Abstract
Sustained identification of innovative chemical entities is key for the success of chemical biology and drug discovery. We report the fragment-based, computer-assisted de novo design of a small molecule inhibiting Helicobacter pylori HtrA protease. Molecular binding of the designed compound to HtrA was confirmed through biophysical methods, supporting its functional activity in vitro. Hit expansion led to the identification of the currently best-in-class HtrA inhibitor. The results obtained reinforce the validity of ligand-based de novo design and binding-kinetics-guided optimization for the efficient discovery of pioneering lead structures and prototyping drug-like chemical probes with tailored bioactivity.
Keywords: biophysical methods, chemical biology, computer-assisted drug design, drug design, receptor–ligand interactions
The success of future drug discovery heavily relies on the sustainable identification of new chemical entities (NCEs).[1] By complementing high-throughput compound screening and conventional in silico methods, computer-assisted de novo molecular design is emerging as a promising technology.[2] For example, it has recently been applied to the rational design of prototypical kinase inhibitors.[3] Herein we disclose the de novo design of an innovative Helicobacter pylori high-temperature requirement A (HtrA) serine protease inhibitor. The computationally designed compound displays suitable properties for further development and chemical biology studies. It blocked functional HtrA-mediated cleavage of E-cadherin and inhibited H. pylori transmigration in an in vitro pathogenesis model. Surface plasmon resonance (SPR) and saturation-transfer difference (STD) NMR data further confirmed specific binding of the NCE to H. pylori HtrA. Hit expansion on the designed small molecule led to the currently best-in-class HtrA inhibitor with favorable physicochemical properties, binding kinetics and functional activity. The results obtained validate automated ligand-based de novo design as a prime concept for the pioneering discovery of tailored chemical matter for new macromolecular targets.
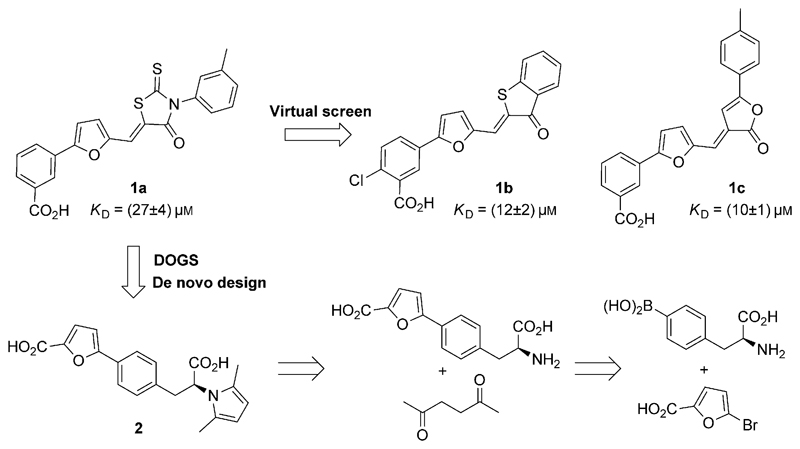
H. pylori is a Gram-negative bacterial pathogen colonizing about 50% of the world’s population and constitutes a risk factor for developing gastric/duodenal ulcers, as well as gastric cancer.[4] With increasing resistance against our conventional antibiotic armamentarium, there is an unmet need for novel therapeutic agents capable of preventing disease onset and progression. Additionally, host–pathogen interactions need to be further understood by use of small-molecule probes to enable the design of efficacious drugs.[5] HtrA is a virulence factor secreted by H. pylori and other pathogens, and plays an important role in cleaving E-cadherin and disrupting epithelial cell-to-cell adhesion.[6] Though, the adequate study of target biology and validation of HtrA as a relevant clinical drug target has been precluded by the lack of proper chemical tools. Current state of the art relies on rhodanine derivative 1a (Figure 1),[7] a poorly soluble, aggregating chemical entity with potential for conjugation with macromolecules through Michael addition. In a first attempt at finding analogues of 1a with amended structural features, we performed computational similarity searching in a collection of 3255 508 commercially available compounds. Using 1a as query, we identified 11 potential HtrA inhibitors (Table S2 in the Supporting Information). We used SPR for compound profiling and assessing their binding affinities to HtrA. While the query 1a has a KD value of (27 ± 4) μm, compounds 1b [(12 ± 2) μm] and 1c [(10 ± 1) μm] showed the highest affinities among the 11 selected analogues (Figure S22).
