterol) requires achieving a high ratio of competitor to photolabel. This can be accomplished by decreasing the concentration of the photolabeling reagent or increasing the concentration of competitor. Decreasing concentration of LKM38 prevents reproducible quantitation of photolabeled peptides. Increasing cholesterol concentration is limited by its poor solubility in solvents used for mass spectrometric analysis.
Figure 3: Cross competition assay showing competitive prevention of sterol labeling at both neurosteroid binding sites.
(A) Photolabeling efficiency of intrasubunit site (TM4) by 10 μM 5α−6AziP in the presence of no competitor (Cntrl), 100 μM Allopregnanolone (AlloP, n=3), or 100 μM Cholesterol (Chol, n=3). Efficiencies are normalized to percent of control. **, p<0.01. (B) Photolabeling efficiency of intersubunit site (TM3) by 10 μM LKM38 in the presence of no competitor (Cntrl), 100 μM Allopregnanolone (AlloP, n=3), or 100 μM Cholesterol (Chol, n=3). Efficiencies are normalized to percent of control. * p<0.05. (C) Photolabeling efficiency of intersubunit site (TM3) by 10 μM 5α−6AziP in the presence of no competitor (Cntrl), 100 μM Allopregnanolone (AlloP, n=3), or 100 μM Cholesterol (Chol, n=3). Efficiencies are normalized to percent of control. **p<0.01.
3.3. Mapping the orientation of cholesterol labeling in GLIC
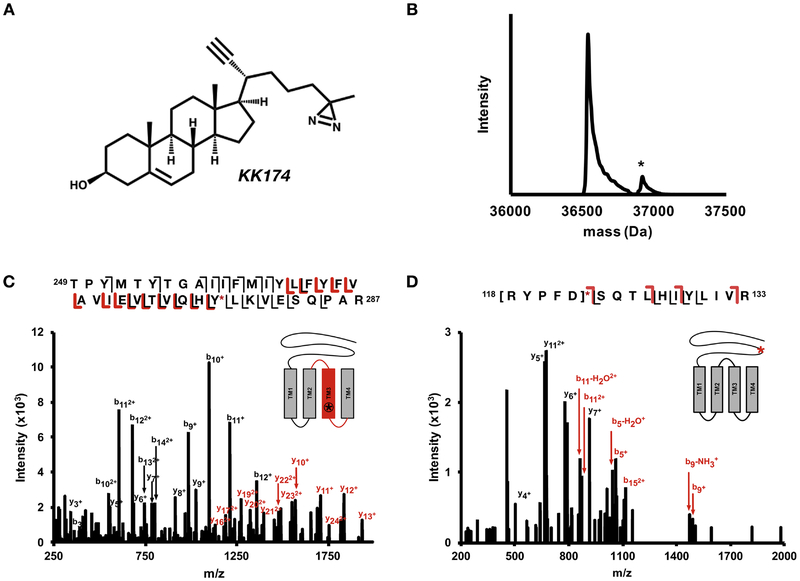
A second analogue of cholesterol, KK174, which contains a diazirine on the aliphatic tail, was used to map the orientation of cholesterol within each pocket (Fig. 4A) (30). KK174 labels GLIC with a much lower efficiency than LKM38. Even labeling at saturating concentrations of KK174, sequential labeling with 100 μM KK174 three times (total of 300 μM), resulted in a low labeling efficiency (Fig. 4B), preventing us from determining whether KK174 also labels GLIC at two sites using an intact mass measurement. We were also unable to identify pairs when KK174 was clicked to FLI-tag, likely due to this lower labeling efficiency. Therefore, we used XICs of both the labeled and unlabeled tryptic peptides to identify the photolabeled sites. We used accurate mass and retention time (i.e. the photolabeled peptide was expected to elute after the unlabeled peptide) as additional criteria for confident identification of photolabeled peptides. Through this analysis, we identified labeling in TM3, TM1+TM2, and a peptide in the β6-β7 loop (supplementary table 1). Analysis of the fragmentation (product-ion) spectra localized labeling to Y278 in TM3 (Fig. 4C), residues 118–122 in the β6-β7 loop (Fig. 4D), and residues I190–I198 at the N-terminus of TM1 (Supplementary Fig. 1). The low efficiency of labeling prevented localization to a single amino acid in the TM1 + TM2 peptide, as well as the β6-β7 loop peptide (Fig. 4). The same N-terminal region of TM1 was photolabeled by neurosteroids, and we favor labeling of Y194 or Y197, given that these are the strongest nucleophiles in this region and thus most likely to react with the aliphatic diazirine in KK174 (38). Regardless of the exact photolabeled residues, the data indicate that KK174 labels two distinct sites in GLIC: an intrasubunit site between TM1 and TM4 (Fig. 5A), and an intersubunit site between TM1 and TM3 of adjacent subunits (Fig. 5C).
