SUMMARY
In Bacillus subtilis, sporulation requires that the 26-aa protein SpoVM embeds specifically into the forespore membrane, a structure with convex curvature. How this nanometer-sized protein can detect curves on a micron scale is not well understood. Here, we report that SpoVM exploits a “dash-and-recruit” mechanism to preferentially accumulate on the forespore. Using time-resolved imaging and flow-cytometry, we observe that SpoVM exhibits a faster adsorption rate onto membranes of higher convex curvature. This preferential adsorption is accurately modeled as a two-step process: first, an initial binding event occurs with a faster on-rate, then cooperative recruitment of additional SpoVM molecules follows. We demonstrate that both this biochemical process and effective sporulation in vivo require an unstructured and flexible SpoVM N-terminus. We propose that this two-pronged strategy of fast adsorption followed by recruitment of subsequent molecules is a general mechanism that allows small proteins to detect subtle curves with a radius 1000-fold their size.
Keywords: ArfGAP1, PtdIns3KC3, autophagy, BAR domain, amphipathic helix, septins, ALPS, SpoIVA, DivIVA, geometric cue
INTRODUCTION
Despite a general absence of membrane-bound compartments, bacterial cells can nonetheless display a high level of intracellular organization. Beyond sorting proteins to chemically distinct locations, such as the plasma or outer membranes, cell wall, periplasm, or extracellular milieu (Siegel et al., 2016; Tsirigotaki et al., 2017; Wilson and Bernstein, 2016), some bacterial proteins also localize to distinct subcellular regions within these destinations (Rudner and Losick, 2010). One example of this phenomenon is the non-uniform localization of membrane-associated proteins at particular sites within the cell. Unlike eukaryotic cells, which harbor protein transport machinery, proteins in bacteria largely move by diffusion (Deich et al., 2004), and subcellular protein localization is achieved when, over time, a specific localization factor at a particular cellular site captures a diffusing protein that is destined to reside there (Rudner et al., 2002). A longstanding challenge in the field of bacterial cell biology has been to identify these localization factors, particularly those that do not require other localization factors of their own (Dworkin, 2014; Shapiro et al., 2009).
One proposed localization factor is membrane curvature, on the micron-scale, defined by the local shape of the cell itself (Updegrove and Ramamurthi, 2017). For example, in a rod-shaped bacterium, the poles display two-dimensional concave hemispherical curvature, whereas the membranes along the lateral edge of the cell are more curved in the circumferential compared with the longitudinal dimension. During cell division, the region at which the division septum meets the lateral edge can display high two-dimensional concave curvature. Several reports have identified concave membrane curvature as a beacon that recruits certain shape-sensing proteins to reside preferentially at the poles, division septum, or along the lateral surface (Govindarajan et al., 2013; Lenarcic et al., 2009; Ramamurthi and Losick, 2009; Renner et al., 2013; Strahl et al., 2015; Ursell et al., 2014).
Bacterial cells are typically devoid of internal organelles, and therefore rarely display positive (convex) membrane curvature in their cytosol, but exceptions do exist. One notable instance is during endospore (spore) formation in Bacillus subtilis where, upon starvation, the rod-shaped bacterium elaborates a spherical double membrane-enclosed internal organelle (termed the forespore) that harbors a copy of the cell’s genetic material (Higgins and Dworkin, 2012; Tan and Ramamurthi, 2014) (Figure 1A). After forespore elaboration, the rod-shaped outer cell (termed the mother cell) deposits a thick protein shell (termed the coat (Driks and Eichenberger, 2016)) on the convex forespore surface. Ultimately, when the mother cell lyses to release the mature, dormant spore, the coat helps to protect the spore’s genetic material from environmental insults (Setlow, 2014).
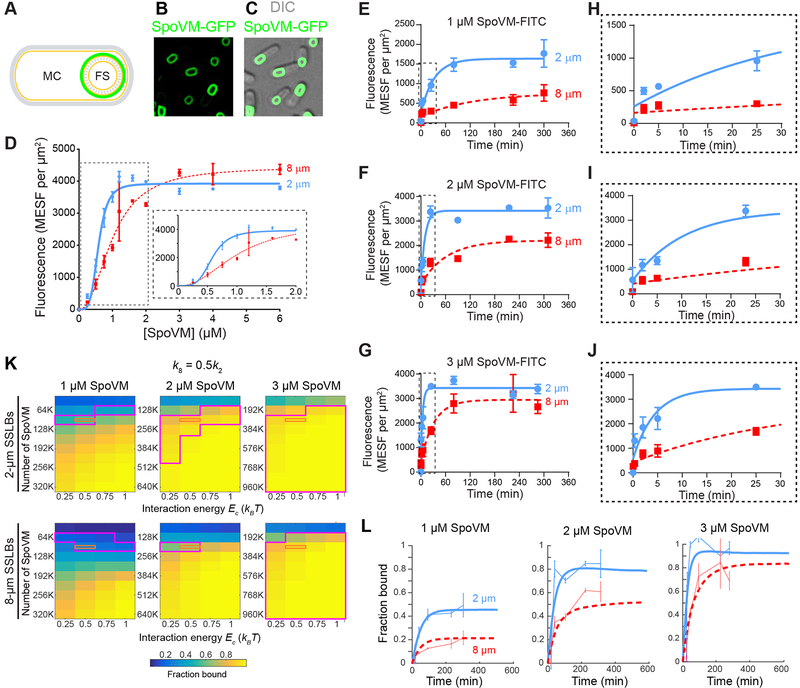
Figure 1: Preferential in vitro adsorption of SpoVM-FITC onto more positively curved membranes using a flow cytometry-based assay.
(A) Schematic of sporulating B. subtilis showing the mother cell (MC) and forespore (FS). Membranes are shown in yellow and cell wall in gray; site of SpoVM localization on the forespore is depicted in green. (B-C) Fluorescence micrographs of sporulating B. subtilis cells (strain CVO1195) producing SpoVM-GFP (B, GFP fluorescence; C, overlay of GFP fluorescence and DIC image). Strain genotypes are listed in STAR Methods. (D) Adsorbance of SpoVM-FITC onto differently curved membranes in vitro. Fluorescence intensities (MESF, Molecules of Equivalent Soluble Fluorochrome) were measured using flow cytometry at varying concentrations of SpoVM-FITC incubated with a mixed population of 2-μm (blue) and 8-μm (red) SSLBs. Insets: magnification of the initial part of the binding curve highlighted by dashed box. Data were fit with an allosteric sigmoidal model. (E-J) Association kinetics of SpoVM-FITC onto differently curved membranes. Mixture of 2-μm (blue) and 8-μm (red) SSLBs incubated with (E) 1 μM, (F) 2 μM, or (G) 3 μM SpoVM-FITC. Fluorescence intensity associated with SSLBs was measured at times indicated using flow cytometry. (H-J) Magnification of the initial part of the adsorption curves in (E-G), respectively, indicated by dashed box. Data were fit with a one-phase association model. Data points and error bars represent mean values and standard deviation, respectively, from three independent replicates. (K-L) Monte Carlo simulations predict that a faster on-rate coupled to mild cooperativity is sufficient to recapitulate experimental binding kinetics. In the simulations, SpoVM molecules in solution can bind to surfaces representing 2- and 8-μm SSLBs at equal total surface area, with on-rates k2 and k8, respectively, in the absence of other molecules bound at nearby sites (Methods). The on-rate is enhanced on all surfaces by exp(Ec/kBT) for each neighboring binding site containing a SpoVM molecule. The off-rate is the same for both surfaces. (K) Across a wide range of values of the total number of SpoVM molecules (N) and the cooperativity energy Ec, we computed the normalized steady-state binding with k8/k2 = 0.5 and identified regions of parameter space (magenta) that were consistent with our experimental data (within one standard deviation of the mean) on 2- and 8-μm SSLBs for simulations with N, 2N, and 3N molecules to mimic 1, 2, and 3 μM SpoVM concentrations. The orange rectangle denotes parameters for which simulations agreed with our experimental data for both SSLB sizes and across the 3-fold concentration range. (L) Simulations of binding kinetics (thick lines) using one of the parameter sets within the orange rectangle (Ec = 0.5kBT, N = 76,800) displayed excellent agreement with our experimental data (thin lines, error bars represent one standard deviation). Blue and red lines represent 2- and 8-μm SSLBs, respectively. See also Figure S1 and S2.