Figure 1.
Structures of 1a, its analogues 1b,c, and the de novo designed entity 2 with its retrosynthetic analysis.
Noteworthy, no key scaffold hops were achieved by similarity searching, as the selected compounds, like query 1a, still contain potentially reactive moieties. This result points to limitations of virtual screens as a feasible solution for the multidimensional exploration of chemical space. Thus, we hypothesized that computer-assisted de novo design might be better suited for this purpose, and provide chemotypes that mitigate the liabilities of 1a–c. While a similar approach had previously been pursued for the generation of innovative chemotypes with predictable target affinities,[8] scaffold hops have rarely been attempted with the goal of fulfilling multiple objectives, including the improvement of physicochemical properties.
Using 1a as design template, we applied the reaction- and fragment-based de novo design software DOGS (design of genuine structures)[9] to computationally generate new HtrA inhibitor candidates. The algorithm suggested 1707 unique molecules, among which 826 had computed logS>–6 (MOE v2011.10). We further processed the obtained candidates with a 3D pharmacophore model generated with LigandScout (v3.02) that captured features we considered as critical for bioactivity (see the Supporting Information). From the remaining 65 molecules, we selected compound 2 (Figure 1) for synthesis based on its ranking position, the absence of a potential Michael acceptor, profound structural dissimilarity to the template (Tanimoto index=0.26; ECFP4), and unreported chemotype (according to SciFinder). We obtained compound 2 through a two-step synthetic route. Suzuki coupling of l-(4-boronophenyl)alanine with 5-bromo-2-furoic acid afforded the required intermediate for subsequent Paal–Knorr pyrrole synthesis (Figure 1). Importantly, 2 is chemically stable unlike related fragments reported elsewhere.[10]
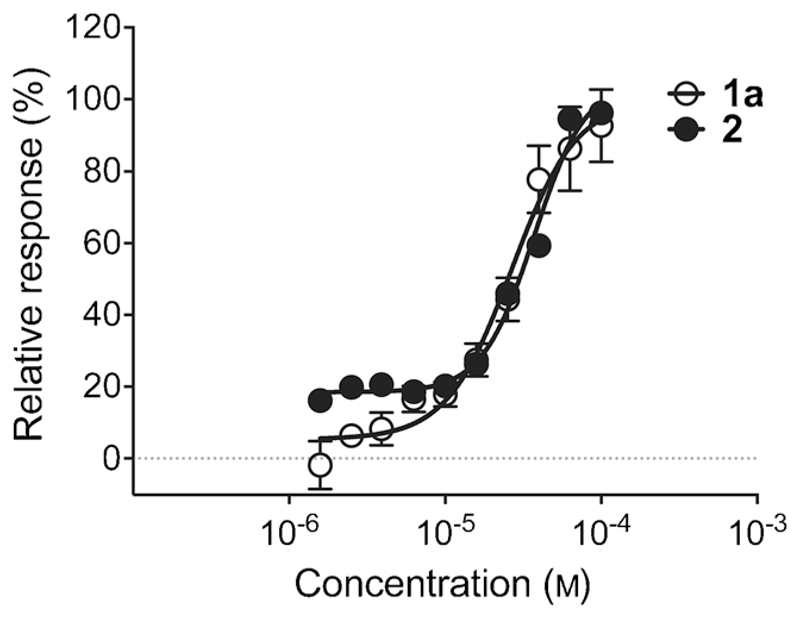
Colloidal aggregation of small molecules often yields false-positive target-engagement readouts in early drug-discovery programs.[11] We did not detect colloidal aggregates of 2 in concentrations up to 100 μm. Conversely, dynamic light-scattering measurements of 1a revealed critical colloidal aggregation in concentrations between 25 and 50 μm. Hence, the poor aqueous solubility of 1a in bioactive concentrations may cause unspecific ligand–receptor interactions. We employed SPR to further profile 2. Compared to 1a, the de novo designed compound 2 exhibited similar affinity (KD=(37 ± 4) μm, Figure 2), but a higher ligand efficiency (Table 1).