Figure 4: KK174 photolabeling sites.
(A) Structure of KK174. (B) Deconvoluted spectra of intact GLIC photolabeled with 3 × 100 μM KK174 showing an unlabeled GLIC and GLIC with one KK174 bound (*). (C) HCD fragmentation spectra of TM3 + KK174 with site defining ions placing KK174 on Tyr278. (D) HCD fragmentation spectra of β6−β7 loop + KK174. Fragment ions localize labeling to residues 118–112 within the β6−β7 loop.
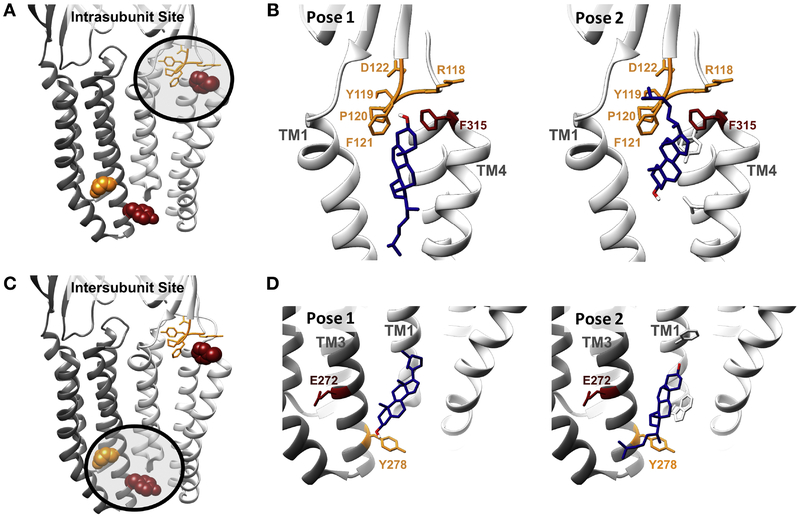
Figure 5: Cholesterol docking in GLIC.
(A) The residues labeled by LKM38 (red*) and KK174 (orange) in the intrasubunit site. (B) The two predominant clusters of cholesterol poses in the intrasubunit site from AutoDock. Pose 1 (left) has a binding energy of 5.6 kJ/mol and constituted 78% of AutoDock poses in this site. Pose 2 (right) has a binding energy of 5.7 kJ/mol and constituted 22% of AutoDock poses in this site. Our photolabeling data is most consistent with pose 2, where the aliphatic tail of KK174 labels the β6−β7 loop and the 7 position of LKM38 labels the C-terminus of TM4. (C) The residues labeled by LKM38 (red) and KK174 (orange) in the intersubunit site. (D) The two predominant clusters of cholesterol poses from AutoDock in the intersubunit site. Pose 1 (left) has a binding energy of 7.8 kJ/mol and constituted 34% of AutoDock poses in this site. Pose 2 (right) has a binding energy of 7.7 kJ/mol and constituted 66% of AutoDock poses in this site. Our photolabeling data is consistent with pose 2, where the aliphatic tail of KK174 labels Tyr268, about two turns below Glu272, labeled by LKM38 * Phe317 is labeled by LKM38, but is not resolved in the crystal structure. The residue highlighted in red in the intrasubunit site is Phe315.