The coat is an architecturally complex structure composed of ~80 different proteins. Assembly of the coat initiates with the localization of a 26-aa protein, SpoVM (Levin et al., 1993), that embeds in the outermost membrane surrounding the forespore (Figure 1B–C). Previously, we proposed that SpoVM preferentially adsorbs onto positively curved membranes (Ramamurthi et al., 2009), such as that found on the forespore surface, and that membrane curvature is sufficient to drive its localization to the forespore surface in a manner largely independent of lipid composition. For example, SpoVM produced artificially in mutant Escherichia coli or Saccharomyces cerevisiae adsorbed onto convex surfaces that resembled the curvature of the forespore. Additionally, when incubated with a population of giant unilamellar vesicles of mixed sizes composed of phosphatidylcholine, purified SpoVM preferentially adsorbed onto the most convex vesicles available (Ramamurthi et al., 2009). After adsorption, SpoVM recruits a structural protein, SpoIVA (Ramamurthi et al., 2006; Roels et al., 1992), which hydrolyzes ATP to irreversibly polymerize into a platform encasing the forespore on top of which the rest of the coat assembles (Castaing et al., 2013; Ramamurthi and Losick, 2008; Wu et al., 2015). SpoVM mislocalization not only prevents the proper assembly of this basement layer, but also the rest of the spore coat (Ramamurthi et al., 2006).
Here, we examine the mechanistic basis of SpoVM localization onto micron-scale positively curved membranes. We developed an in vitro assay to measure the kinetics of SpoVM adsorption onto differently curved membranes and discovered that SpoVM adsorbs faster onto membranes of higher convex curvature, which we herein define as “preferred curvature”. Simulations based on a biophysical model that invoke a faster on-rate of SpoVM onto membranes of preferred curvature and assume a weak cooperativity between SpoVM molecules in the lipid bilayer accurately replicated the experimental kinetic data. In vitro data suggested that the observed cooperativity between SpoVM molecules requires an unstructured N-terminus. In vivo, SpoVM variants with a structured N-terminus failed to localize properly during sporulation and failed to promote proper coat assembly. Taken together, we propose a “dash-and-recruit” mechanism in which SpoVM molecules initially accumulate more rapidly on membranes of preferred curvature and then positively influence the recruitment of subsequent SpoVM molecules to drive the aggregate localization of SpoVM onto positively curved membranes.
RESULTS
An in vitro assay to quantify SpoVM adsorption onto differently curved membranes
To investigate the preferential adsorption of SpoVM onto convex membrane surfaces, we developed in vitro assay to quantify SpoVM adsorption kinetics onto differently curved surfaces. We adapted our previous work on spherical supported lipid bilayers (SSLBs) (Gill et al., 2015), which consist of a single phospholipid bilayer encasing a silica bead of defined diameter and generate membranes of a defined positive (convex) curvature (Bayerl and Bloom, 1990; Gopalakrishnan et al., 2009), to measure the adsorption of fluorescently-labeled SpoVM onto a mixture of differently curved SSLBs using flow cytometry. In contrast to our previous, microscopy-based approach, flow cytometry increased throughput by approximately three orders of magnitude, with an acquisition rate of over a thousand particles per second. This essentially eliminated the problem of photobleaching and allowed us to label SpoVM with the small molecule, FITC, instead of the larger GFP used in previous work.
We used this flow cytometry-based assay to quantitate the binding of SpoVM-FITC to curved membranes of different diameters. We prepared SSLBs of 2-μm diameter (to represent membranes with curvature similar to that of B. subtilis endospores, which are ~1 μm in diameter) and 8-μm diameter (membranes with a four-fold reduction in curvature) as previously described (Gill et al., 2015). Increasing concentrations of SpoVM-FITC were incubated with a mix of 2-μm and 8-μm SSLBs present in a molar ratio that ensured equal total surface area provided by each type of SSLB. Flow cytometry was then used to distinguish the two bead sizes based on forward and side scatter and to quantify fluorescence values for each bead in the population (Figure S1). Fitting the median fluorescence for each population as a function of protein concentration (Figure 1C) to an allosteric sigmoidal model revealed differences in the parameters that describe the adsorption of SpoVM onto the 2-μm or 8-μm SSLBs. The fitted Bmax (maximal binding) values were similar for adsorption of SpoVM-FITC onto both populations of SSLBs (Table 1), indicating that the number of SpoVM-FITC binding sites per square micron was similar on both curved surfaces. While the density of SpoVM-FITC at saturation was similar between the two different curved beads, we found that SpoVM-FITC bound to 2-μm SSLBs at a half maximal concentration of 0.58 ± 0.03 μM, versus 1.0 ± 0.07 μM for 8-μm SSLBs, suggesting a ~1.7-fold increase in affinity for the more curved surface (Table 1). We also observed that SpoVM-FITC displays cooperativity even on the 8-μm SSLBs, with a Hill coefficient of 2.2 ± 0.3. However, on 2-μm SSLBs we measured a Hill coefficient of 4.3 ± 0.8, suggesting an even higher degree of cooperative binding onto more highly curved surfaces. These measurements are in qualitative agreement with our previous, microscopy-based work on SpoVM-GFP (see Table 1 of Gill et al., 2015 for comparison). Together, these data suggest that the adsorption of SpoVM-FITC to more highly curved SSLBs is driven both by increased affinity to these membranes and apparent cooperative interactions between SpoVM molecules within these membranes.
Table 1: Parameters for the allosteric sigmoidal fit of adsorption of SpoVMWT and SpoVM variants onto differently curved surfaces as measured by flow cytometry.