Figure 2.
Affinity of compounds 1a and 2 to H. pylori HtrA from SPR measurements.
Table 1.
Summary of the binding-kinetics parameters measured for H. pylori HtrA, and physicochemical properties of compounds 1a and 2.
| Compound | KD [μm] | koff [s−1] | t1/2 [s] | LE[a] | logP[b] | logS[b] |
|---|---|---|---|---|---|---|
| 1a | 27 ± 4 | 1.4 | 0.5 | 0.22 | 4.8 | −8.7 |
| 2 | 37 ± 4 | 1.8 | 0.4 | 0.24 | 3.5 | −4.3 |
Ligand efficiency (= −1.4 logKD/(number of non-hydrogen atoms)).
Octanol–water partition coefficient P and aqueous solubility S calculated with MOE 2011.10.
Understanding the kinetics of drug binding to macromolecular targets is of vital importance for lead optimization, as both the on- (kon) and off-rates (koff) determine the duration of a given direct pharmacological effect.[12] The residence time (t1/2) within a binding pocket is chiefly governed by the ligand's off-rate, with key implications for drug efficacy and selectivity. Compounds 1a and 2 showed fast koff rates and short residence times (Table 1), suggesting only few strong and directed interactions within the binding pocket, that is, moderate enthalpic contribution to binding. These observations are in agreement with allosteric inhibition kinetics, as reported elsewhere.[13] Altogether, the SPR data obtained for 2 advocate an innovative mechanism of action warranting further investigation.
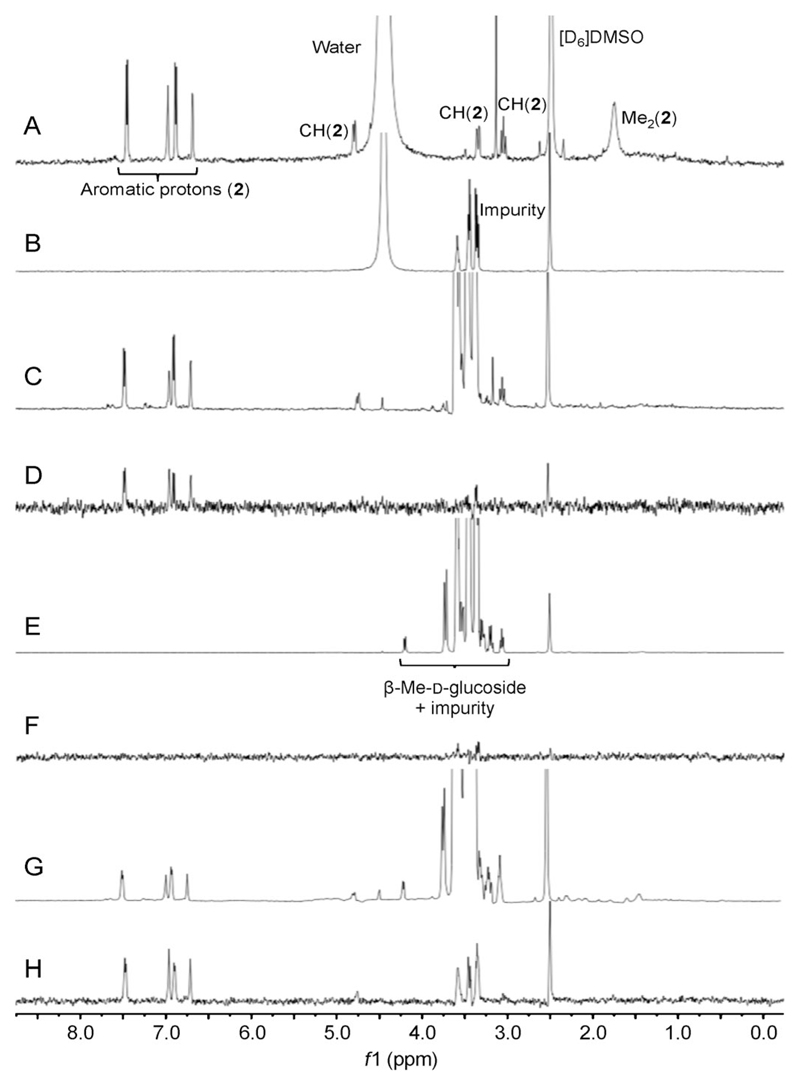
We used 1H STD NMR spectroscopy as an orthogonal method to gain insight into the molecular basis of binding of 2 to HtrA (see the Supporting Information). 1H NMR reference spectra were recorded for compound 2 and HtrA separately (Figure 3A,B), as well as the off-resonance reference (Figure 3C) and STD spectrum (Figure 3D) for the mixture of compound 2 and HtrA. Saturation transfer could be best identified for the phenylfuranyl moiety (6.75–7.50 ppm) and, to some minor extent, the methylene protons of compound 2. Together with an appropriate negative control (β-methyl-d-glucoside, Figure 3E–H), this result not only confirms direct binding, as