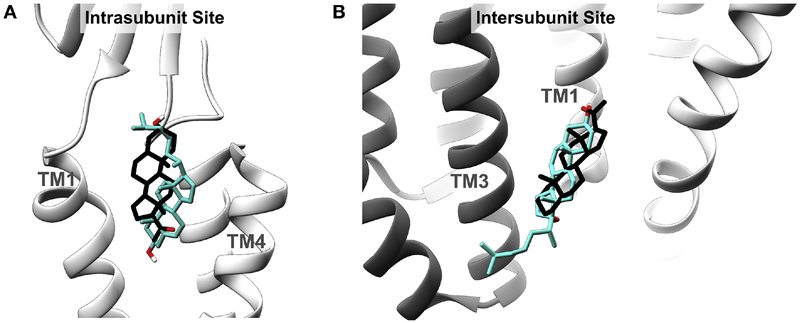
To further explore the orientation of cholesterol binding in these sites, AutoDock simulations were performed docking cholesterol to the GLIC crystal structure (PDB: 4F8H) (39). Cholesterol was constrained by a 30 Å × 30 Å × 30 Å box containing the photolabeled residues at either the intrasubunit or intersubunit sites and 1000 poses were obtained and clustered with a 2 Å RMSD. At either site, two clusters of poses were obtained with the cholesterol aliphatic tail oriented toward the center or the periphery of the TMDs (Fig 5B, D). For the intrasubunit site, the cluster of poses where the aliphatic tail points to the periphery of the TMDs (Pose 2) is most consistent with the photolabeling data, with the 7-position of cholesterol pointing toward TM4 and the aliphatic tail proximal to the β6-β7 loop and the N-terminus of TM1. In poses where the aliphatic tail faces to the center of the TMDs, the 7-position points towards TM1, which is inconsistent with the photolabeling results. For the intersubunit site, a cluster of poses (Pose 2) where the aliphatic tail points to the periphery of the TMDs is most consistent with our photolabeling data. Interestingly, the orientation of cholesterol photolabeling in both sites is the opposite compared to the previously identified preferred poses for allopregnanolone binding identified using photolabeling (i.e., the 3-hydroxy is rotated approximately 180 degrees) (Fig. 6) (28).
Figure 6: Overlapping sterol sites in GLIC.
Cholesterol pose consistent with photolabeling data (teal) and the previously investigated (28) allopregnanolone binding pose (black) in the Intrasubunit Site (A) and Intersubunit Site (B).
3.4. Effect of sterols on GLIC thermal stability
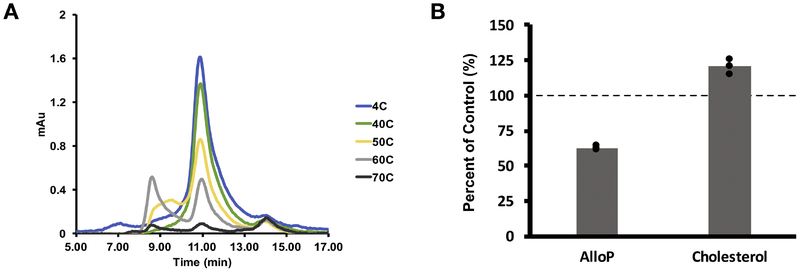
Steroids can affect the stability of detergent-solubilized pLGICs (27). To test the effect of cholesterol and neurosteroids on GLIC thermal stability, we first determined the thermal stability of GLIC by incubating the protein at temperatures ranging from 4° to 70 °C and then measuring the height of the monodisperse pentamer peak detected by UV on size exclusion FPLC (SEC). 55°C was identified as the approximate temperature at which half denaturation occurs (Fig. 7A), We then heated GLIC to 55 °C in the presence or absence of 100 μM cholesterol or 100 μM allopregnanolone and analyzed by SEC (Supplementary Fig. 2). A decrease in ratios indicates destabilization of the pentamer upon heating, and an increase indicates stabilization of the pentamer. Interestingly, cholesterol stabilized (p=0.02, n=3) whereas neurosteroids destabilized (p=0.003, n=3) the GLIC pentamer (Fig. 7B).
Figure 7: Cholesterol and neurosteroids have opposite effects on GLIC thermal stability.