Bmax, maximal binding value; h, Hill coefficient; K1/2, concentration of protein producing half-maximal binding; MESF, molecules of equivalent soluble fluorochrome. Errors are SEM (n=3 independent trials, each trial containing data from >30,000 SSLBs). “N/D” indicates “not determinable”.
| Protein | Bead diam. (μm) | Bmax (MESF/μm2) | h | K1/2 (μM) |
|---|---|---|---|---|
| SpoVM-FITC | 2 | 3,920 (± 100) | 4.3 (± 0.8) | 0.58 (± 0.03) |
| 8 | 4,450 (± 200) | 2.2 (± 0.3) | 1.0 (± 0.07) | |
| SpoVMP9A-FITC | 2 | 3,040 (± 70) | 1.5 (± 0.1) | 0.58 (± 0.03) |
| 8 | 3,340 (± 480) | 1.1 (± 0.3) | 0.84 (± 0.26) | |
| SpoVML8P,P9A-FITC | 2 | 3,640 (± 190) | 1.9 (± 0.2) | 1.67 (± 0.13) |
| 8 | 5,140 (± 750) | 2.4 (± 0.4) | 3.40 (± 0.53) | |
| SpoVMP9A,K10P-FITC | 2 | 4,350 (± 310) | 1.8 (± 0.2) | 1.81 (± 0.20) |
| 8 | N/D | 1.1 (± 0.3) | N/D | |
| SpoVMP9A,F11P-FITC | 2 | 4,150 (± 160) | 2.5 (± 0.3) | 1.48 (± 0.07) |
| 8 | N/D | 1.2 (± 0.2) | N/D | |
| SpoVMP9G-FITC | 2 | 2,950 (± 80) | 3.9 (± 0.9) | 0.41 (± 0.03) |
| 8 | 3,090 (± 560) | 1.1 (± 0.4) | 0.83 (± 0.33) |
SpoVM adsorbs faster to membranes of preferred curvature
Next, we asked whether these differences in affinity for 2-μm SSLBs and 8-μm SSLBs could reflect differences in on-rate. Various concentrations of SpoVM-FITC were incubated with a mix of 2-μm and 8-μm SSLBs for increasing amounts of time, and SSLBs associated with SpoVM-FITC were quantified using flow cytometry. For all concentrations tested, the t1/2 values were greater for binding onto 8-μm beads compared to 2-μm beads, indicating faster adsorption of SpoVM onto more positively curved membranes (Figure 1E–J). Due to very slow dissociation of SpoVM from membranes, we were unable to quantitatively measure dissociation rates of SpoVM-FITC from 2-μm and 8-μm beads. Nonetheless, assuming a similar, low dissociation of SpoVM from all membranes, a faster association onto more positively curved membranes alone could account for the higher apparent affinity of the protein for convex surfaces. This provides experimental support for the idea that, assuming an equal off-rate from differently curved membranes, a faster kinetic association rate onto more convex membranes could, together with cooperative interactions, achieve preferential localization onto more positively curved membranes. Previous Monte Carlo simulations have also demonstrated the plausibility of this model (Gill et al., 2015).
The experimentally determined adsorption rate, which was on the minute time scale, may be modeled as an aggregate of on-rate, off-rate, and cooperativity, which ultimately results in stable adsorption of SpoVM. To determine on-rates and cooperative energy that would account for the overall faster adsorption of SpoVM, we performed Monte Carlo simulations in which a prescribed number of SpoVM molecules were able to bind and unbind from surfaces representing 2-μm and 8-μm SSLBs of equal combined area (Methods). The surfaces were modeled as a periodic lattice for which each binding site had four neighbors, and the presence of a SpoVM molecule at a neighboring site increased the on-rate by where is an energy denoting the strength of cooperativity. We fixed the on-rate for 2-μm SSLBs k2 (Methods), and varied the values of the 8-μm SSLB on-rate k8 from 0.4k2 to 0.6k2 (Figure 1K, k8 = 0.5k2; Figure S2, k8 = 0.4k2 and k8 = 0.6k2), the cooperativity energy Ec from 0.25kBT to 1kBT, and the number of particles over an order of magnitude spanning estimates of the density of SpoVM at saturated binding levels in our experimental assay (Figure 1K). To directly compare with our experimental measurements of binding kinetics at 1, 2, and 3 μM SpoVM (Figure 1E–J), we performed simulations with 1-, 2-, and 3-fold each value of the number of SpoVM molecules.
Our simulations yielded predictions of the kinetics of binding for each set of parameter values (Figure 1K), from which we extracted the steady-state fractions of sites on 2- and 8-μm SSLBs bound by SpoVM molecules. We then compared these binding fraction estimates to the values from our experimental data after subtracting the background value at time t = 0 (Figure 1L). We identified a region of parameter space for k8 = 0.5k2 and Ec = 0.5kBT that produced bound fractions in good agreement with our experimental data (Figure 1K, orange box), and closer examination of the binding kinetics agreed with our experimental curves (Figure 1L). Taken together, we conclude that the increased affinity of SpoVM for positively curved membranes can be explained by an approximately two-fold faster on-rate of the protein, which is likely due to faster folding and/or insertion of the protein into more highly curved membranes, combined cooperativity between SpoVM molecules. Although this cooperativity is weak (on the order of 1kBT) when considering microscopic interactions between individual SpoVM molecules, it is appreciable (Hill coefficient≈4) in bulk, at steady state.
Structural flexibility near position 9 is critical for SpoVM membrane curvature discrimination
The proline at residue 9 of SpoVM is completely conserved among SpoVM orthologs (van Ooij and Losick, 2003). Previous work demonstrated that substitution of Pro9 with alanine degrades the specificity of SpoVM binding (van Ooij and Losick, 2003), resulting in its absorption to both the mother cell and forespore membranes in vivo (Fig 2C). Accordingly, in vitro, SpoVMP9A-GFP bound promiscuously to membranes regardless of curvature (Ramamurthi et al., 2009) and we observed that SpoVMP9A-FITC was also less able to discriminate between differently curved membranes as measured via flow cytometry (Figure 2K, Table 1).
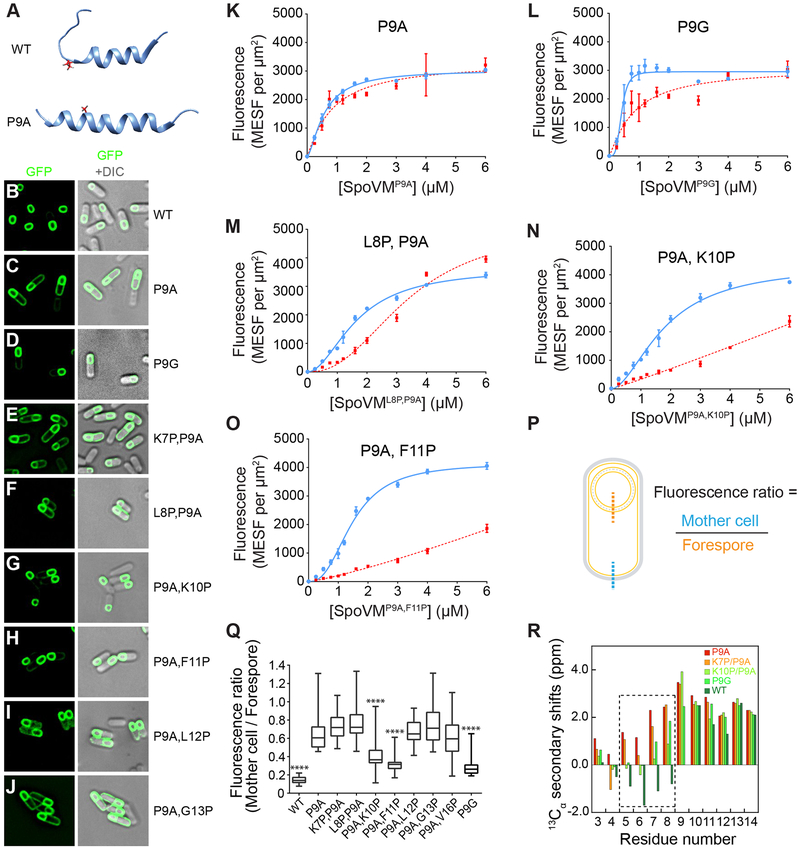
Figure 2: An unstructured N-terminus is required for proper localization of SpoVM in vivo and in vitro.
