Abstract
Slow-wave activity (SWA) in the electroencephalogram during slow-wave sleep (SWS) varies as a function of sleep-wake history. A putative sleep-active population of neuronal nitric oxide synthase (nNOS)-containing interneurons in the cerebral cortex, defined as such by the expression of Fos in animals euthanized after protracted deep sleep, may be a local regulator of SWA. We investigated whether electrophysiological responses to activation of these cells are consistent with their role of a local regulator of SWA. Using a Cre/loxP strategy, we targeted the population of nNOS interneurons to express the light-activated cation channel Channelrhodopsin2 and the histological marker tdTomato in mice. We then performed histochemical and optogenetic studies in these transgenic mice. Our studies provided histochemical evidence of transgene expression and electrophysiological evidence that the cerebral cortex was responsive to optogenetic manipulation of these cells in both anesthetized and behaving mice. Optogenetic stimulation of the cerebral cortex of animals expressing Channelrhodopsin2 in nNOS interneurons triggered an acute positive deflection of the local field potential that was followed by protracted oscillatory events only during quiet wake and slow wave sleep. The response during wake was maximal when the electroencephalogram (EEG) was in a negative polarization state and abolished when the EEG was in a positive polarization state. Since the polarization state of the EEG is a manifestation of slow-wave oscillations in the activity of underlying pyramidal neurons between the depolarized (LFP negative) and hyperpolarized (LFP positive) states, these data indicate that sleep-active cortical neurons expressing nNOS function in sleep slow-wave physiology.
Keywords: evoked potential, electroencephalogram, mice, optogenetic, slow-wave activity
Introduction
High-amplitude slow-wave activity (SWA; < 4 Hz) in the cerebral cortical electroencephalogram (EEG) is a defining feature of slow-wave sleep (SWS) and is thought to mediate functional consequences of SWS (Tononi and Cirelli, 2006; Puentes-Mestril and Aton, 2017; Siclari and Tononi, 2017; Walsh et al., 2006; Tasali et al., 2008; Aton et al., 2014). We previously demonstrated that type I neuronal nitric oxide synthase (nNOS) cells in the cerebral cortex are activated during episodes of SWS associated with increased SWA (Gerashchenko et al., 2008). Further, we performed injections of neurokinin 1 (NK1) receptor agonists and antagonists into the cerebral cortex and found that SWA was locally enhanced by NK1 receptor agonists and reduced by NK1 receptor antagonists (Zielinski et al., 2015). Because NK1 receptors are expressed exclusively in nNOS neurons in the cerebral cortex (Dittrich et al., 2012), this result suggests that modulation of the SWA production is caused by changes in the activity of nNOS neurons. Collectively, these results suggest the important role of nNOS neurons in the cerebral cortex in the production of SWA and emphasize the importance of studying the mechanisms by which the sleep-active cells affect SWA production. Herein, we used a pharmacologically-activated double transgenic mouse model to measure the effects of activating these cells optogenetically on neuronal activity at the EEG level. We report that a Channelrhodopsin2 (ChR2) construct (Feng, 2012) driven by the nNOS promoter (Taniguchi et al., 2011) is expressed in cortical nNOS-positive cells and that the optogenetic activation of this cell population induces EEG potential fluctuations more robustly and reliably during wake than during SWS. The magnitude of evoked EEG responses is polarization-state dependent and is abolished when the cortex is at a positive potential, akin to its state during a physiological slow wave. These observations are compatible with a role for type I nNOS cells in the cortex regulating local SWA homeostatic dynamics.
Experimental Procedures
All in vivo experimentation was approved by the institutional animal care and use committee and was performed in accordance with National Institutes of Health guidelines. Immunohistochemical studies were performed at the Harvard Medical School in the West Roxbury VA Medical Center. Optogenetic studies were performed at Elson S. Floyd College of Medicine and Department of Integrative Physiology and Neuroscience in Washington State University. Thirty five adult male mice were used in these studies.
Transgenic mouse generation
A loxP-regulated genetic construct was previously reported as a tool for selectively targeting a neuronal population of interest for optogenetic excitation with the light-sensitive ion channel, ChR2 (Feng, 2012). These mice of the B6;129-Gt(ROSA)26Sortm1(CAG-COP4*E123T*H134R,-tdTomato)Gfng/J strain are referred to hereafter as loxP-ChR2. The transgene construct contains a CAG promoter, a loxP site-flanked STOP fragment and pGK-NEO-pA cassette, 2 copies of ChR2 with the E123T mutation (ChETA variant) and the H134R mutation, a P2A oligopeptide sequence, the tdTomato variant of enhanced red fluorescent protein (RFP), and the WPRE (Woodchuck Hepatitis Virus (WHP) Posttranscriptional Regulatory Element) sequence at the Gt(ROSA)26Sor locus. In the absence of in vivo recombination, expression of ChR2 and tdTomato is blocked by the loxP-flanked STOP fragment inserted between the Gt(ROSA)26Sor promoter and the ChR2/tdTomato sequence. Mice that are homozygous for the targeted mutation are viable, fertile, normal in size and do not display any gross physical or behavioral abnormalities (http://jaxmice.jax.org/strain/017455.html).
In order to target ChR2 expression to a population of interest in loxP-ChR2 mice, it is necessary to activate ChR2 expression by Cre recombinase-mediated excision of the loxP stop site. Cre recombinase-mediated recombination removes the loxP-flanked STOP fragment and results in functional ChR2 and tdTomato expression (separately, not as a fusion protein). A line of mice (Taniguchi et al., 2011) has been made public through JAXMice.org (B6;129S-Nos1tm1.1(cre/ERT2)Zjh/J; JAX stock # 14541; genotype referred to hereafter as nNOS-CreER) in which Cre recombinase expression is driven by the nNOS promoter. The transgene construct is integrated at the endogenous nNOS locus as a ‘knock-in’ and accordingly, homozygous nNOS-Cre mice are nNOS-deficient. The animals used in the current study were heterozygous for the transgene. The transgene construct is a Cre recombinase coding sequence fused to a triple mutant human estrogen receptor (ER). In the absence of an activating ligand, the ER-dependent construct is silent. The three mutations collectively render the receptor non-responsive to estradiol at physiological concentrations but inducible by the ER ligand tamoxifen. Tamoxifen was mixed with corn oil (20 mg/mL) and administered by oral gavage at a dose of 200 mg/kg/day for 3 days. Mice undergoing this protocol are referred to hereafter as nNOS-ChR2 mice—this protocol has been used with no side effects (Taniguchi et al., 2011). Control mice (referred to hereafter as non-expressor) received equivalent volumes of corn oil via gavage.
Male B6.Cg-Tg (Thy1-COP4/eYFP)18Gfng/J mice (JAX strain #7612) were used only in the anesthetized preparation studies. In these mice, the “mouse thymus cell antigen 1 (thy1)” promoter drives expression of ChR2 in cortical pyramidal cells (Arenkiel et al., 2007). We therefore refer to them hereafter as pyramidal-ChR2 mice. Founders were bred with CD1 mice to produce hemizygous transgenic pyramidal-ChR2 males, as previously described (Wisor et al., 2013). Non-expressor and nNOS-ChR2 mice used in the anesthetized preparation studies were also males.
Local field potentials in anesthetized mice
nNOS-ChR2 mice, non-expressor mice and pyramidal-ChR2 mice, were used in the experiments in which local field potentials were measured under isoflurane anesthesia (5% induction, 1.5% maintenance) with perfluoroalkoxy alkane (PFA) coated tungsten electrodes (A-M Systems, Sequim, WA, catalog No. 797000, 0.008” bare diameter, AWG 32). A 0.5 mm ball burr bit was used to drill an initial hole through the skull for placement of the electrodes. Holes were placed in left vibrissal motor cortex (0.86 mm anterior/1.5 mm lateral from bregma) and frontal association cortex (2 mm anterior/2 mm lateral from bregma). A 90 degree bend was made in each electrode wire 2.0 mm from the tip. Electrodes were inserted until the bend rested on the skull. Insulation was removed only from the tip of the electrode, thus field potentials were measured at a depth of approximately 1.5 mm into the brain, corresponding to infragranular cortex. A screw placed in the contralateral parietal cortex served as a cable ground. A screw placed in contralateral occipital cortex and connected by wire to the surgical table served as a body ground. Fast curing, 2-part orthodontic acrylic resin was used to secure and insulate all electrodes. The electrophysiological signal was collected at 5,000 Hz, amplifier gain 1000X, input voltage range = +/− 819.2 mV (Multichannel Systems, Reutlingen, Germany; products # MPA81, SC8x8; MC_Rack software version 4.4.8). The signal was processed through a Butterworth 2nd Order filter with a high pass of 0.5 Hz. Data files were converted to raw binary files for data analysis in the MATLAB programming environment (Mathworks, Natick, MA, USA).