(A) UV absorbance traces from SEC showing the GLIC pentamer peak at 10.8 min. Increasing temperature destabilized the pentamer. (B) GLIC was heated to 55 °C with and without sterol. The height of the monodisperse pentamer peak was compared for control (no ligand), AlloP (100 μM Allopregnanolone), and Chol (100 μM Cholesterol) (n = 3). A lower percent of control indicates destabilization of the pentamer and a higher percent indicates stabilization.
4. Discussion
Cholesterol is important in maintaining the structure and function of mammalian pLGICs. In this study, we used GLIC as a model to investigate cholesterol binding sites in pLGICs. We mapped two cholesterol binding sites in GLIC: an intrasubunit site near the extracellular domain, and an intersubunit site towards the intracellular side of GLIC. Site identification was enhanced with MS1 doublets from FLI-tag, and stoichiometry was confirmed with intact protein MS. This data is consistent with previous studies that suggest cholesterol can bind to specific sites in pLGICs, such as GLIC (29), nAchR (22,23), and GABAAR (21). Molecular dynamic (MD) simulation studies of cholesterol binding to GLIC (29) or nAchR (22) identified a similar intrasubunit site; in the case of the nAchR, multiple binding orientations were identified, one of which is consistent with our photolabeling data in GLIC. A PAL study of nAchR using [3H] 6-azicholesterol identified labeled residues at the N-terminus (intracellular side) of TM4, as well as the C-terminus (extracellular side) of TM4 (23). These labeled residues do not delineate cholesterol binding pockets, but are consistent with the pockets we mapped in GLIC for an intersubunit and intrasubunit site, respectively. [3H]6-azicholesterol labeling was also identified in TM1 and TM3 of nAchR subunits, but the specific residues of ligand incorporation could not be determined. Our PAL data is consistent with and expands on this study by more precisely mapping the location and binding orientation within similar sites in GLIC. In contrast, MD and docking studies of the nAchR (22) and GABAAR (21) proposed an intersubunit cholesterol site that is more extracellular and distinct from the site identified in this study by PAL. This discrepancy at the intersubunit site may be due to the different approaches taken (computation vs. experimental) or the different pLGICs studied.
GLIC is a bacterial pLGIC that is widely used as a structural model of mammalian pLGICs (40), and provides multiple advantages for PAL studies including the ease of high-level expression and performing intact protein MS analysis to determine the stoichiometry of steroid labeling (28). Bacterial membranes do not contain sterols such as cholesterol but do contain hopanoids, which are bacterial pentacyclic sterol-like molecules. Hopanoids have been proposed to be endogenous modulators of GLIC activity, and dock favorably to the same intrasubunit site in GLIC identified in this study (29). In addition, eukaryotic sterols such as cholesterol (41) and neurosteroids (28) have been shown to inhibit GLIC channel activity. In light of these findings, it has been proposed that sterol binding pockets evolved in bacterial pLGICs to accommodate hopanoids (29). Both nAChRs and GLIC contain a CARC, cholesterol recognition, motif in an intrasubunit pocket. Cholesterol analogues label this site but the cholesterol orientation identified in photolabeling sites is opposite to that predicted for a CARC motif (29,42). The cholesterol binding pockets identified in this study are on both the extracellular and intracellular sides of the TMDs, with the intrasubunit site in a CARC domain on the extracellular end of the TMDs, and the intersubunit site on the intracellular end of the TMDs. In mammalian homologs, cholesterol binding to non-annular sites within both leaflets may be important in localizing pLGICs to cholesterol-rich domains, as has been discussed for proteins that contain double CARC/CRAC domains (43).
Consistent with the hypothesis that pLGICs contain binding pockets that can accommodate different sterols, the intersubunit and intrasubunit cholesterol binding sites identified in this study are the same sites as previously identified for neurosteroids in GLIC (28). Further evidence for common sterol binding sites is provided by a cross-competition assay. Excess cholesterol significantly reduced the labeling efficiency of the neurosteroid photolabeling reagent, 5α−6-AziP, at the intrasubunit site. Similarly, excess neurosteroid significantly reduced the labeling efficiency of our cholesterol photolabeling reagent, LKM38, at the intersubunit site, supportive of two common sterol pockets in the model pLGIC, GLIC. Interestingly, our photolabeling studies identified cholesterol and neurosteroids in opposite orientations within these pockets. It has been shown that small changes in ligand structure can lead to differences in binding orientation (44). Different binding orientations may be a mechanism by which structurally different sterols are able to bind to the same hydrophobic pockets.