previously measured by SPR, but also corroborates directed interactions of 2 with HtrA. Furthermore, the STD NMR spectra support the fast off-rate of 2 and provide a rationale for advanced lead-structure optimization. Head-to-head comparison of the de novo designed compound 2 to its template 1a through STD NMR spectroscopy was unsuccessful because of poor aqueous solubility of the latter (logS(1a) = −8.7 vs. logS(2) = −4.3, Table 1).
Figure 3.
STD NMR experiments revealing the direct interaction between compound 2 and H. pylori HtrA. A) 1H NMR spectrum of HtrA ligand 2 (1 μm). B) 1H NMR spectrum of H. pylori HtrA (10 μm). C) Off-resonance reference NMR spectrum of compound 2 (1 mm) in the presence of HtrA (10 μm). D) STD NMR spectrum of compound 2 (1 mm) in the presence of HtrA (10 μm). E) Off-resonance reference NMR spectrum of β-methyl-d-glucoside (1 μm) in the presence of HtrA. F) STD NMR spectrum of β-methyl-d-glucoside (1 μm) as a negative control; 10 μm HtrA. G) Off-resonance reference NMR spectrum of compound 2 (1 mm) together with β-methyl-d-glucoside (1 mm) in the presence of HtrA (10 μm). H) STD NMR spectrum of compound 2 (1 mm) together with β-methyl-d-glucoside (1 mm) in the presence of HtrA (10 μm). In B), C), E), G), and H) an impurity in the protein batch was observed at 3.25–3.75 ppm. In addition to residual water, the expected signals, and residual protons of deuterated solvent ([D6]DMSO), are also observable. The impurity is not visible in the STD spectrum in (D) indicating that it does not interact with HtrA. This spectrum clearly shows saturation transfer from HtrA to the aromatic protons of compound 2 (6.75–7.50 ppm) and, to some minor extent, a methylene proton (3.25 ppm). The spectra in (E) and (F) demonstrate that no saturation transfer occurs between HtrA and β-methyl-d-glucoside. The spectrum in (H) shows the observed saturation transfer from HtrA to the aromatic protons of compound 2, in the mixture between compound 2 and β-methyl-d-glucoside with HtrA, indicating the selective saturation transfer to the binding ligand.