(A) NMR structures of SpoVM and SpoVMP9A (PDB IDs 2MVH and 2MVJ, respectively). (B-J) Fluorescence micrographs of sporulating B. subtilis cells producing SpoVM-GFP (B; strain CVO1195), SpoVMP9A (C; CVO1395), SpoVMP9G-GFP (D; EYKS904), SpoVMK7P,P9A-GFP (E; EYKS869), SpoVML8P,P9A-GFP (F; EYKS870), SpoVMP9A,K10P-GFP (G; EYKS871), SpoVMP9A,F11P-GFP (H; EYKS872), SpoVMP9A,L12P-GFP (I; EYKS873), or SpoVMP9A,G13P-GFP (J; EYKS874). (K-O) Adsorbance of SpoVM-FITC variants, as measured by flow cytometry onto differently curved membranes in vitro. Fluorescence intensities were measured using flow cytometry for (K) SpoVMP9A-FITC, (L) SpoVMP9G-FITC, (M) SpoVML8P,P9A-FITC, (N) SpoVMP9A,K10P-FITC, or (O) SpoVMP9A,F11P-FITC at varying concentrations incubated with a mixed population of 2-μm (blue) and 8-μm (red) SSLBs. Data points and error bars represent mean values and standard deviation, respectively, from three independent replicates. (P) Schematic of sporulating B. subtilis and representative line scans across the mother cell (blue) and forespore (orange) membranes. (Q) Ratio of mother cell to forespore membrane maximum fluorescence intensities from line scans as shown in (P) in sporulating B. subtilis cells producing the indicated SpoVM-GFP variants. **** indicates p < 0.0001 compared to SpoVMP9A. See also Figure S3. (R) 13Cα secondary chemical shifts for residues 3 to 14 of SpoVM and variants are shown. Similar 13Cα secondary shifts of N-terminal residues of SpoVMP9A and SpoVMK7P,P9A (red and orange, respectively) indicate their structured nature. Conversely, the 13Cα secondary shifts of SpoVMP9G and SpoVMP9A,K10P (fluorescent and light green, respectively) reflect their unstructured N-terminus, similar to the wildtype protein (dark green). Additional supports by NMR NOESY and circular dichroism are shown in Figures S4 and S5.
We asked if these effects P9A on the function of SpoVM could be rationalized in terms of its structure. SpoVM inserts spontaneously into convex membranes, parallel to the plane of the membrane, and assumes a structure with an N-terminal unstructured region that extends from residue 1–10, and a C-terminal amphipathic α-helix (Gill et al., 2015). In contrast to wild-type SpoVM (Fig. 2A, top), which contains an unstructured N-terminus and an α-helical C-terminus (Gill et al., 2015), the NMR structure of SpoVMP9A is α-helical along its entire length (Fig. 2A, bottom). Given that substitution of Pro9 resulted in mislocalization of the protein in vivo (van Ooij and Losick, 2003) and an entirely α-helical structure that was unable to discriminate between more and less convex membranes in vitro (Gill et al., 2015; Ramamurthi et al., 2009), this suggests that the helix breaking propensity of Pro9 is functionally important and that the unstructured region of SpoVM plays an active role in specifying its localization in vivo.
To test this hypothesis, we asked whether substitution of Pro9 with glycine, another residue known to break α-helices, would phenocopy SpoVMWT localization. In sporulating B. subtilis cells, SpoVMP9G-GFP localized largely to the forespore surface, similar to wildtype (Figure 2D). To quantify the extent of localization of SpoVM variants to the forespore, we measured the maximum fluorescence intensity along a line scan of a patch of forespore membrane and a patch of mother cell membrane distal to the forespore, and calculated the ratio of the mother cell membrane intensity to that of the forespore membrane (Figure 2P, for controls, see Figure S3A). The median fluorescence ratio for SpoVM-GFP was 0.14, whereas that of SpoVMP9A-GFP was 0.61, consistent with the preferential localization of SpoVM-GFP to the forespore and the less forespore-specific localization of SpoVMP9A-GFP (Figure 2Q). In comparison, the median fluorescence ratio for SpoVMP9G-GFP was 0.26 (Figure 2Q), suggesting that a glycine at position 9 was largely able to restore the preferential localization of SpoVM to the forespore surface compared to SpoVMP9A-GFP (p < 0.0001). Accordingly, quantification of the in vitro binding parameters for SpoVMP9G-FITC showed preferential binding to the more highly curved 2-μm beads (Figure 2L), and the fitted parameters were similar to wildtype, albeit with a slightly higher binding affinity and a lower Bmax (Table 1). These data demonstrate that Gly at position 9 can partially substitute for Pro with respect to the ability of SpoVM to discriminate between differently curved membranes in vivo and in vitro. We hypothesize that it is glycine’s ability to break the alpha-helix and thereby delineate between the unstructured N-terminal and α-helical C-terminal regions of SpoVM, not the identity of the amino acid at position 9 per se, that is important for SpoVM function in vivo and in vitro.
The unstructured N-terminal region of SpoVM mediates cooperative behavior in vitro
If this interpretation is correct, then altering the N-terminal structure by moving the relative position of the Pro residue could affect SpoVM function. To this end, we constructed variants that initially harbored the P9A substitution, and then re-introduced Pro substitutions at positions 7–13, thereby presumably altering the unstructured N-terminus, and examined the localization of these variants fused to GFP in sporulating B. subtilis cells using fluorescence microscopy (Figure 2E-J,Q). We identified two variants (SpoVMP9A,K10P and SpoVMP9A,F11P) with fluorescence localization ratios significantly lower than SpoVMP9A in a two-tailed unpaired Student’s t-test (Figure 2G-H,Q), indicating that they largely localized properly in vivo. In contrast, all other variants largely failed to preferentially localize to the forespore surface.
NMR characterization of 13C, 15N-labeled SpoVM, SpoVMP9A, SpoVMP9G, SpoVMK10P,P9A, and SpoVMK7A,P9A in the presence of micelles provided direct structural evidence that variants with impaired localization retained α-helical structure in their N-termini, whereas variants that localized properly retained an unstructured N-terminus (Figure 2R, S4). In addition, all variants displayed characteristic local minima at 208 and 222 nm in their circular dichroism (CD) spectra, indicating adoption of an α-helical region within the protein (Figure S5).