The Multichannel Systems STG4002 TTL generator and MC_Stimulus II stimulus generator software were used to generate a TTL signal, which was split into one recording channel and one output signal to a blue light emitting diode (LED) generator (Doric Lenses, Quebec, Canada, product # D480–1003). Light generated by the LED (wavelength 465 nm, full width at half-maximum 25 nm, 0.018 W/mm2) was projected through a 200 μM diameter fiber optic cable described elsewhere (Clegern et al., 2012), the end of which was inserted into the somatosensory barrel cortex [1.7 mm posterior/2.5 mm lateral relative to bregma; a source of axons that project monosynaptically to ipsilateral vibrissal motor cortex (Aronoff et al., 2010)] at a depth of 1 mm relative to dura for optogenetic stimulation. Stimuli of 10 msec durations were delivered at 7 Hz over a 10- to 15- minute recording session.
EEG/EMG recordings and optogenetic stimulation in freely behaving animals Experiment 1- optogenetic stimulation at frequencies of 1–40 Hz
Mice underwent surgical implantation of bilateral fronto-parietal EEG and electromyographic (EMG) leads [as in (Wisor et al., 2011a; Wisor et al., 2011b; Wisor et al., 2013; Clegern et al., 2012)] subsequent to tamoxifen or corn oil treatment. One of the two frontal EEG leads was glued to the shaft of a guide cannula inserted in the skull to the level of the dura, and extended 1 mm beyond the tip of that guide cannula into the infragranular frontal cortex (the proximal lead); the other lead rested on the surface of the cerebral cortex (the distal lead). Signal from the distal lead was used for state classification. The effects of optogenetic stimulation on evoked potentials and on EEG power spectra are based on the signal from the proximal lead.
At least two weeks after surgery, mice were subjected to a 3-day optogenetic stimulation protocol, with individual experimental days separated by at least one non-experimental day. A fiber optic cable was inserted into the guide cannula, such that its tip penetrated 0.5 mm into the cerebral cortex, 0.5 mm dorsal to the exposed tip of the proximal EEG lead. The stimulus generator, laser and fiber optic cable used to apply optogenetic stimuli in freely behaving animals are described elsewhere (Clegern et al., 2012; Wisor et al., 2013). Each 6-hr EEG/EMG recording session began at least two hours into the daily light phase of the LD12:12 cycle and ended at least two hours before the end of the daily light phase. The three sessions included a baseline, a low-intensity optogenetic stimulus protocol (3.14 mW total; 0.025 W/mm2) previously shown to reliably induce an evoked response in pyramidal-ChR2 animals (Clegern et al., 2012; Wisor et al., 2013), and a high-intensity optogenetic stimulus protocol (31.6 mW total; 0.250 W/mm2), which was applied in anticipation that the low-intensity optogenetic stimulus might not induce a detectable response in nNOS-ChR2 animals expressing ChR2 in the much more uncommon nNOS-positive cell type. The order of the three recording sessions was randomized individually for each animal studied. Laser stimuli applied at a low wattage (3.14 mW) did not yield detectable evoked responses to stimulation in nNOS-ChR2 mice (F49,196=0.81, P =0.797). A ten-fold increase in wattage to 31.6 mW resulted in statistically significant responses. Hence data from the latter measurements are reported. Due to the relative rarity of rapid-eye movement sleep (REMS) epochs, a majority of animals failed to undergo stimulation during REMS. Therefore, the effects of stimulation during REMS are not reported here.
During the baseline recording, the optogenetic fiber cable was inserted into the guide cannula but optogenetic stimuli were not applied throughout the duration of the 6-hr recording. During the low- and high-intensity stimulus application sessions, optogenetic stimuli of 10-ms pulse duration were applied in each of four stimulus train types: 1 Hz, 10 Hz, 20 Hz and 40 Hz. Each of these four 1-hr stimulus trains was preceded by a 30-min interval, during which no stimulation was applied. This protocol was exploratory; it was necessary to apply a broad range of stimulation parameters due to the absence of information on the electrophysiological behavior of this cell type when harboring ChR2. For each stimulus train type, trains were delivered during the first 10-sec epoch of each minute over an interval of 1 hr. Therefore, during 1 Hz stimulation, each animal received a total 600 stimuli (1 per second during the first 10-seconds of each minute over a 60-min interval); during 10 Hz stimulation, each animal received a total of 6000 stimuli; during 20 Hz stimulation, each animal received a total 12000 stimuli; during 40 Hz stimulation, each animal received a total 24000 stimuli. Stimulation sessions took place in a light-controlled, sound attenuated chamber.
Experiment 2- optogenetic stimulation at 1 Hz during spontaneous sleep and sleep deprivation
Having established in experiment 1 that stimulation at frequencies of 1 to 40 Hz does not alter sleep states and that 1 Hz stimulation allows for detection of evoked potentials, a second experiment was performed to assess evoked potentials across both spontaneous sleep cycles and continuous sleep deprivation. Nine male mice were treated with tamoxifen and subjected to instrumentation as described above. Each mouse underwent two 6-hr recording sessions and received 10 msec optogenetic stimuli at 1 Hz throughout the duration of both sessions. The laser output was varied by cycling through ten minutes at each of five laser power values (0, 6.2, 12.2, 18.9, 25.0 mW at the end of the fiber cable inserted into the brain) and a ten minute interval of TTL signal without laser power during each hour. Thus, the animal received 600 stimuli of each intensity (60 stimuli per minute over ten minutes) within each hour, for a total of 36 00 1 Hz stimuli over 6 hrs. In practice, the lowest two voltage values did not produce any laser output, thus, evoked responses to only four values are reported. In one 6-hr session, the mice were allowed to sleep spontaneously throughout. In the other 6-hr session, wakefulness was maintained continuously by a rotating bar at the base of the cage. One animal of nine did not undergo sleep deprivation due to signal deterioration. Sleep deprivation and spontaneous sleep sessions occurred in counterbalanced order within a one-week interval across the eight animals that were subjected to both sessions. Sleep states were defined as wake, SWS or REMS based on standard EEG/EMG criteria (Wisor et al., 2011a; Wisor et al., 2011b; Wisor et al., 2013; Clegern et al., 2012). Wake was further divided into active wake (AW; those epochs of wake in which integrated EMG values were equal to or greater than the 66th percentile of all wake epochs) and quiet wake (those epochs of wake in which integrated EMG values were less than or equal to the 33rd percentile of all wake epochs).
Evoked potentials in freely behaving animals
The acute EEG response to optogenetic stimulation of the nNOS population was assessed by custom algorithms designed and applied within the MATLAB programming environment. Event-triggered potentials were averaged over all stimulus applications delivered at 1Hz frequency within each animal’s record to generate an averaged curve representing the evoked response for that animal. Grand means ± standard error of the means (SEM) of the averaged curves for evoked responses to 1 Hz stimulation within the nNOS-ChR2 and Non-expressor groups are reported. For control data, an equal number of data segments drawn at random times within the same recording were averaged within each animal’s record. Since post hoc analyses indicated electrophysiological responses lasting longer than 100 msec, the responses to individual stimuli delivered at frequencies of >10 Hz could not be discriminated. Consequently, evoked potential data are shown from the 1 Hz stimulation sessions only.