Sterols have been shown to alter the stability of detergent-solubilized pLGICs (26,27). We therefore compared the effects of cholesterol and neurosteroids on the thermal stability of GLIC pentamers, and found that their effects were opposite. We speculate that this may be a result of differences in binding orientation or specific protein-ligand interactions unique to each sterol. It is likely that hydrophobic pockets analogous to those in GLIC are also present in mammalian pLGICs. As such, sterols, such as neurosteroids, may compete for their binding and effect with bulk sterols, such as cholesterol, that are found in mammalian membranes. In the GABAAR, increased cholesterol content has been shown to reduce the potentiating effects of neurosteroids (12). One could speculate that a higher cholesterol content results in a higher occupancy of cholesterol in these sites, and therefore more competition with neurosteroids for their binding and effect. If the differences in sterol binding orientation observed in GLIC are also present in mammalian pLGICs, this may be a mechanism by which structurally different sterols are able to exert different or biased functional effects from the same binding pocket. Understanding binding modes, including the sites and orientation of binding is important for designing steroid-based therapeutics that target specific sites and have specific functional effects. Future studies are needed to confirm the presence of these cholesterol binding sites in mammalian pLGICs, where cholesterol is an endogenous, abundant component of the cell membrane.
Supplementary Material
Highlights:
Cholesterol binds to two sites in a model pentameric ligand gated ion channel: an intersubunit site and an intrasubunit site
Both cholesterol binding sites are shared sterol pockets
Cholesterol labels both sites in the opposite orientation as neurosteroids
Cholesterol and neurosteroids have opposite effects on the thermal stability of Gloeobacter ligand gated ion channel
Acknowledgements
We would like to thank Raimund Dutzler for the gift of the pET26-MBP-GLIC vector and James Janetka for his guidance with FLI-tag synthesis. We would also like to thank Brad Manion and Ailing Tong for their assistance in purifying GLIC.
Funding: This work was supported by the National Institutes of Health grants R01GM108799 (to A. S. E. and D. F. C.), T32GM108539 (to A. S. E. and W. W. C.) and K08GM126336 (to W. W. C.), an International Anesthesia Research Society (IARS) mentored research award (to W. W. C.), The National Science Foundation grant DGE-1143954 (to M. M. B.) and the Taylor Family Institute for Innovative Psychiatric Research. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Althoff T, Hibbs RE, Banerjee S, and Gouaux E (2014) X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maksay G, Laube B, and Betz H (2001) Subunit specific modulation of glycine receptors by neurosteroids. Neuropharmacology, 369–376 [DOI] [PubMed] [Google Scholar]
- 3.Bertrand D, Valera S, Bertrand S, Ballivet M, and Rungger D (1991) Steroids inhibit nicotinic acetylchollinereceptors. Synaptic Transmission 2 [DOI] [PubMed] [Google Scholar]
- 4.Baenziger JE, Morris M, Darsaut TE, and Ryan SE (2000) Effect of membrane lipid composition on the nicotinic acetylcholine receptor. J Biol Chem 2275, 777–784 [DOI] [PubMed] [Google Scholar]
- 5.Baenziger JE, Henault CM, Therien JP, and Sun J (2015) Nicotinic acetylcholine receptor-lipid interactions: Mechanistic insight and biological function. Biochim Biophys Acta 1848, 1806–1817 [DOI] [PubMed] [Google Scholar]
- 6.Barrantes FJ (2015) Phylogenetic conservation of protein-lipid motifs in pentameric ligand-gated ion channels. Biochim Biophys Acta 1848, 1796–1805 [DOI] [PubMed] [Google Scholar]
- 7.Sunshine C, and McNamee MG (1994) Lipid modulation of nicotinic acetylcholine receptor function: the roleof membrane lipid composition and fluidity. Biochim Biophys Acta, 59–64 [DOI] [PubMed] [Google Scholar]