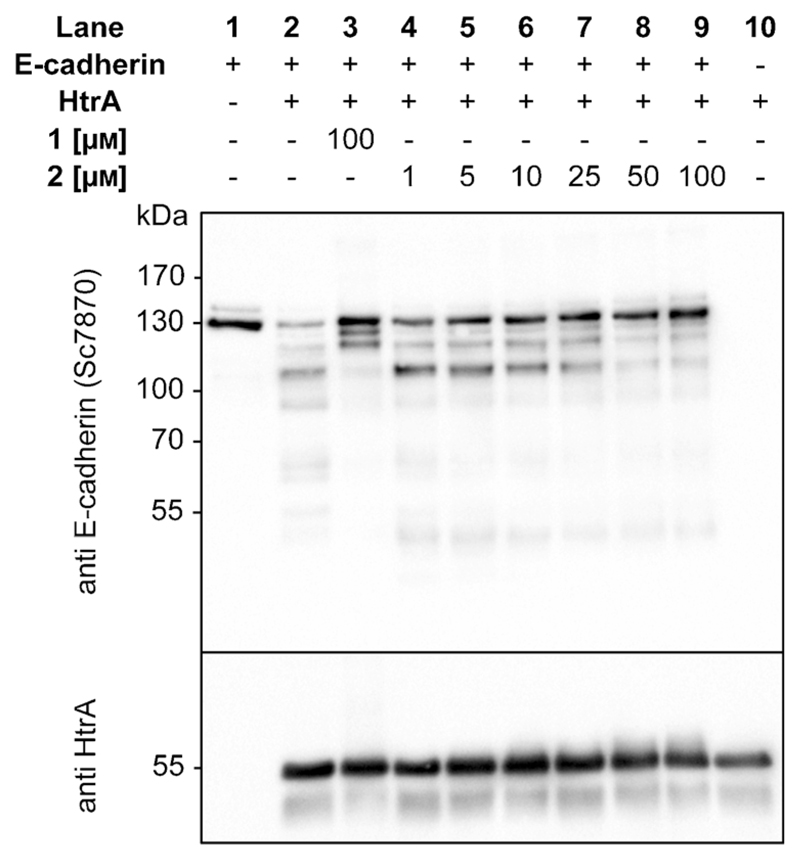
Following the thorough assessment of 2 by biophysical methods, we evaluated whether binding of the de novo designed entity translated into functional inhibition of H. pylori HtrA. Full-length 130 kDa E-cadherin is an HtrA substrate and the soluble E-cadherin fragment of about 90 kDa has been associated with a broad range of cancer types.[14] Efficient inhibition of HtrA-mediated E-cadherin cleavage might therefore provide an original means for preventing H. pylori infection. Western blot analysis (Figure 4) revealed that compound 2 shows concentration-dependent behavior and starts inhibiting cleavage of recombinant E-cadherin at a concentration of 10 μm (Figure 4, lane 6). At 50 μm the proteolytic activity of HtrA is blocked by 2 (Figure 4, lane 8), thereby supporting de novo design as a chief tool for obtaining innovative chemotypes with potent bioactivity profiles.
Figure 4.
Western blot analysis of recombinant E-cadherin cleavage by H. pylori HtrA in the presence of compounds la and 2 at different concentrations.