Immunohistochemical markers of Fos-positive, nNOS-positive, and transgene-positive cells in the cerebral cortex
Thirteen nNOS-ChR2 mice were used for immunohistochemical analysis. The mice were sleep deprived using an automated method (a rotating bar in the base of the cage) as described previously (Wisor et al., 2011a). Seven of the 13 mice were euthanized immediately at the end of a 6-hr interval of sleep deprivation ending 8 hrs into the light phase of the LD12:12 cycle (SD). Six of the 13 mice were also sleep deprived for the same 6-hr interval, but were then allowed to sleep spontaneously for a 2-hr interval before being euthanized (SD-Rec). In addition to the above 13 nNOS-ChR2 mice, three non-expressor (corn oil-treated) mice were euthanized and subjected to perfusion to verify selective transgene expression in tamoxifen-treated animals. Animals were euthanized by intraperitoneal pentobarbital injection (50 mg/kg). They were perfused transcardially with 20 ml of PBS followed by 20 ml of 10% phosphate-buffered formalin. Brains were then post-fixed overnight in 10% phosphate-buffered formalin. Brains were stored in 30% sucrose saline freezing solution until further processing. Histological procedure was derived from the protocol first used to identify nNOS-positive cortical interneurons as a sleep-active population (Gerashchenko et al., 2008). Brains were sliced into 40 μm thick coronal sections at the location between 0 mm and −3.2 mm from bregma using a freezing microtome and collected in separate sets for subsequent immunostaining. Dilutions of all antibodies was done in the blocking buffer containing 5% donkey serum (Jackson ImmunoResearch, West Grove, PA), PBS (Sigma-Aldrich), and 1% Triton X-100 (Sigma-Aldrich).
Red Flourescent Protein Immunostaining
To analyze the distribution of the red fluorescent protein (RFP)-positive neurons, brain tissue sections were treated with 1% H2O2 (Sigma-Aldrich) for 15 min to quench endogenous peroxidases, incubated in the blocking buffer for at least 2 hours, and then incubated overnight in rabbit-anti-RFP antisera (1:2,000; Rockland Immunochemicals, Gilbertsville, PA) at room temperature. Sections were then rinsed in PBS, incubated in biotinylated donkey anti-rabbit IgG (1:500; Jackson ImmunoResearch, West Grove, PA) for 2 hours, incubated with peroxidase-conjugated avidin-biotin complex (1:200; ABC, Vector Laboratories, Burlingame, CA) for 1 hour, followed by the addition of 3, 3’-diaminobenzidine tetrahydrochloride, H2O2 and NiSO4 (DAB Peroxidase Substrate Kit, Vector Laboratories, Burlingame, CA) to visualize the reaction product.
nNOS/RFP immunofluorescent double staining
For the double-label study, brain sections were processed for nNOS/RFP double-fluorescence labeling. After two hours of incubation in the blocking buffer, the sections were incubated overnight in a combination of mouse anti-nNOS (1:1,000; Sigma-Aldrich) and rabbit anti-RFP (1:1,500; Rockland Immunochemicals, Gilbertsville, PA) antibody. The sections were then rinsed in PBS, incubated in a mixture of donkey DyLight 488 anti-mouse IgG (1:500; Jackson ImmunoResearch, West Grove, PA) and biotinylated donkey anti-rabbit IgG (1:500; Jackson ImmunoResearch, West Grove, PA), again rinsed in PBS, and incubated in DyLight 594 streptavidin conjugate (1:500; Jackson ImmunoResearch, West Grove, PA). After rinsing in PBS, all sections were mounted on gelatin-coated slides and coverslipped using Fluoromount mounting media (Electron Microscopy Sciences, Hatfield, PA).
nNOS/c-Fos double staining
To determine Fos expression in nNOS neurons in the cerebral cortex in mice of the SD and SD-Rec groups, one set of tissue sections from each of six mice from the SD group and seven mice from the SD-Rec group was processed for immunohistochemistry with Fos and nNOS antisera as described previously (Gerashchenko et al., 2008). Sections were treated with 1% H2O2 (Sigma-Aldrich) for 15 min, incubated in the blocking buffer for at least 2 hours, and then incubated overnight in rabbit-anti-cFos antisera (1:2,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at room temperature. After washing, the sections were placed in the secondary antibody for 2 hours (1:500; biotinylated donkey anti-rabbit IgG; Jackson ImmunoResearch, West Grove, PA) followed by incubation in avidin–biotin complex for 2 hours (1:200; ABC, Vector Laboratories, Burlingame, CA). The DAB Peroxidase Substrate Kit (1:200; Vector Laboratories, Burlingame, CA) was used to produce a black reaction product in cell nuclei. The sections were then incubated in mouse anti-nNOS monoclonal antibody (1:2,000; Sigma-Aldrich, N2280), rinsed in PBS, incubated in biotinylated donkey anti-mouse IgG (1:500; Jackson ImmunoResearch, West Grove, PA) for 2 hours and then incubated with biotin-conjugated alkaline phosphatase (ABC-AP) for 2 hours (Vector Laboratories, Burlingame, CA), washed again, and reacted in a working solution of Vector Red substrate (Vector Red AP Substrate Kit; Vector Laboratories) to produce a red reaction product.
nNOS/RFP/cFos triple staining
The purpose of this experiment was to assess Fos expression in those cortical interneurons that expressed RFP but did not express nNOS. We used the mouse anti-nNOS antiserum to identify RFP neurons that co-localized with nNOS and eliminated those cells from subsequent analysis. Thus, brain sections were initially incubated with mouse-anti-nNOS antisera (1:2,000; Sigma-Aldrich) and stained using DAB-Ni (black label). Then, the sections were incubated in rabbit anti-cFos antisera (1:2,000; Santa Cruz Biotechnology, Santa Cruz, CA) and also stained using DAB-Ni to produce a black reaction product in cell nuclei. Finally, the sections were incubated in rabbit-anti-RFP antisera (1:2,000; Rockland Immunochemicals, Gilbertsville, PA) and stained using DAB (brown label).
Cell counts
A single examiner, blind to treatment conditions, counted all of the nNOS and RFP immunoreactive somata on both sides of the brain as previously described (Gerashchenko et al., 2001). Five brain sections located 0 mm, −0.8 mm, −1.6 mm, −2.4 mm and −3.2 mm from bregma were examined. Cells were counted in the entire cerebral cortex of the section except for the areas adjacent to the EEG electrodes or anchor screws. nNOS-positive and Fos/nNOS double-labeled neurons were counted in one set of tissue, and RFP-positive and RFP/nNOS double-labeled neurons were counted in another set of tissue. Neurons were considered to be Fos positive if they contain black Ni+/DAB reaction product that was darker than a visually established threshold. The total numbers of cells were determined for each animal, percentages of Fos in nNOS- and RFP-immunoreactive cells determined and used for further statistical analysis.
Statistical analyses
For evoked potential analyses, local field potentials collected from anesthetized animals at 5000 Hz were processed in the MATLAB programming environment. To account for drift in the signal, data were subjected to normalization, first by subtracting the mean of all sampled values across the recording from each individual sampled value in the recording. Next, the data were subjected to a smoothing algorithm, in which the mean of the potential across a 3 sec window centered on each data point was subtracted from the value at that data point. A single vector consisting of 15000 data points (3-seconds of data) per recording was generated from these transformed potential values by averaging values across 3-second segments of data centered on every TTL-triggered stimulus onset in the file. Data were then downsampled 10-fold to 500 Hz, by averaging across 10 consecutive values, for statistical analysis. The data from a 90 msec window beginning 10 msec before the 10 msec optogenetic stimulus application, and an equal number of randomly-timed segments of data from the same recording were subjected to repeated measures analysis of variance (ANOVA) within each genotype group.
In studies of sleep assessment without anesthesia, EEG and EMG were collected at 400 Hz and were not downsampled. Evoked potentials were not normalized in the manner described above for the anesthetized state; raw evoked potentials were averaged across all 1 Hz stimulation trials within each animal’s record. These averaged value and an equal number of randomly-timed segments of data from the same recording were subjected to repeated measures ANOVA within each genotype group. Evoked potential parameters were extracted from each animal’s averaged curve based on empirical observations. p50 was defined as the maximum value detected 40–80 msec after stimulus onset. n100 was defined as the minimum value detected 80–120 msec after stimulus onset. p200 was defined as the maximum value detected 150–250 msec after stimulus onset. n300 was defined as the minimum value detected 350–400 msec after stimulus onset. The root mean square was calculated from signal collected 100–500 msec after stimulus onset (201 data points).