- 8.Dalziel AW, Rollins ES, and McNamee MG (1980) The effect of cholesterol on agonist-induced flux in reconstituted acetylcholine receptor vesicles. FEBS Lett 122, 193–196 [DOI] [PubMed] [Google Scholar]
- 9.Fantini J, and Barrantes FJ (2009) Sphingolipid/cholesterol regulation of neurotransmitter receptor conformation and function. Biochim Biophys Acta 1788, 2345–2361 [DOI] [PubMed] [Google Scholar]
- 10.Rankin SE, Addona GH, Marek AA, Bugge B, and Miller KW (1997) The cholesterol dependence of activation and fast desensitization of the nicotinic acetylcholine receptor. Biophysical Journal 73, 2446–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrantes FJ (2004) Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Res Brain Res Rev 47, 71–95 [DOI] [PubMed] [Google Scholar]
- 12.Sooksawate T, and Simmonds MA (1998) Increased membrane cholesterol reduces the potentiation of GABAA currents by neurosteroids in dissociated hippocampal neurones. Neuropharmacology 37, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 13.Bristow DR, and Martin IL (1987) Solubilisation of the y-Aminobutyric Acid/Benzodiazepine Receptor from Rat Cerebellum: Optimal Preservation of the Modulatory Responses by Natural Brain Lipids. J Neurochem 49, 1386–1393 [DOI] [PubMed] [Google Scholar]
- 14.Dunn SM, Martin CR, Agey MW, and Miyazaki R (1989) Functional Reconstitution of the Bovine Brain GABAa Receptor from Solubilized Components. Biochemistry 28, 2545–2551 [DOI] [PubMed] [Google Scholar]
- 15.Sooksawate T, and Simmonds MA (2000) Effects of membrane cholesterol on the sensitivity of the GABAA receptor to GABA in acutely dissociated rat hippocampal neurones. Neuropharmacology, 178–184 [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Kenworthy AK, and Sanders CR (2014) Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci 23, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albi E, Tomassoni ML, and Viola-Magni M (1997) Effect of Lipid Composition on Rat Liver Nuclear Membrane Fluidity. Cell biochemistry and function 15, 181–190 [DOI] [PubMed] [Google Scholar]
- 18.Leibel W, Firestone LL, Legler DC, Braswell LM, and Miller KW (1987) Two pools of cholesterol in acetylcholine receptor-rich membranes from Torpedo. Biochim Biophys Acta 897, 249–260 [DOI] [PubMed] [Google Scholar]
- 19.Lecuyer H, and Dervichian DG (1969) Structrue of aqueous mixtures of lecithin and cholesterol. J Mol Biol 45, 39–57 [DOI] [PubMed] [Google Scholar]
- 20.Brown DA (2006) Lipid Rafts, Detergent-Resistant Membranes, and Raft Targeting Signals. Physiology 21, 430–439 [DOI] [PubMed] [Google Scholar]
- 21.Henin J, Salari R, Murlidaran S, and Brannigan G (2014) A predicted binding site for cholesterol on the GABAA receptor. Biophys J 106, 1938–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brannigan G, Henin J, Law R, Eckenhoff RG, and Klein ML (2008) Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A 105, 14418–14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamouda AK, Chiara DC, Sauls D, Cohen JB, and Blanton MP (2006) Cholesterol interacts with transmembrane alpha-helices M1, M3, and M4 of the Torpedo nicotinic acetylcholine receptor: photolabeling studies using [3H]Azicholesterol. Biochemistry 45, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert JJ, Belelli D, Hill-Venning C, and Peters JA (1995) Neurosteroids and GABA, receptor function. Trends Pharmacol Sci 16, 295–303 [DOI] [PubMed] [Google Scholar]