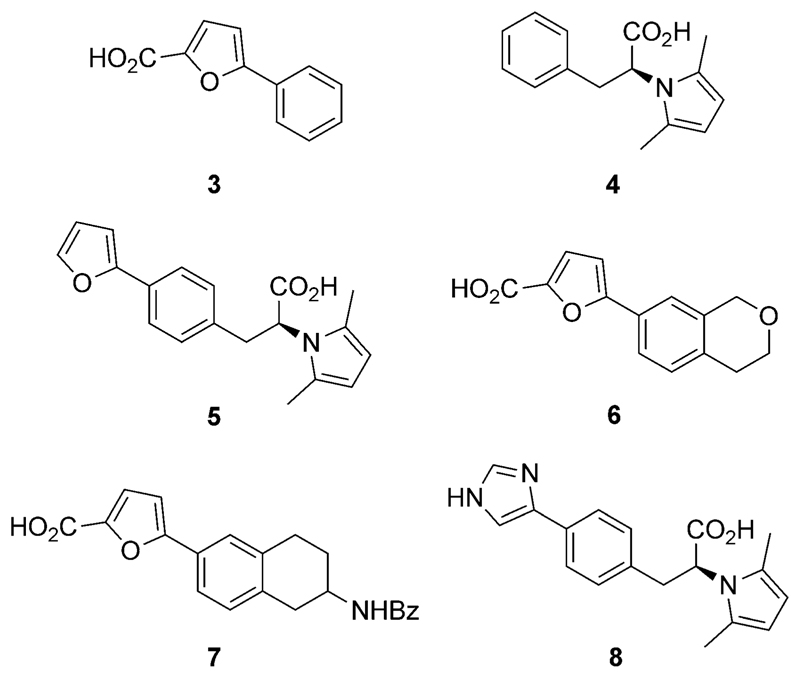
Having unveiled 2 as a lead structure with a tractable profile as an HtrA inhibitor, we engaged in a hit-expansion program. Specifically, we set out to: 1) acquire preliminary structure–activity relationship (SAR) data; 2) probe key pharmacophore features for HtrA inhibition; and 3) optimize the ligand efficiency of 2 through the modulation of its binding kinetics. Hence, we synthesized compounds 3–8(Figure 5). In 3–5 we truncated different moieties of 2 to assess the importance of the carboxylic acid groups and the steric bulk, in both the eastern and western regions of 2, on HtrA affinity and in vitro functional effect. Furthermore, with 6 and 7 we studied how the absence of the eastern carboxylic acid but presence of different bulky substituents affects bioactivity. With 8 we probed the effect of diverging physicochemical properties of the heteroaromatic moiety on target engagement. We conducted SPR and Western blot experiments to profile all compounds. The collected data (see the Supporting Information) suggest that the eastern carboxylic acid group is important for HtrA affinity and prevention of E-cadherin cleavage. Conversely, an ionizable heteroaromatic system, for example, 8, is apparently detrimental, resulting in a sharp loss of activity. Of note, the SPR sensorgrams obtained for compound 8 advocate a different mode of binding compared to 2–7 as no steady state was observed (Figure S23). Similar findings have been reported for two-step calcium-dependent protein–protein interactions.[15]
Figure 5.
Analogues of 2 obtained in hit expansion.
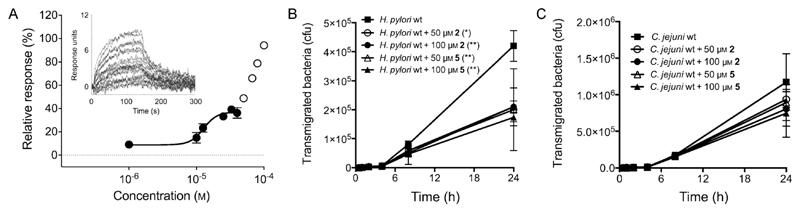
As determined by SPR, our new lead compound 5 has a KD value of (13 ± 2) μm, that is, a 3-fold affinity improvement over 2, and appropriate ligand efficiency (LE = 0.30; Figure 6 A). This improvement was chiefly governed by a slower off-rate and longer residence time (koff = 8 × 10−3 s−1; t1/2 = 89 s). Interestingly, an additional binding event occurs at concentrations higher than 40 μm, which may deserve further studies. These results were corroborated by inhibition of E-cadherin cleavage at concentrations as low as 5 μm (see the Supporting Information) and the absence of colloidal aggregation at 194 μm over the course of 1 hour (kinetic solubility > 60 mg mL-1),[16] suggesting specific target binding and suitable solubility. For further development it should be kept in mind that H. pylori HtrA is an extracellular protein and efficient inhibitors will require solubility in the gastric microenvironment without membrane permeation. The designed compounds have the required physicochemical profile.
Figure 6.