Statistics were performed with Statistica 9.0 (StatSoft; Tulsa, OK). Parametric tests have been used because we found normal distributions and equal variances in the sampled distributions.
Results
Expression of transgene markers in the sleep active cells of the cerebral cortex
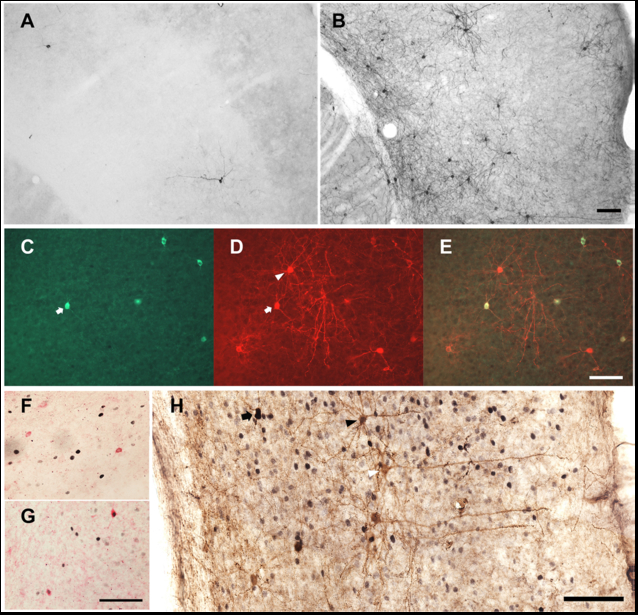
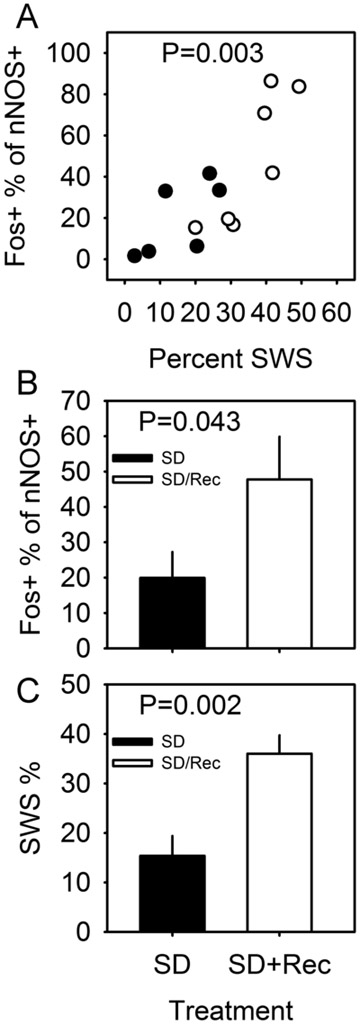
In a double transgenic mouse line (see Materials and Methods for details), tamoxifen treatment triggers Cre recombinase-dependent expression of Channelrhodopsin 2 (ChR2) and the histological marker RFP. Corn oil-treated double transgenic mice did not express RFP in the cerebral cortex (Figure 1A); tamoxifen-treated double transgenic mice expressed RFP in a small number of cells in the cerebral cortex (Figure 1B). All nNOS-positive cells expressed RFP (Figure 1 C, D and E), indicating that the pattern of recombination targeted the type I nNOS neuron profile. In order to verify that ChR2 expression in nNOS-positive cells of the cerebral cortex does not disrupt the sleep-active profile of those cells, we performed dual immunohistochemical labeling for Fos and nNOS (Figure 1 F and G) in nNOS-ChR2 mice under two conditions. Mice were euthanized after either 8-hrs continuous sleep deprivation (SD) or 6-hrs SD followed by 2 hrs of spontaneous sleep (SD-Rec). Mice subjected to 8-hr SD spent approximately 15% of time in SWS in the last 2 hrs of SD. SD-Rec animals allowed to sleep spontaneously exhibited a 2.5 fold increase in SWS as a percentage of time relative to SD animals (Figure 2C; t11 =3.77, P=0.003). Fos expression was elevated in SD-Rec mice relative to SD mice (Figure 2B; t11 =1.88, P=0.043). Multiple regression indicated a significant main effect for SWS% on Fos expression in nNOS cells (Figure 2A; F2,10=11.01, P=0.003). These data demonstrate that nNOS cells of the cerebral cortex are a sleep-active population when harboring a ChR2 construct. About a one half of RFP+ cells were nNOS-negative (Figure 1 C-E, H). However, the percentage of Fos-positive RFP+/nNOS- neurons was not different between the SD and SD-Rec groups of mice (SD mice: 3.8±0.3%; SD-Rec mice: 4.0±0.6%), indicating that nNOS-negative RFP cells in the cerebral cortex are not sleep-active.
Figure 1.
Genetic targeting of nNOS cells in the cerebral cortex of nNOS-ChR2 mice. (A) RFP-containing cells are rare in the nNOS-CreERmice treated with corn oil. (B) Tamoxifen treatment induces RFP expression in neurons throughout the cerebral cortex in these mice. (C) nNOS immunofluorescence. (D) RFP immunofluorescence. (E) Merged images of the nNOS and RFP stained cells. All nNOS-immunoreactive cells also contain RFP immunoreactivity in the cerebral cortex (white arrow), but some RFP-containing cells do not contain nNOS (white triangle). (F) Fos expression (black nuclei) in nNOS neurons (red) of a representative mouse from the SD group. Most nNOS cells did not express Fos in this group of animals. (G) Approximately half of nNOS cells expressed Fos in the SD-Rec group of animals. (H) nNOS (black cytoplasm), Fos (black nuclei) and RFP (brown cytoplasm) immunostaining in the cerebral cortex of an animal from the SD-Rec group. nNOS cells were stained in black to discriminate them from nNOS-negative RFP cells (black arrow). Some nNOS-negative RFP-containing cells expressed Fos (black triangle), but the majority of these cells did not express Fos (white triangle). Scale bars = 100 µm. Scale bar in B also applies to A, scale bar in E also applies to C and D, scale bar in G also applies to F.
Figure 2.
Sleep-dependent Fos expression in the nNOS-positive cells of the cerebral cortex of nNOS-ChR2 mice. (A) Percentage of nNOS-positive cells that are also Fos-positive in the cerebral cortex of nNOS-ChR2 mice euthanized after 8-hr SD (black) or 6-hr SD+2-hr spontaneous sleep (white), plotted as a function of percent time spent in SWS in the 2-hr interval immediately prior to euthanasia. P value refers to main effect of SWS % on cell count in multiple regression. (B) Percentage of nNOS cells also positive for Fos is elevated in SD-Rec mice relative to SD mice. P value refers to one-tailed T test comparing groups. (C) Percentage of time in SWS in the 2 hrs prior to euthanasia. P value refers to one-tailed T test comparing groups.