Next, we asked whether the two identified localization-competent double mutants recapitulated wild type behavior in vitro. SpoVMP9A,K10P-FITC and SpoVMP9A,F11P-FITC displayed preferential binding to the more highly curved 2-μm beads across all concentrations tested (Figure 2N–O), albeit with reduced overall affinities for both membranes (indicated by the fitted K1/2 values; Table 1) as compared to wildtype. Additionally, although the Hill coefficients decreased for binding of both double mutants onto both surfaces, SpoVMP9A,K10P-FITC and SpoVMP9A,F11P-FITC maintained a significantly higher cooperativity for binding onto 2-μm beads. By contrast, the saturation binding curve for SpoVML8P,P9A, which did not localize to the forespore in vivo, showed reduced preference for 2-μm beads and displayed a three-fold decrease in affinity compared to wildtype for both 2-μm and 8-μm beads. Furthermore, unlike SpoVMP9A,K10P and SpoVMP9A,F11P, binding of SpoVML8P,P9A to 2-μm and 8-μm beads displayed similar Hill coefficients, suggesting that disrupting the N-terminus of SpoVM reduced the difference in cooperativity displayed on differently curved membranes. Taken together, these observations demonstrate that while moving Pro9 closer to the C-terminus (which allows retention of an unstructured N-terminus) does not abolish curvature-based localization, moving Pro9 closer to the N-terminus results in a structured N-terminus and specifically reduces preferential cooperative binding on more convex surfaces. Moreover, it is relative, not absolute, differences in affinity and cooperativity that allow SpoVM to discriminate between membranes of different curvature.
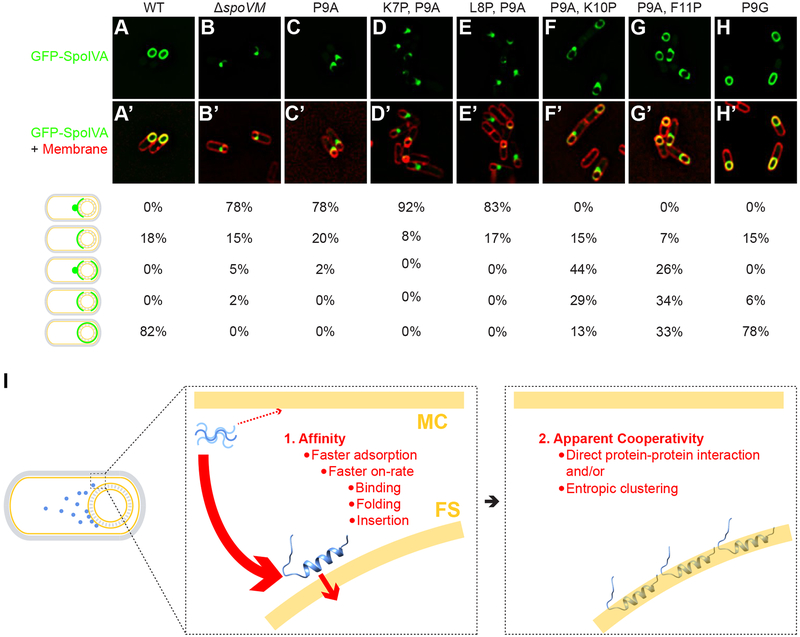
We had identified two sets of variants: those with behavior that is qualitatively similar to wild-type but quantitatively different, and those that fail to specifically localize to the forespore. Next, we asked whether the functional variants could recapitulate wild-type function in the broader context of sporulation. We first tested the ability of SpoVM variants in recruiting SpoIVA, the structural component of the basement layer of the spore coat (Roels et al., 1992) that is tethered to the forespore surface by SpoVM (Ramamurthi et al., 2006; Wu et al., 2015). In sporulating B. subtilis producing SpoVMWT, GFP-SpoIVA localized evenly around the forespore in 82% of cells (Figure 3A) (Price and Losick, 1999). In the absence of SpoVM, or in the presence of SpoVMP9A, GFP-SpoIVA did not uniformly localize around the forespore, but instead mislocalized as puncta on the mother cell-proximal face of the forespore in the majority of cells (Figure 3B,C) (Ramamurthi et al., 2006). For cells producing the variants that are localization-competent in vitro, 13% (SpoVMP9A,K10P), 33% (SpoVMP9A,F11P), or 78% (SpoVMP9G) of cells displayed proper, wild-type localization of GFP-SpoIVA. This trend was also reflected in the ability of these cells to produce viable spores. SpoVMP9A,K10P and SpoVMP9A,F11P were largely unable to sporulate (Table S1, strains G–H), indicating that simple forespore localization of SpoVM and SpoIVA is not sufficient for initiation of cortex assembly. However, cells producing SpoVMP9G, which mimics the kink provided by Pro at position 9, sporulated at near wildtype levels. (Table S1, strain I). Taken together, these results indicate that the forespore localization and SpoIVA recruitment functions of SpoVM require a flexible N-terminus, and that full in vivo function (including the ability to signal initiation of cortex assembly) requires a helix break specifically at position 9, which may be provided by Pro or, to a slightly lesser extent, Gly.
Figure 3: Proper localization of GFP-SpoIVA in vivo requires properly localized SpoVM.
Localization of GFP-SpoIVA in sporulating B. subtilis cells in the presence (A; strain KR165) or absence (B; KR178) of SpoVM, or in the presence of SpoVMP9A (C; SE50), SpoVMK7P,P9A (D; EYKS919), SpoVML8P,P9A (E; EYKS920), SpoVMP9A,K10P (F; EYKS892), SpoVMP9A,F11P (G; EYKS893), or SpoVMP9G (H; EYKS929). (A’-H’) Overlay of GFP fluorescence (green) and membranes (red) visualized with fluorescent dye FM4–64 from panels A-H, respectively. Fraction of cells displaying the indicated localization pattern is shown to the right (n > 55 cells for each). (I) Model for the affinity/cooperativity-driven localization of SpoVM onto positively curved membranes. Left: depiction of sporulating B. subtilis in which SpoVM molecules (blue) synthesized in the mother cell (MC) adsorb onto the convex membrane surface (yellow) of the forespore (FS). Center: unordered SpoVM molecule (blue, upper left) has a higher intrinsic affinity for the convex forespore surface (thick red arrow). Higher affinity is mediated by a faster on-rate, which is composed of three steps: initial binding, protein folding, and membrane insertion. We propose that folding and/or insertion occur more quickly on membranes of preferred curvature. Right: successful insertion of the first SpoVM molecule positively influences the subsequent insertion of other SpoVM molecules, mediated by direct protein-protein interaction and/or entropic clustering.