A) KD determination for compound 5 by SPR. Un unstudied binding event occurs at concentrations higher than 40 μm (N = 3 independent experiments). B) Inhibition of wild-type H. pylori transmigration across an epithelial layer of human gastric adenocarcinoma MKN-28 cells over a period of 24 h. Compounds 2 and 5 were tested at concentrations of 50 and 100 μm. C) Inhibition of wild-type Campylobacter jejuni transmigration over a period of 24 h. Compounds 2 and 5 were tested at concentrations of 50 and 100 μm (N = 3 independent experiments). Significance at 24 h was analyzed with one-way ANOVA (Dunnett test), in comparison to untreated control (solid boxes): (*) p < 0.05; (**) p < 0.01; cfu = colony-forming units.
Finally, we assessed H. pylori transmigration in vitro in the presence of compounds 2 and 5 at 50 and 100 μm (Figure 6B). A transwell migration assay[17] served as a model of microbial pathogenesis by measuring pathogen transmigration across a polarized epithelial layer of human gastric adenocarcinoma MKN-28 cells (see the Supporting Information). In accordance with the Western blot analyses, significant effects were detected at 50 μm over 24 h. A strikingly dissimilar outcome was obtained for both compounds against Campylobacter jejuni HtrA, suggesting critical differences between the HtrA homologues of the two pathogens that may explain selective blockage of H. pylori transmigration across the model gastric epithelium by 2 and 5 (Figure 6 C).
The results of the present fragment-based de novo design study leverage chemical-biology and early drug-discovery programs applied to HtrA. Particularly, we revealed the best-in-class HtrA inhibitor to date through multi-objective prioritization and binding-kinetics-guided optimization. The outcome of this study corroborates computer-assisted de novo design as a prime technology for obtaining innovative chemical probes with desired properties.
Supplementary Material
Supporting information for this article (experimental details, additional and raw data) is available on the WWW under http://dx.doi.org/10.1002/anie.201504035.
Contributor Information
Thomas P. Schmidt, Department of Molecular Biology, Universität Salzburg (Austria)
Dr. Manja Böhm, Department of Biology, Universität Erlangen-Nürnberg (Germany)
Prof. Dr. Steffen Backert, Department of Biology, Universität Erlangen-Nürnberg (Germany)
Prof. Dr. Silja Wessler, Department of Molecular Biology, Universität Salzburg (Austria)
Prof. Dr. Gisbert Schneider, Department of Chemistry and Applied Biosciences, ETH Zurich Vladimir-Prelog-Weg 4, 8093 Zürich (Switzerland)
References
- [1].Bennani YL. Drug Discovery Today. 2011;16:779–792. doi: 10.1016/j.drudis.2011.06.004. [DOI] [PubMed] [Google Scholar]
- [2].a) Schneider G, editor. De Novo Molecular Design. Wiley-VCH; Weinheim: 2013. [Google Scholar]; b) Rodrigues T, Schneider G. Synlett. 2014:170–178. [Google Scholar]
- [3].a) Rodrigues T, Kudoh T, Roudnicky F, Lim YF, Lin YC, Koch CP, Seno M, Detmar M, Schneider G. Angew Chem Int Ed. 2013;52:10006–10009. doi: 10.1002/anie.201304847. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2013;125:10190–10193. [Google Scholar]; b) Spänkuch B, Keppner S, Lange L, Rodrigues T, Zettl H, Koch CP, Reutlinger M, Hartenfeller M, Schneider P, Schneider G. Angew Chem Int Ed. 2013;52:4676–4681. doi: 10.1002/anie.201206897. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2013;125:4774–4779. [Google Scholar]; c) Rodrigues T, Roudnicky F, Koch CP, Kudoh T, Reker D, Detmar M, Schneider G. Chem Sci. 2013;4:1229–1233. [Google Scholar]