The evoked potential responses to optogenetic stimulation of pyramidal cells and nNOS cells are of opposite polarity
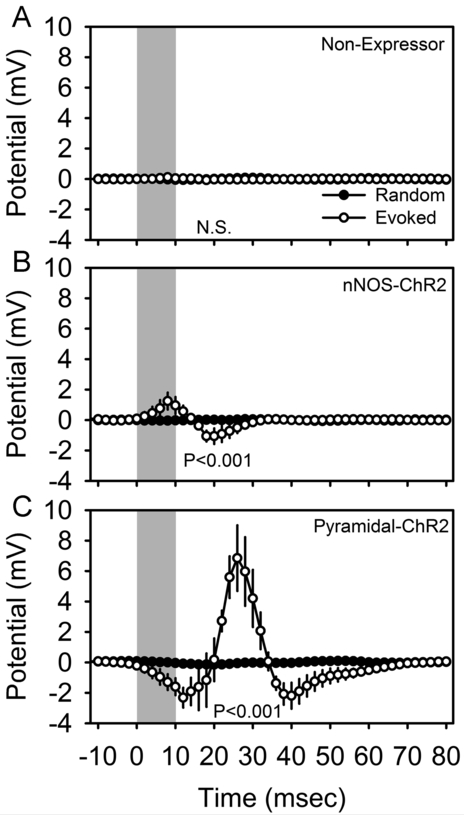
Optogenetic stimuli of 10 msec duration applied in the somatosensory barrel cortex of isoflurane-anesthetized mice triggered reliable changes in electrical potential in the motor cortex, a monosynaptic target of barrel cortex axons, in mice harboring ChR2 in nNOS-positive cells (Figure 3B; F49,147 = 2.58, P < 0.001) and in a distinct transgenic line of mice (‘pyramidal-ChR2’ mice; see Materials and Methods) harboring ChR2 in pyramidal cells (Figure 3C; F49,147 = 5.97, P < 0.001), but not non-expressing corn oil-treated mice (Figure 3A; F49,196 = 1.06, P = 0.375). In pyramidal-ChR2 mice, the acute response to optogenetic activation was a negative deflection (Figure 3C), which in the local field potential coincides with depolarization at the cellular level. This response is to be expected, as pyramidal cells projecting from the barrel cortex to the site of recording in the motor cortex are directly activated in these mice. This acute response was followed by a positive deflection of much greater amplitude. In nNOS-ChR2 mice, however, the acute evoked response was a positive deflection (Figure 3B), which in the local field potential is indicative of hyperpolarization of the majority cell population (i.e., ChR2-negative pyramidal cells) at the site of recording. These data confirm that the pyramidal-ChR2 and nNOS-ChR2 lines confer functional expression of ChR2 to distinct cell types. That non-expressor mice failed to exhibit evoked responses (Figure 3A) demonstrates that the expression of the construct is dependent on tamoxifen induction.
Figure 3.
Optogenetically-evoked potentials in the cerebral cortex of isoflurane anesthetized non-expressor (A; n=5), nNOS-ChR2 (B; n=4) and pyramidal-ChR2 (C; n=4) transgenic mice. Local field potentials were averaged across 4200-5000 trials per animal. Peristimulus histograms for optogenetic stimuli are shown in white and those from an equal number of randomly timed segments of data in the same recording are shown in black. Gray shading indicates the timing of 10 msec optogenetic stimulation. Data shown are grand mean of these averaged curves across subjects. P values denote main effect of time in repeated measures ANOVA.
Evoked responses to 10-msec optogenetic stimulation of nNOS-positive cells are modulated by sleep state and local electrical potential at stimulus onset
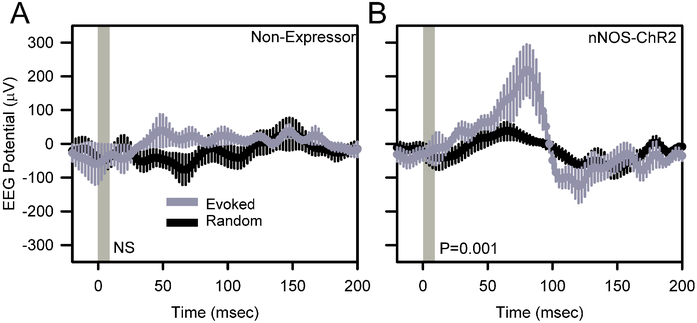
Comparison of event-triggered potentials against random curves generated from the same data stream indicated a significant local evoked response to 1 Hz stimulation at the stimulation site in awake, behaving nNOS-ChR2 (Figure 4B; F199,796=3.81, P<0.001) mice but not non-expressors (Figure 4A; F199,796=0.75, P=0.993). Evoked responses to 1 Hz stimulation in nNOS-ChR2 mice during wake (Figure 4B) consisted of a positive deflection of 186±67 μV, peaking at 80 msec after stimulus onset, and a subsequent more modest negative deflection. The amplitude of the positive peak was dependent on wakefulness. While the EEG response to stimulation was maximal 80 msec after the onset of stimulation during SWS just as it was during wake, the response was not significant, peaking at 82±101 μV (n=4 mice; data not shown).
Figure 4.
Optogenetically-evoked potentials in the cerebral cortex of awake, unanesthetized non-expressor (A) and nNOS-ChR2 (B) transgenic mice. EEG traces were averaged across 600 trials per animal. Data shown are grand mean of these averaged curves across subjects. Peristimulus histograms for optogenetic stimuli are shown in gray and those from an equal number of randomly timed segments of data in the same recording are shown in black. Vertical gray shading indicates the timing of 10 msec optogenetic stimulation. P values denote main effect of time in repeated measures ANOVA.
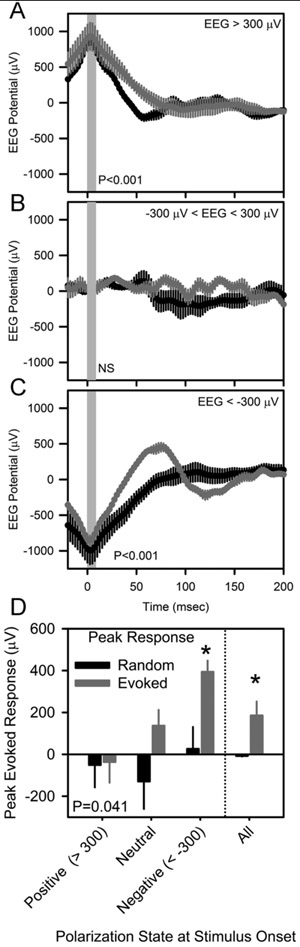
Variability in the magnitude of the peak evoked response during wakefulness and the inconsistent response during SWS led us to ask whether the response might be dependent on ongoing EEG dynamics at the time of stimulation. We therefore sorted, post-hoc the evoked potentials during wakefulness based on the EEG polarization state at the time of stimulus onset (Figure 5). The amplitude of the peak evoked response varied as a function of the polarization state at the time of stimulus onset (Figure 5D; F3,12 =3.75, P=0.041). The amplitude of the evoked response was greatest when stimulation was applied to the cerebral cortex at a negative (< −300 μV) potential (Figure 5C). Evoked responses were not significant when stimulation was applied to the cerebral cortex at a neutral (potential between −300 μV and + 300 μV) or positive (>300 μV) potential (Figure 5A,B).
Figure 5.
Optogenetically-evoked potentials in nNOS-ChR2 transgenic mice, categorized according to local EEG polarization state at the time of stimulus onset. Peristimulus histograms for optogenetic stimuli are shown in gray and those from an equal number of random segments of data beginning at the same EEG potential are shown in black. Vertical gray shading indicates the timing of 10 msec optogenetic stimulation. P values in A-C denote main effect of time in repeated measures ANOVA. D, EEG potential 80 msec after the onset of stimulation, derived from the curves in A-C. P value in D indicates effect of EEG potential at stimulus onseton peak response. Asterisks in D indicate difference between evoked and random traces, Fisher’s LSD.