DISCUSSION
The number of reported bacterial proteins whose subcellular localization appears mediated by detection of micron-scale differences in membrane curvature, dictated by the local shape of the cell, has steadily increased (Updegrove and Ramamurthi, 2017). It was recently reported that eukaryotic septins may also exploit membrane curvature to drive protein localization, suggesting that curvature-driven protein localization may be a widely conserved strategy (Bridges et al., 2016). However, the underlying mechanistic details are just beginning to be understood, underscoring the importance of relatively simple model proteins whose geometric sensing is amenable to interrogation by multiple cytological, structural, and biophysical approaches. We have investigated the localization of a small bacterial protein, SpoVM, that preferentially embeds in the slightly convex surface of the ~1-μm forespore during sporulation in B. subtilis. Previously, we reported that membrane curvature sensing by SpoVM requires it to assume a structure that harbors a flexible N-terminus and an α-helical C-terminus, and that this structure mediates an affinity/cooperativity-driven preferential adsorption onto convex membranes. Here, we demonstrated that SpoVM adsorbs faster onto more convex membranes, which at steady state is revealed as a higher intrinsic affinity for more convex membranes. The faster adsorption of SpoVM can be explained by an approximately two-fold higher on-rate, an equal and slow off-rate, and weak cooperativity between SpoVM molecules, which is mediated by the unstructured N-terminus.
Our data suggest a model in which the relatively simple structure of SpoVM plays a central role in determining its subcellular localization (Figure 3I). In this model, SpoVM is synthesized in the mother cell cytosol as a largely unstructured 26-aa protein (Prajapati et al., 2000) and then adsorbs onto convex membranes. The on-rate component of this adsorption (as with other amphipathic helices that spontaneously insert into membranes) can be deconstructed into three distinct steps (Seelig, 2004). First, electrostatic forces, mediated by positive charges on the unstructured protein and negative charges of phospholipid head groups of a membrane, drive an initial reversible binding of the protein on the membrane surface. We hypothesize that this reversible step occurs equivalently on membranes regardless of curvature and that this allows SpoVM to sample different membranes before committing to one surface. This step is followed by folding and stable insertion of the peptide into the membrane bilayer. We propose that membranes of preferred curvature accelerate one or both of these steps. The stable insertion of a properly folded SpoVM molecule then positively influences subsequent SpoVM binding events in a manner that requires a flexible N-terminus, which is reflected in steady state measurements as apparent cooperativity, although the molecular nature of the apparent cooperativity is currently unclear. In one model, the flexible N-terminus may mediate direct protein-protein interactions between SpoVM molecules. Alternatively, the N-terminus could drive entropic clustering of SpoVM molecules in a lipid bilayer without requiring physical contact between SpoVM molecules, similar to proposed mechanisms for aggregation of mechanosensitive channels (Grage et al., 2011; Ursell et al., 2007). In our simulations, a relatively low cooperative energy of 0.5kBT was sufficient to drive preferential localization of SpoVM, which is consistent with membrane-mediated indirect interactions between SpoVM molecules. Regardless, the model provides a mechanism by which intrinsic affinity (faster on-rate) and apparent cooperativity (mediated by the flexible N-terminus) can result in the aggregate accumulation of SpoVM molecules onto membranes of preferred curvature.
Despite the diminutive size of SpoVM, appending its 26 amino acids appears sufficient to direct unrelated proteins such as GFP to convex membranes, similar to how appending localization signals to heterologous proteins is sufficient to drive their localization to particular subcellular sites (Blobel, 1980). It is therefore tempting to speculate that there may exist proteins in other systems that harbor localization domains resembling SpoVM, wherein a flexible N-terminus is followed by an amphipathic helix, separated by a residue such as Gly or Pro that confers a region of flexibility. Due to the small size of SpoVM, we have been unable to identify such a motif using bioinformatic approaches. Moreover, given the underrepresentation of hydrophobic, membrane-associated domains in the protein database, it is not surprising that we have been unable to identify a protein whose structure has been solved that harbors a SpoVM-like motif, since amphipathic alpha helices are usually unstructured in the absence of a phospholipid bilayer. Perhaps structural determinations of such domains will reveal a common mechanism by which proteins preferentially embed in membranes that are curved at the micron scale.
STAR METHODS
Contact for Reagent and Resource Sharing
Please contact K.S.R. (ramamurthiks@mail.nih.gov) for reagents and resources generated in this study.
Experimental Model and Subject Details
All bacterial strains used in this study are derivatives of Bacillus subtilis PY79 (Youngman et al., 1984).
Method Details
Strain construction and general methods
Genotypes of strains used are provided in STAR Methods. Strains expressing variants of spoVM at the amyE locus, driven by the native PspoVM promoter, were created by site-directed mutagenesis from integration vector pKR55, and variants of spoVM-gfp from integration vector pKC2 (Ramamurthi et al., 2006). Expression of gfp-spoIVA at the thrC locus was driven by the native PspoIVA promoter. Plasmids expressing His6-SUMO-spoVMK7P,P9A, His6-SUMO-spoVMP9A,K10P, and His6-SUMO-spoVMP9Gwere created by site-directed mutagenesis from plasmid pKR209, harboring His6-SUMO-spoVMP9A described previously (Gill et al., 2015). SpoVM-FITC and variants used for the SSLB flow-cytometry assay were synthesized (Biomatik) and purified by HPLC to >95% purity. The FITC label was conjugated via the addition of Lys-FITC at the C-terminus.
Fluorescence microscopy
Cells were induced to sporulate by resuspension in SM medium (Sterlini and Mandelstam, 1969). For microscopy, cells were harvested from 1 ml culture by centrifugation and resuspended in 100 μl SM medium. If required, the membrane dye FM4–64 was added at 15 μg/ml and incubated for at least 15 min at room temperature. 3 μl cell suspension were placed on glass-bottom culture dishes (Mattek Corp). A pad made of 1% agarose (in ultrapure water) was cut to size and placed over the cells. Cells were viewed at room temperature with a DeltaVision Core microscope system (GE Healthcare/Applied Precision) equipped with a Photometrics CoolSnap HQ2 camera as described previously (Eswaramoorthy et al., 2014). Ten z-planes were acquired at 200-nm intervals and the data were deconvolved using SoftWorx (ver. 5.5, Applied Precision/GE Healthcare). Analysis of fluorescence intensities along line scans was performed using ImageJ. Images shown are representatives of three independent experiments.
SSLB preparation
SSLBs were made as described previously (Bayerl and Bloom, 1990; Gopalakrishnan et al., 2009). Briefly, liposomes were produced by sonication using 100 μl (25 mg/ml) E. coli polar lipid extract (Avanti), which were first evaporated under vacuum overnight at room temperature onto the bottom of 16 mm × 100 mm borosilicate tubes, then hydrated with 1 ml ultrapure water at 42 °C for 1 h with occasional vortexing. Resuspended lipids were subjected to five cycles of freezing in an ethanol/dry ice bath and thawing at 42 °C, followed by bath sonication for 1 h, at which point the suspension became transparent. Debris was removed by centrifugation at 13,000g for 10 min, and the cleared supernatant, which contained unilamellar vesicles, was retained. Next, 10 mg of 2 μm-diameter silica beads (Polysciences, Inc.) or 8 μm-diameter silica beads (Cospheric LLC) were prepared for coating by successive washing, three times each, in ultrapure water, methanol, 1 M NaOH, and ultrapure water. The final resuspension volume for the beads was 200 μl in ultrapure water, to which 200 μl of the prepared unilamellar vesicles were added. CaCl2, at a final concentration of 1 μM, was added and the suspension was vortexed for 2 min, then incubated at 42 °C for 1 h with occasional vortexing. The SSLBs were then collected by centrifugation at 13,000g, washed three times in ultrapure water, and resuspended in 1 ml of ultrapure water.