- [4].a) Wroblewski LE, Peek RM, Jr, Wilson KT. Clin Microbiol Rev. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Singh N, Kuppili RR, Bose K. Arch Biochem Biophys. 2011;516:85–96. doi: 10.1016/j.abb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- [5].a) Amieva MR, El-Omar EM. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]; b) Hoy B, Geppert T, Boehm M, Reisen F, Plattner P, Gadermaier G, Sewald N, Ferreira F, Briza P, Schneider G, Backert S, et al. J Biol Chem. 2012;287:10115–10120. doi: 10.1074/jbc.C111.333419. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Backert S, Boehm M, Wessler S, Tegt-meyer N. Cell Commun Signaling. 2013;11:72. doi: 10.1186/1478-811X-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Posselt G, Backert S, Wessler Cell Commun Signaling. 2013;11:77. doi: 10.1186/1478-811X-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoy B, Lower M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schroder P, Sewald N, Backert S, Schneider G, Wessler S. EMBO Rep. 2010;11:798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Löwer M, Geppert T, Schneider P, Hoy B, Wessler S, Schneider G. PLoS One. 2011;6:e17986. doi: 10.1371/journal.pone.0017986. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Klenner A, Hähnke V, Geppert T, Schneider P, Zettl H, Haller S, Rodrigues T, Reisen F, Hoy B, Schaible AM, Werz O, et al. Mol Inf. 2012;31:21–26. doi: 10.1002/minf.201100147. [DOI] [PubMed] [Google Scholar]
- [8].a) Reutlinger M, Rodrigues T, Schneider P, Schneider G. Angew Chem Int Ed. 2014;53:4244–4248. doi: 10.1002/anie.201310864. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2014;126:4330–4334. [Google Scholar]; b) Rodrigues T, Hauser N, Reker D, Reutlinger M, Wunderlin T, Hamon J, Koch G, Schneider G. Angew Chem Int Ed. 2015;54:1551–1555. doi: 10.1002/anie.201410201. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2015;127:1571–1575. [Google Scholar]
- [9].Hartenfeller M, Zettl H, Walter M, Rupp M, Reisen F, Proschak E, Weggen S, Stark H, Schneider G. PLoS Comput Biol. 2012;8:e1002380. doi: 10.1371/journal.pcbi.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhu W, Groh M, Haupenthal J, Hartmann RW. Chem Eur J. 2013;19:8397–8400. doi: 10.1002/chem.201301289. [DOI] [PubMed] [Google Scholar]
- [11].Seidler J, McGovern SL, Doman TN, Shoichet BK. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- [12].Copeland RA, Pompliano DL, Meek TD. Nat Rev Drug Discovery. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- [13].a) Siarheyeva A, Senisterra G, Allali-Hassani A, Dong A, Dobrovetsky E, Wasney GA, Chau I, Marcellus R, Hajian T, Liu F, Korboukh I, et al. Structure. 2012;20:1425–1435. doi: 10.1016/j.str.2012.06.001. [DOI] [PubMed] [Google Scholar]; b) Urbaniak MD, Collie IT, Fang W, Aristotelous T, Eskilsson S, Raimi OG, Harrison J, Navratilova IH, Frearson JA, van Aalten DM, Ferguson MA. ACS Chem Biol. 2013;8:1981–1987. doi: 10.1021/cb400411x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Han Z, Yang J-L, Jiang SX, Hou S-T, Zheng R-Y. PLoS One. 2013;8:e64894. doi: 10.1371/journal.pone.0064894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grabowska MM, Day ML. Front Biosci. 2012;17:1948–1964. doi: 10.2741/4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Seeger C, Gorny X, Reddy PP, Seidenbecher C, Danielson UH. J Mol Recognit. 2012;25:495–503. doi: 10.1002/jmr.2215. [DOI] [PubMed] [Google Scholar]
- [16].Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- [17].Boehm M, Hoy B, Rohde M, Tegtmeyer N, Bæk KT, Oyarzabal OA, Brøndsted L, Wessler S, Backert S. Gut Pathog. 2012;4:3. doi: 10.1186/1757-4749-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.