Evoked potential waveforms vary across sleep states
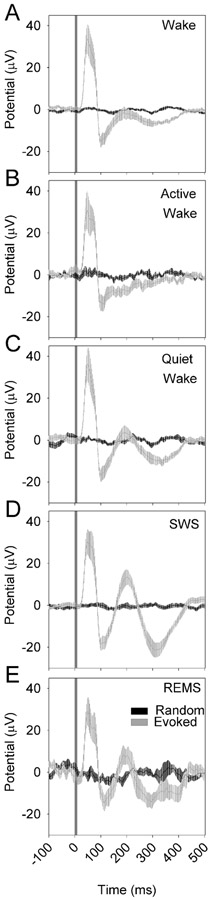
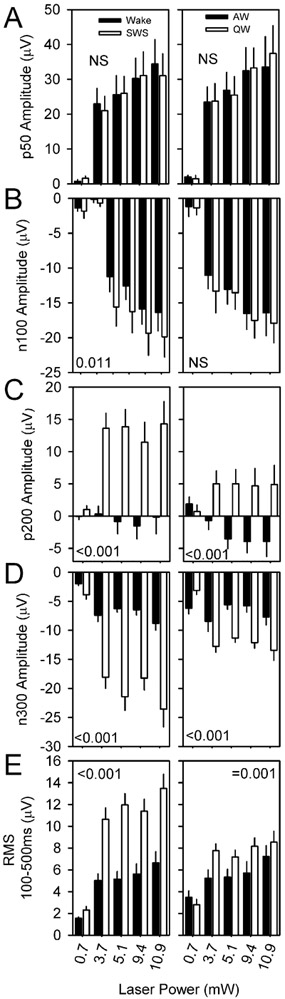
In a second experiment, nine nNOS-ChR2 mice were subjected to 10 msec pulses of blue light administered at 1 Hz continuously over an interval of 6 hours, during which they were allowed to sleep spontaneously. Optogenetic stimuli were administered at incrementing intensities to establish an intensity-response relationship for evoked curves across sleep states of wake, SWS and REMS. Wake was additionally subdivided based on muscle tone into AW and QW. Stimuli were delivered in all states in all 9 animals (Figure 6). In all states, there was a significant interaction of time and condition (evoked vs random) in affecting EEG potential (P < 0.001). This effect indicates cortical responsiveness is intact across all states. An acute positive deflection emerging within 50 msec of the stimulus (p50; Figure 7A) occurred in response to stimuli delivered in all states. The amplitude of the p50 event did not significantly differ between wake and SWS. However, secondary oscillatory events were potentiated in SWS relative to wake, including a negative event at 100 msec (n100; Figure 7B, left panel), a positive deflection at approximately 200 msec (p200; Figure 7C, left panel) and a negative deflection at 300 msec (n300; Figure 7D, left panel). As a result of the prolonged response to stimulation in SWS, the RMS of the evoked curve from 100 to 500 msec after the stimulus onset was elevated in SWS relative to wake (Figure 7E, left panel). Prolonged oscillatory behavior was also seen in response to stimuli delivered during QW, albeit at a lower amplitude. P200, n300 and the RMS of the evoked curve from 100 to 500 msec were all elevated in QW relative to AW (figure 7, right panels). Thus, the cerebral cortex exhibits prolonged oscillatory behavior lasting at least 300 msec when the animal is in slow-wave sleep or QW.
Figure 6.
Evoked responses to optogenetic activation of nNOS-positive cells vary as a function of state. A 10-msec optogenetic stimulus was applied from time 0 to time 10 msec. Gray curves indicate the grand mean±SEM of averaged evoked responses to pulses (n=9 mice); black lines indicate the grand mean±SEM of randomly selected snippets of LFP trace data from the same recordings. Data are from wake (A), active wake (B), quiet wake (C) SWS, (D) and REMS (E). In all states, there was a significant interaction of time and conditon (evoked vs random) in affecting EEG potential (P < 0.001).
Figure 7.
Evoked potential parameters are affected by state. Evoked potential parameters p50 (A), n100 (B), p200 (C), n300 (D) and the root mean square (RMS) of the ERP trace from 100 msec to 500 msec post-stimulus are compared for stimuli of four intensities during wake vs SWS (black and white, respectively, in left panels) and AW vs QW (black and white, respectively, in right panels). P values denote interaction of state and laser intensity.
p200 of the evoked potential is potentiated by sleep deprivation
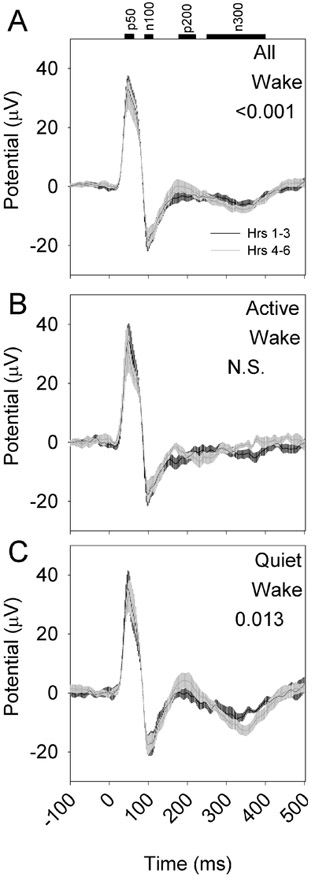
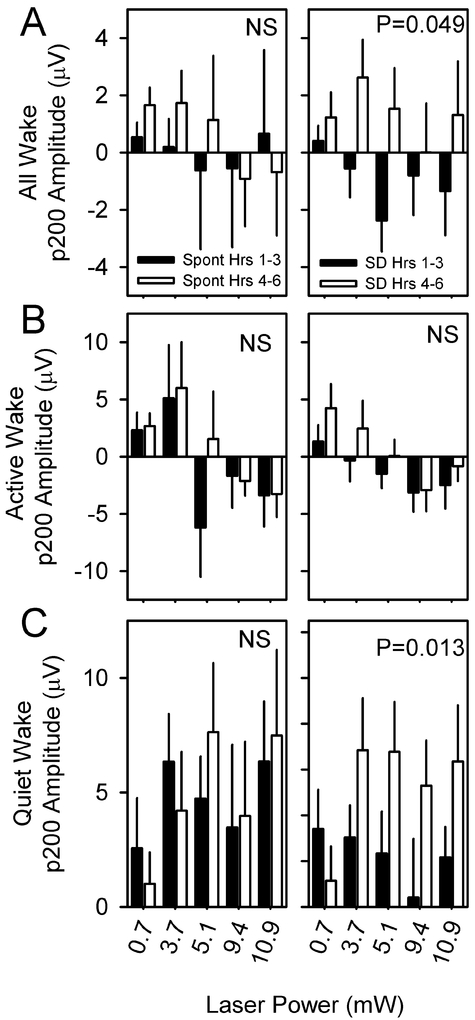
To determine whether the evoked response to optogenetic activation is influenced by sleep need, eight nNOS-ChR2 mice were subjected to 10 msec pulses of blue light administered at 1 Hz continuously over an interval of 6 hours, during which wakefulness was enforced by a rotating bar at the base of the cage. Evoked responses during the first three hours of continuous sleep deprivation differed from evoked responses during hours 4–6 of continuous sleep deprivation (Figure 8A). However, when responses were assessed in QW and AW separately, the effect of sleep deprivation was significant only in QW (Figure 8B) and not AW (Figure 8C). Specifically, the amplitude of p200 was potentiated during hrs 4–6 relative to hrs 1–3 in QW and not in AW (Figure 9). Other evoked potential parameters (p50, n100, p200, n300, RMS) were not significantly affected by sleep deprivation.
Figure 8.
Evoked responses to optogenetic activation of nNOS-positive cells vary as a function of duration of sleep deprivation (n=8 mice). A 10-msec optogenetic stimulus was applied from time 0 to time 10 msec. Black curves indicate the grand mean±SEM of averaged evoked responses during the first three hours of continuous sleep deprivation; gray curves indicate the grand mean±SEM of averaged evoked responses during hours 4-6 of continuous sleep deprivation. Data are from wake (A), active wake (B), quiet wake (C). P values denote interaction of time relative to stimulus onset and SD phase (hrs 1-3 vs hrs 4-6) in affecting EEG potential.
Figure 9.
p200 amplitude is potentiated by sleep deprivation. p200 values from hours 1-3 (black bars) or 4-6 (white bars) of spontaneous sleep (left panels) and sleep deprivation (right panels) are compared within states of wake (A), AW (B) or QW (C) for stimuli of four intensities. P value denotes main effect of time (hours 1-3 vs hours 4-6). Other evoked potential parameters (p50, n100, p200, n300, RMS) were unaffected by sleep deprivation.
Discussion
The occurrence of SWA, the defining EEG feature of SWS, is regulated by the antagonistic interactions of subcortical wake-promoting and sleep-promoting nuclei (Jones, 2004; Saper et al., 2005; Siegel, 2009; Szymusiak, 2010). Yet the homeostatic regulation of sleep SWA is not manifested uniformly across the cerebral cortex. Intensive use of a functional unit in the cerebral cortex causes an increase in SWA within that unit (Kattler et al., 1994; Vyazovskiy et al., 2000), whereas experimental attenuation of the activity in a functional unit during wake reduces SWA in subsequent sleep (Huber et al., 2006). These data demonstrate that sleep homeostatic regulation is a local, use-dependent property of the cerebral cortex. It is possible that intrinsic cells of the cerebral cortex regulate the local use-dependence of SWA. The sleep-active population of nNOS-positive neurons in the cerebral cortex, so named based solely on Fos expression patterns (Zielinski et al., 2013; Gerashchenko et al., 2008), are a potential local regulator of sleep SWA homeostatic dynamics. The observations in the current study are compatible with that hypothesized role. The targeted population, at least when manipulated optogenetically on a local basis in the cerebral cortex, does not regulate sleep state globally, but only affects EEG waveforms locally.