Flow cytometry
Varying concentrations of synthesized SpoVM-FITC (or variants) were incubated in Tris buffer (50 mM Tris at pH 7.5, 400 mM NaCl, and 10 mM MgCl2) containing 1 μl (10 mg/ml) 2-μm SSLBs and 4 μl (10 mg/ml) 8-μm SSLBs to ensure equal surface area representation by both types of SSLBs (40 μl total reaction volume). For equilibrium binding experiments, the reactions were incubated for 4 h at 25 °C following a program of alternating shaking (700 rpm) and resting every 5 min to ensure that the beads did not settle. For kinetic binding experiments, the reactions were incubated for the reported amount of time using the same program of shaking, or continuous shaking for time points under 5 min. The beads were then diluted to a total volume of 1 ml in Tris buffer and immediately analyzed on a FACSCanto II (BD Biosciences, San Jose, CA) flow cytometer using a 488-nm laser and 530/30 bandpass filter to detect FITC fluorescence. The following voltage values were used: 200 V for forward scatter, 250 V for side scatter, and 400 V for the FITC channel. For each sample, data from approximately 40,000 particles were acquired. Gates were drawn around the 2-μm and 8-μm beads based on forward and side scatter (Supporting Information, Figure S1), and the median FITC fluorescence intensity values were recorded. The arbitrary fluorescence units were normalized to units of Molecules of Equivalent Soluble Fluorochrome (MESF) using Quantum FITC-5 MESF beads (Bangs Laboratories, Inc.), which were analyzed on the same day for each experiment. Three replicates of each sample were analyzed using the above method for each condition, and the reported values are the average and standard deviations of these three independent replicates.
Sporulation assay
Sporulation was induced by nutrient depletion in cultures grown in 2 ml Difco sporulation medium (DSM; KD Medical) for at least 24 h at 37 °C. At least three independent cultures were subjected to heat treatment at 80 °C for 20 min, then serially diluted in ultrapure water and plated onto LB agar. The LB agar plates were incubated at 37 °C, and the sporulation efficiency was reported as the number of colony forming units (CFUs) relative to the number of CFUs obtained from a parallel culture of B. subtilis PY79.
Monte Carlo simulations
Simulations were performed similar to our previous study (Gill et al., 2015). In brief, to model the binding of SpoVM molecules onto 2- and 8-μm SSLBs, two sets of square lattices were initialized as empty to represent available binding sites on the beads. The 2-μm beads were represented by sixteen 80 × 80 lattices, and an 8-μm bead was represented by a single 320 × 320 lattice, so that there was an equal fraction of binding sites on each bead size. All lattices had the same spacing and had periodic boundary conditions. We assumed that all lattice sites are equally accessible to unbound SpoVM molecules, corresponding to the limit of rapid diffusion. In the absence of cooperativity, during each time step dt, an empty site on a 2- or 8-μm surface became bound with probability k2Nunbounddt or k8Nunbounddt, respectively, where Nunbound is the number of unbound SpoVM molecules. To represent cooperativity in binding when a site was occupied by a SpoVM molecule, the probability for SpoVM binding to its four nearest-neighbor sites was increased by a factor exp(Ec/kBT). The probability for a bound SpoVM to be released was determined by an off-rate koff that was the same for 2- and 8-μm SSLBs. For the simulations in Figure 3, k2 = 10−7 min−1 and koff = 0.0025 min−1. The time step dt = 1 min was chosen to be sufficiently small so that all probabilities were <1. We verified that the kinetics were virtually unchanged for shorter time steps. Simulations were terminated when the density of molecules bound to the 2- and 8-μm beads had reached a steady-state value.
NMR spectroscopy
Expression and initial purification of isotope-labeled proteins were performed as described previously (Gill et al., 2015). To cleave the N-terminal His6-Sumo tag from SpoVM variants, H-Ulp1-His6-Sumo protease was added to the eluate of a Ni-NTA column for 3–10 h with agitation at room temperature. The solution was then diluted with 20 mM Tris at pH 8.0 to achieve a final concentration of 50 mM imidazole. The sample was then re-applied three times to a clean Ni-NTA column, and washed with buffer A (20 mM Tris at pH 8.0, 500 mM NaCl, 80 mM guanidine-HCl, and 20 mM 1,2 dihexanoyl-sn-glycero-3-phosphocholine (DHPC)), then buffer B (20 mM Tris at pH 8.0, 2 M NaCl, 80 mM guanidine-HCl, and 20 mM DHPC). Both fractions contained the target protein, and were concentrated using an Amicon Ultra centrifugal filter (Millipore) to a volume of 300 μl for loading into the NMR tube. Typical NMR samples contained 0.5 to 1 mM protein in a buffer of ~100 mM DHPC, 80 mM NaCl, and 16 mM phosphate at pH 6.0. Protein backbone resonance assignments were carried out using traditional triple resonance experiments as described previously (Gill et al., 2015). All NMR data were collected on a Bruker 600-MHz spectrometer equipped with a cryoprobe. Data were processed using NMRPipe and analyzed using NMRView.
Quantification and Statistical Analysis
For flow cytometry experiments in Fig. 1D–J and Fig. 2K–O, average values and standard deviations were calculated from three independent experiments. These data were fit with a sigmoidal model using GraphPad Prism to determine equilibrium binding and kinetic parameters. For determining significance in Fig. 2Q, line scans from N > 20 cells were used to perform a two-tailed Student’s t test. **** indicates p-value < 0.0001. For Fig. 3A–H, the percentage of cells found in each morphological category was determined from N > 55 cells. Sporulation efficiencies in Table S1 were determined by counting colonies from three independent experiments.