- 25.Chen ZW, Manion B, Townsend RR, Reichert DE, Covey DF, Steinbach JH, Sieghart W, Fuchs K, and Evers AS (2012) Neurosteroid analog photolabeling of a site in the third transmembrane domain of the beta3 subunit of the GABA(A) receptor. Mol Pharmacol 82, 408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laverty D, Thomas P, Field M, Andersen OJ, Gold MG, Biggin PC, Gielen M, and Smart TG (2017) Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nat Struct Mol Biol 24, 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller PS, Scott S, Masiulis S, De Colibus L, Pardon E, Steyaert J, and Aricescu AR (2017) Structural basis for GABAA receptor potentiation by neurosteroids. Nat Struct Mol Biol 24, 986–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng WWL, Chen ZW, Bracamontes JR, Budelier MM, Krishnan K, Shin DJ, Wang C, Jiang X, Covey DF, Akk G, and Evers AS (2018) Mapping Two Neurosteroid Modulatory Sites in the Prototypic Pentameric Ligand Gated Ion Channel GLIC. J Biol Chem 293, 3013–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrantes FJ, and Fantini J (2016) From hopanoids to cholesterol: Molecular clocks of pentameric ligand-gated ion channels. Prog Lipid Res 63, 1–13 [DOI] [PubMed] [Google Scholar]
- 30.Budelier MM, Cheng WWL, Bergdoll L, Chen ZW, Janetka JW, Abramson J, Krishnan K, Mydock-McGrane L, Covey DF, Whitelegge JP, and Evers AS (2017) Photoaffinity labeling with cholesterol analogues precisely maps a cholesterol-binding site in voltage-dependent anion channel-1. J Biol Chem 292, 9294–9304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budelier MM, Cheng WW, Bergdoll L, Chen ZW, Abramson J, Krishnan K, Qian M, Covey DF, Janetka JW, and Evers AS (2017) Click Chemistry Reagent for Identification of Sites of Covalent Ligand Incorporation in Integral Membrane Proteins. Anal Chem 89, 2636–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basak S, Chatterjee S, and Chakrapani S (2016) Site Directed Spin Labeling and EPR Spectroscopic Studies of Pentameric Ligand-Gated Ion Channels. J Vis Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle REN, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, Perera RM, Covey DF, Nomura DK, Ory DS, and Zoncu R (2017) Lysosomal cholesterol activates mTORC1 via an SLC38A9–Niemann-Pick C1 signaling complex. Science 355, 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, and Marshall AG (1998) A Universal Algorithm for Fast and Automated Charge State Deconvolution of Electrospray Mass-to-Charge Ratio Spectra. J Am Soc Mass Spectrom 9, 225–233 [DOI] [PubMed] [Google Scholar]
- 35.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, and Hutchison GR (2011) Open Babel: An open chemical toolbox. J Cheminform 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuttelkopf AW, and van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 37.Miller PS, and Aricescu AR (2014) Crystal structure of a human GABAA receptor. Nature 512, 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das J (2011) Aliphatic diazirines as photoaffinity probes for proteins: recent developments. Chem Rev 111, 4405–4417 [DOI] [PubMed] [Google Scholar]
- 39.Sauguet L, Poitevin F, Murail S, Van Renterghem C, Moraga-Cid G, Malherbe L, Thompson AW, Koehl P, Corringer PJ, Baaden M, and Delarue M (2013) Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J 32, 728–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.daCosta CJ, and Baenziger JE (2013) Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure 21, 1271–1283 [DOI] [PubMed] [Google Scholar]
- 41.Velisetty P, and Chakrapani S (2012) Desensitization mechanism in prokaryotic ligand-gated ion channel. J Biol Chem 287, 18467–18477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Scala C, Baier CJ, Evans LS, Williamson PTF, Fantini J, and Barrantes FJ (2017) Relevance of CARC and CRAC Cholesterol-Recognition Motifs in the Nicotinic Acetylcholine Receptor and Other Membrane-Bound Receptors. Curr Top Membr 80, 3–23 [DOI] [PubMed] [Google Scholar]
- 43.Fantini J, Di Scala C, Evans LS, Williamson PT, and Barrantes FJ (2016) A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes. Sci Rep 6, 21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruepp MD, Wei H, Leuenberger M, Lochner M, and Thompson AJ (2017) The binding orientations of structurally-related ligands can differ; A cautionary note. Neuropharmacology 119, 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.