The evoked response to nNOS cell stimulation was a positive deflection of the EEG peaking 50–80 msec after the onset of a 10 msec stimulus. EEG potential fluctuations are a consequence of the synchronized firing and hyperpolarization of cells in the vicinity of the EEG lead and are largely reflective of the polarization state of the majority population of cerebral cortical cells, pyramidal cells (Nunez and Katznelson, 1981; Buzsaki et al., 2012). It is likely that the evoked response to nNOS cell stimulation that we detected in the EEG is a secondary hyperpolarizing response of pyramidal cells rather than a direct consequence of the change in membrane potential of the very sparse nNOS cell population. Polarization state in the extracellular milieu is inverse to that of individual cells (Buzsaki et al., 2012). Thus, the positive deflection in the EEG peaking 80 msec after optogenetic stimulation of nNOS cells is indicative of a synchronized hyperpolarization of pyramidal cells, precisely the type of event that one would expect to see in response to the activation of a local slow-wave-promoting population of GABAergic interneurons. This contention is supported by the fact that (at least under isoflurane anesthesia) direct optogenetic stimulation of pyramidal cells in the pyramidal-ChR2 transgenic line resulted in a negative deflection of the LFP. Furthermore, the evoked response to nNOS cell stimulation was dependent on the excitation state of the cortex at the time of stimulus application. When the cortex was in a positive deflection (as occurs when the pyramidal cells at the site of stimulation are uniformly hyperpolarized), there was no evoked response in nNOS-ChR2 mice. Slow waves occur locally during wakefulness (Vyazovskiy et al., 2011); it may be the case that stimuli applied when the cortex was in a positive potential were, in fact, applied during physiological slow waves. The lack of an evoked response may have been either due to ongoing activity in the targeted nNOS population, or the already hyperpolarized pyramidal cells being refractory to the hyperpolarizing effect of an optogenetically-activated nNOS population.
In the neocortex, nitrergic neurons are generally divided into type I and type II neurons (Perrenoud et al., 2012). Type I cells are more intensely stained in nNOS immunohistochemical or NADPH diaphorase staining procedures and have larger somata than type II cells (Perrenoud et al., 2012). In rodents, type I cells are found in all cortical layers (Oermann et al., 1999), although they are more frequently observed in infragranular layers (Vercelli et al., 2000; Lee and Jeon, 2005). In the animal model used in the present study (nNOS-ChR2 transgenic mice), we observed that about a one half of RFP-positive neurons were intensely stained by nNOS immunohistochemistry. These neurons belong to the type I population of nNOS neurons. Fos was highly induced during recovery sleep after a period of sleep deprivation in this type of cells (Figure 1 G). Another half of RFP neurons were not intensely stained by nNOS immunohistochemistry (Figure 1 C-E, H). Because RFP expression in these neurons is also driven by nNOS promoter, they are expected to belong to the type II nNOS neurons. However, some of these neurons displayed pyramidal cell morphology (Figure 1B and H) that is not characteristic of type II neurons. These nNOS-negative neurons expressed little Fos (about 4%), and the percentage of Fos positive neurons was similar in SD and SD-Rec groups of animals. Therefore, it is unlikely that the state-dependent evoked responses observed in the present study were mediated by the nNOS-negative RFP cells.
In a recent study, chemogenetic activation of somatostatin-positive cells in the cerebral cortex increased SWA, slope of individual slow waves, and non-REM sleep duration, whereas chemogenetic inhibition of these cells decreased SWA and slow wave incidence without changing time spent in non-REM sleep (Funk et al., 2017). Since all type I nNOS-positive cells are also somatostatin-positive cells (Kilduff et al., 2011), it is likely that the changes in SWA observed in this study depended, at least partially, on the change in the activity of nNOS cells. Nevertheless, the relative contribution of nNOS-positive vs. nNOS-negative somatostatin cells in the regulation of SWA is not known.
How is the activity of the sleep-active nNOS-positive cells regulated in vivo? In rodents, these cells are histochemically-enriched for neurokinin 1 (NK1) receptors and are depolarized by the NK1 receptor ligand substance P (Dittrich et al., 2012). Substance P is shown to have varying effects on non-REM sleep and SWA in humans and rodents (Andersen et al., 2006; Lieb et al., 2002; Zhang et al., 2004; Zielinski and Gerashchenko, 2017). Whereas changes in non-REM sleep timing may be mediated by effects of substance P on subcortical circuitry, the effect on SWA is likely due to effects on cortical type 1 nNOS sleep active cells. Indeed, NK1 receptor agonists injected into the cortex of mice enhance SWA locally, while NK1 receptor antagonists have an opposing effect (Zielinski et al., 2015). Nevertheless, it is not known whether substance P release in the cerebral cortex is elevated during SWS relative to wake. nNOS cells receive serotonergic innervation (Cauli et al., 2004), and serotonin concentration is elevated in the cerebral cortex during wake relative to SWS (Portas et al., 1998; Bjorvatn et al., 2002). Bath application of serotonin hyperpolarizes via 5-HT1A receptors nNOS neurons located along the external capsule of the amygdala (Bocchio et al., 2016). Because the neurochemical profile of these neurons is similar to cortical nNOS neurons (Wang et al., 2017), it is possible that type I nNOS neurons in the cerebral cortex are also inhibited by serotonin. Therefore, a SWS-dependent serotonergic disinhibition of the type 1 nNOS-positive cells is plausible.
We conclude that the evoked response of the EEG to nNOS cell activation in the cerebral cortex resembles a physiological slow wave. This response is modulated as a function of EEG polarization state. Collectively, these properties are compatible with those of a putative sleep regulatory neuronal population.
Acknowledgments
We thank Svetlana Karpova, John M. Loft and William C. Clegern for technical assistance. This work was supported by the National Institutes of Health (RO1NS078498, R21NS092926 and RO3NS082973) and the Department of Veterans Affairs (IK2BX002823).