Supplementary Material
Key Resources Table
| Bacillus subtilis strains used in this study | ||
|---|---|---|
| Strain | Genotype | Reference |
| PY79 | Prototrophic derivative of B. subtilis 168 | (Youngman et al., 1984) |
| KR94 | ΔspoVM::tetR | (Ramamurthi et al., 2006) |
| EYKS883 | ΔspoVM::tet, amyE::spoVM cat | |
| EYKS884 | ΔspoVM::tet, amyE::spoVMP9A cat | |
| EYKS917 | ΔspoVM::tet, amyE::spoVMK7P,P9A cat | |
| EYKS918 | ΔspoVM::tet, amyE::spoVML8P,P9A cat | |
| EYKS885 | ΔspoVM::tet, amyE::spoVMP9A,K10P cat | |
| EYKS886 | ΔspoVM::tet, amyE::spoVMP9A,F11P cat | |
| EYKS909 | ΔspoVM::tet, amyE::spoVMP9G cat | |
| KR160 | thrC::gfp-spoIVA spec | (Ramamurthi and Losick, 2008) |
| KR178 | ΔspoVM::tet, thrC::gfp-spoIVA spec | (Ramamurthi and Losick, 2008) |
| KR165 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVM cat | (Ramamurthi and Losick, 2008) |
| SE50 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVMP9A cat | (Ebmeier et al., 2012) |
| EYKS919 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVMK7P,P9A cat | |
| EYKS920 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVML8P,P9A cat | |
| EYKS892 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVMP9A,K10P cat | |
| EYKS893 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVMP9A,F11P cat | |
| EYKS929 | ΔspoVM::tet, thrC::gfp-spoIVA spec, amyE::spoVMP9G cat | |
| CVO1195 | amyE::spoVM-gfp cat | (van Ooij and Losick, 2003) |
| CVO1395 | amyE::spoVMP9A-gfp cat | (van Ooij and Losick, 2003) |
| EYKS869 | amyE::spoVMP9A,K7P-gfp cat | |
| EYKS870 | amyE::spoVMP9A,L8P-gfp cat | |
| EYKS871 | amyE::spoVMP9A,K10P-gfp cat | |
| EYKS872 | amyE::spoVMP9A,F11P-gfp cat | |
| EYKS873 | amyE::spoVMP9A,L12P-gfp cat | |
| EYKS874 | amyE::spoVMP9A,G13P-gfp cat | |
| EYKS875 | amyE::spoVMP9A,V16P-gfp cat | |
| EYKS904 | amyE::spoVMP9G-gfp cat | |
| Item | Manufacturer | Catalog number |
| Peptides | ||
| SpoVM-FITC and variants | Biomatik | Custom |
| Lipids | ||
| E. coli polar lipid extract | Avanti Polar Lipids, Inc. | 100600C |
| Beads | ||
| 2 μm silica microspheres | Polysciences, Inc. | 24328–15 |
| 8 μm silica microspheres | Cospheric, LLC | SiO2MS-1.8 8um |
| Fluorescence bead reference | Bangs Laboratories, Inc. | 555 |
ACKNOWLEDGEMENTS
We thank members of our labs for comments on the manuscript; K. Wolcott (NCI Flow Cytometry Core Facility); and S. Gottesman, S. Wickner, M. Maurizi, and I. Pastan for discussions. This work was funded by a 2016 NCI Director’s Innovation Award (E.Y.K.), NIH NIGMS Grant R01GM105963 (F.T. and K.C.H.), the Allen Discovery Center at Stanford University on Systems Modeling of Infection (K.C.H.), the Pennsylvania Department of Health using Tobacco CURE Funds (F.T.; the Department specifically disclaims responsibility for any analyses, interpretations or conclusions); and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (K.S.R.).
REFERENCES
- Bayerl TM, and Bloom M (1990). Physical properties of single phospholipid bilayers adsorbed to micro glass beads. A new vesicular model system studied by 2H-nuclear magnetic resonance. Biophys J 58, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G (1980). Intracellular protein topogenesis. Proc Natl Acad Sci U S A 77, 1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges AA, Jentzsch MS, Oakes PW, Occhipinti P, and Gladfelter AS (2016). Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J Cell Biol 213, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing JP, Nagy A, Anantharaman V, Aravind L, and Ramamurthi KS (2013). ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc Natl Acad Sci U S A 110, E151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deich J, Judd EM, McAdams HH, and Moerner WE (2004). Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Natl Acad Sci U S A 101, 15921–15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A, and Eichenberger P (2016). The Spore Coat. Microbiol Spectr 4. [DOI] [PubMed] [Google Scholar]
- Dworkin J (2014). Protein Targeting during Bacillus subtilis Sporulation. Microbiol Spectr 2, TBS-0006–2012. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Winter PW, Wawrzusin P, York AG, Shroff H, and Ramamurthi KS (2014). Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet 10, e1004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RL Jr., Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, and Ramamurthi KS (2015). Structural basis for the geometry-driven localization of a small protein. Proc Natl Acad Sci U S A 112, E1908–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan G, Rouiller I, Colman DR, and Lennox RB (2009). Supported bilayers formed from different phospholipids on spherical silica substrates. Langmuir 25, 5455–5458. [DOI] [PubMed] [Google Scholar]
- Govindarajan S, Elisha Y, Nevo-Dinur K, and Amster-Choder O (2013). The general phosphotransferase system proteins localize to sites of strong negative curvature in bacterial cells. MBio 4, e00443–00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grage SL, Keleshian AM, Turdzeladze T, Battle AR, Tay WC, May RP, Holt SA, Contera SA, Haertlein M, Moulin M, et al. (2011). Bilayer-mediated clustering and functional interaction of MscL channels. Biophys J 100, 1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, and Dworkin J (2012). Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36, 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, and Hamoen LW (2009). Localisation of DivIVA by targeting to negatively curved membranes. Embo J 28, 2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Fan N, Ricca E, Driks A, Losick R, and Cutting S (1993). An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol 9, 761–771. [DOI] [PubMed] [Google Scholar]
- Prajapati RS, Ogura T, and Cutting SM (2000). Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim Biophys Acta 1475, 353–359. [DOI] [PubMed] [Google Scholar]
- Price KD, and Losick R (1999). A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol 181, 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, and Losick R (2006). Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62, 1547–1557. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, and Losick R (2009). Geometric cue for protein localization in a bacterium. Science 323, 1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, and Losick R (2008). ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell 31, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, and Losick R (2009). Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A 106, 13541–13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Eswaramoorthy P, Ramamurthi KS, and Weibel DB (2013). Studying biomolecule localization by engineering bacterial cell wall curvature. PLoS One 8, e84143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels S, Driks A, and Losick R (1992). Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol 174, 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, and Losick R (2010). Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol 2, a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, and Losick RM (2002). Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci U S A 99, 8701–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J (2004). Thermodynamics of lipid-peptide interactions. Biochim Biophys Acta 1666, 40–50. [DOI] [PubMed] [Google Scholar]
- Setlow P (2014). Spore Resistance Properties. Microbiol Spectr 2. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, and Losick R (2009). Why and how bacteria localize proteins. Science 326, 1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel SD, Liu J, and Ton-That H (2016). Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol 34, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, and Mandelstam J (1969). Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, Ronneau S, Gonzalez BS, Klutsch D, Schaffner-Barbero C, and Hamoen LW (2015). Transmembrane protein sorting driven by membrane curvature. Nat Commun 6, 8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, and Ramamurthi KS (2014). Spore formation in Bacillus subtilis. Environ Microbiol Rep 6, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigotaki A, De Geyter J, Sostaric N, Economou A, and Karamanou S (2017). Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15, 21–36. [DOI] [PubMed] [Google Scholar]
- Updegrove TB, and Ramamurthi KS (2017). Geometric protein localization cues in bacterial cells. Curr Opin Microbiol 36, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell T, Huang KC, Peterson E, and Phillips R (2007). Cooperative gating and spatial organization of membrane proteins through elastic interactions. PLoS Comput Biol 3, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, and Huang KC (2014). Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A 111, E1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, and Losick R (2003). Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol 185, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MM, and Bernstein HD (2016). Surface-Exposed Lipoproteins: An Emerging Secretion Phenomenon in Gram-Negative Bacteria. Trends Microbiol 24, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, and Ramamurthi KS (2015). A versatile nano display platform from bacterial spore coat proteins. Nat Commun 6, 6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, and Losick R (1984). Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12, 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.