Abbreviations
- ANOVA
analysis of variance
- AW
active wake
- ChR2
channelrhodopsin2
- DAB
3’-diaminobenzidine
- EEG
electroencephalogram
- EMG
electromyogram
- ER
estrogen receptor
- LD
light-dark
- LED
light emitting diode
- LFP
local field potential
- NADPH
nicotinamide adenine dinucleotide phosphate
- NK1
neurokinin 1
- nNOS
neuronal nitric oxide synthase
- PBS
phosphate-buffered saline
- QW
quiet wake
- Rec
recovery sleep
- REMS
REM sleep
- RFP
red fluorescent protein
- SEM
standard error of the means
- SD
sleep deprivation
- SWA
slow-wave activity
- SWS
slow-wave sleep
- WPRE
woodchuck hepatitis virus posttranscriptional regulatory element
References
- Andersen ML, Nascimento DC, Machado RB, Roizenblatt S, Moldofsky H, Tufik S (2006) Sleep disturbance induced by substance P in mice. Behav Brain Res 167: 212–218. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G (2007) In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54: 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronoff R, Matyas F, Mateo C, Ciron C, Schneider B, Petersen CC (2010) Long-range connectivity of mouse primary somatosensory barrel cortex. Eur J Neurosci 31: 2221–2233. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Suresh A, Broussard C, Frank MG (2014) Sleep promotes cortical response potentiation following visual experience. Sleep 37: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorvatn B, Gronli J, Hamre F, Sorensen E, Fiske E, Bjorkum AA, Portas CM, Ursin R (2002) Effects of sleep deprivation on extracellular serotonin in hippocampus and frontal cortex of the rat. Neuroscience 113: 323–330. [DOI] [PubMed] [Google Scholar]
- Bocchio M, Fisher SP, Unal G, Ellender TJ, Vyazovskiy VV, Capogna M (2016) Sleep and Serotonin Modulate Paracapsular Nitric Oxide Synthase Expressing Neurons of the Amygdala. eNeuro 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, Anastassiou CA, Koch C (2012) The origin of extracellular fields and currents--EEG, ECoG, LFP and spikes. Nat Rev Neurosci 13: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Tong XK, Rancillac A, Serluca N, Lambolez B, Rossier J, Hamel E (2004) Cortical GABA interneurons in neurovascular coupling: relays for subcortical vasoactive pathways. J Neurosci 24: 8940–8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegern WC, Moore ME, Schmidt MA, Wisor J (2012) Simultaneous electroencephalography, real-time measurement of lactate concentration and optogenetic manipulation of neuronal activity in the rodent cerebral cortex. J Vis Exp e4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich L, Heiss JE, Warrier DR, Perez XA, Quik M, Kilduff TS (2012) Cortical nNOS neurons co-express the NK1 receptor and are depolarized by Substance P in multiple mammalian species. Front Neural Circuits 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G Direct Data Submission. MGI Ref ID J:179349.Mouse Genome Informatics. 2012. [Google Scholar]
- Funk CM, Peelman K, Bellesi M, Marshall W, Cirelli C, Tononi G (2017) Role of Somatostatin-Positive Cortical Interneurons in the Generation of Sleep Slow Waves. J Neurosci 37: 9132–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko D, Kohls MD, Greco M, Waleh NS, Salin-Pascual R, Kilduff TS, Lappi DA, Shiromani PJ (2001) Hypocretin-2-saporin lesions of the lateral hypothalamus produce narcoleptic-like sleep behavior in the rat. J Neurosci 21: 7273–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko D, Wisor JP, Burns D, Reh RK, Shiromani PJ, Sakurai T, de la Iglesia HO, Kilduff TS (2008) Identification of a population of sleep-active cerebral cortex neurons. Proc Natl Acad Sci U S A 105: 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Ghilardi MF, Massimini M, Ferrarelli F, Riedner BA, Peterson MJ, Tononi G (2006) Arm immobilization causes cortical plastic changes and locally decreases sleep slow wave activity. Nat Neurosci. [DOI] [PubMed] [Google Scholar]
- Jones BE (2004) Activity, modulation and role of basal forebrain cholinergic neurons innervating the cerebral cortex. Prog Brain Res 145: 157–169. [DOI] [PubMed] [Google Scholar]
- Kattler H, Dijk DJ, Borbely AA (1994) Effect of unilateral somatosensory stimulation prior to sleep on the sleep EEG in humans. J Sleep Res 3: 159–164. [DOI] [PubMed] [Google Scholar]
- Kilduff TS, Cauli B, Gerashchenko D (2011) Activation of cortical interneurons during sleep: an anatomical link to homeostatic sleep regulation? Trends Neurosci 34: 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Jeon CJ (2005) Immunocytochemical localization of nitric oxide synthase-containing neurons in mouse and rabbit visual cortex and co-localization with calcium-binding proteins. Mol Cells 19: 408–417. [PubMed] [Google Scholar]
- Lieb K, Ahlvers K, Dancker K, Strohbusch S, Reincke M, Feige B, Berger M, Riemann D, Voderholzer U (2002) Effects of the neuropeptide substance p on sleep, mood, and neuroendocrine measures in healthy young men. Neuropsychopharmacology 27: 1041–1049. [DOI] [PubMed] [Google Scholar]
- Nunez PL, Katznelson RD (1981) Electric fields of the brain. New York: Oxford University Press. [Google Scholar]
- Oermann E, Bidmon HJ, Mayer B, Zilles K (1999) Differential maturational patterns of nitric oxide synthase-I and NADPH diaphorase in functionally distinct cortical areas of the mouse cerebral cortex. Anat Embryol (Berl) 200: 27–41. [DOI] [PubMed] [Google Scholar]
- Perrenoud Q, Geoffroy H, Gauthier B, Rancillac A, Alfonsi F, Kessaris N, Rossier J, Vitalis T, Gallopin T (2012) Characterization of Type I and Type II nNOS-Expressing Interneurons in the Barrel Cortex of Mouse. Front Neural Circuits 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portas CM, Bjorvatn B, Fagerland S, Gronli J, Mundal V, Sorensen E, Ursin R (1998) On-line detection of extracellular levels of serotonin in dorsal raphe nucleus and frontal cortex over the sleep/wake cycle in the freely moving rat. Neuroscience 83: 807–814. [DOI] [PubMed] [Google Scholar]
- Puentes-Mestril C, Aton SJ (2017) Linking Network Activity to Synaptic Plasticity during Sleep: Hypotheses and Recent Data. Front Neural Circuits 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J (2005) Hypothalamic regulation of sleep and circadian rhythms. Nature 437: 1257–1263. [DOI] [PubMed] [Google Scholar]
- Siclari F, Tononi G (2017) Local aspects of sleep and wakefulness. Curr Opin Neurobiol 44: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JM (2009) The neurobiology of sleep. Semin Neurol 29: 277–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymusiak R (2010) Hypothalamic versus neocortical control of sleep. Curr Opin Pulm Med 16: 530–535. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ (2011) A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71: 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasali E, Leproult R, Ehrmann DA, Van Cauter E (2008) Slow-wave sleep and the risk of type 2 diabetes in humans. Proc Natl Acad Sci U S A 105: 1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Cirelli C (2006) Sleep function and synaptic homeostasis. Sleep Med Rev 10: 49–62. [DOI] [PubMed] [Google Scholar]
- Vercelli A, Garbossa D, Biasiol S, Repici M, Jhaveri S (2000) NOS inhibition during postnatal development leads to increased ipsilateral retinocollicular and retinogeniculate projections in rats. Eur J Neurosci 12: 473–490. [DOI] [PubMed] [Google Scholar]
- Vyazovskiy V, Borbely AA, Tobler I (2000) Unilateral vibrissae stimulation during waking induces interhemispheric EEG asymmetry during subsequent sleep in the rat. J Sleep Res 9: 367–371. [DOI] [PubMed] [Google Scholar]
- Vyazovskiy VV, Olcese U, Hanlon EC, Nir Y, Cirelli C, Tononi G (2011) Local sleep in awake rats. Nature 472: 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JK, Randazzo AC, Stone K, Eisenstein R, Feren SD, Kajy S, Dickey P, Roehrs T, Roth T, Schweitzer PK (2006) Tiagabine is associated with sustained attention during sleep restriction: evidence for the value of slow-wave sleep enhancement? Sleep 29: 433–443. [PubMed] [Google Scholar]
- Wang X, Liu C, Wang X, Gao F, Zhan RZ (2017) Density and neurochemical profiles of neuronal nitric oxide synthase-expressing interneuron in the mouse basolateral amygdala. Brain Res 1663: 106–113. [DOI] [PubMed] [Google Scholar]
- Wisor JP, Clegern WC, Schmidt MA (2011a) Toll-like receptor 4 is a regulator of monocyte and electroencephalographic responses to sleep loss. Sleep 34: 1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor JP, Rempe MJ, Schmidt MA, Moore ME, Clegern WC (2013) Sleep slow-wave activity regulates cerebral glycolytic metabolism. Cereb Cortex 23: 1978–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor JP, Schmidt MA, Clegern WC (2011b) Cerebral microglia mediate sleep/wake and neuroinflammatory effects of methamphetamine. Brain Behav Immun 25: 767–776. [DOI] [PubMed] [Google Scholar]
- Zhang G, Wang L, Liu H, Zhang J (2004) Substance P promotes sleep in the ventrolateral preoptic area of rats. Brain Res 1028: 225–232. [DOI] [PubMed] [Google Scholar]
- Zielinski MR, Gerashchenko D (2017) Sleep-inducing effect of substance P-cholera toxin A subunit in mice. Neurosci Lett 659: 44–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski MR, Karpova SA, Yang X, Gerashchenko D (2015) Substance P and the neurokinin-1 receptor regulate electroencephalogram non-rapid eye movement sleep slow-wave activity locally. Neuroscience 284: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski MR, Kim Y, Karpova SA, Winston S, McCarley RW, Strecker RE, Gerashchenko D (2013) Sleep active cortical neurons expressing neuronal nitric oxide synthase are active after both acute sleep deprivation and chronic sleep restriction. Neuroscience 247: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]