Abstract
Images of the retina acquired using optical coherence tomography (OCT) often suffer from intensity inhomogeneity problems that degrade both the quality of the images and the performance of automated algorithms utilized to measure structural changes. This intensity variation has many causes, including off-axis acquisition, signal attenuation, multi-frame averaging, and vignetting, making it difficult to correct the data in a fundamental way. This paper presents a method for inhomogeneity correction by acting to reduce the variability of intensities within each layer. In particular, the N3 algorithm, which is popular in neuroimage analysis, is adapted to work for OCT data. N3 works by sharpening the intensity histogram, which reduces the variation of intensities within different classes. To apply it here, the data are first converted to a standardized space called macular flat space (MFS). MFS allows the intensities within each layer to be more easily normalized by removing the natural curvature of the retina. N3 is then run on the MFS data using a modified smoothing model, which improves the efficiency of the original algorithm. We show that our method more accurately corrects gain fields on synthetic OCT data when compared to running N3 on non-flattened data. It also reduces the overall variability of the intensities within each layer, without sacrificing contrast between layers, and improves the performance of registration between OCT images.
Keywords: Optical coherence tomography, retina, intensity inhomogeneity correction, macular flatspace, registration
1. Introduction
Optical coherence tomography (OCT) is a widely used modality for imaging the retina as it is non-invasive, non-ionizing, provides three-dimensional data, and can be rapidly acquired (Hee et al., 1995). It uses near-infrared light to measure the reflectivity of the retina, producing a clear image of the retinal structure and the cellular layers comprising it. OCT improves upon traditional 2D en-face plane photography by providing depth information, which enables measurements of layer thicknesses that are known to change with certain diseases (Medeiros et al., 2009; Saidha et al., 2011).
Since the analysis of retinal OCT data—measuring layer thicknesses, for example—is a time consuming task when done manually, automated methods are critical for examining populations of patients in large-scale studies. This analysis often relies on image processing to extract information from the images, like layer boundaries and fluid regions, in order to make the measurements of interest. In recent years, many automated methods have been developed for the segmentation of the retinal layers (Garvin et al., 2009; Chiu et al., 2010; Vermeer et al., 2011; Lang et al., 2013, 2015a, 2017) and fluid-filled regions (Wilkins et al., 2012; Chen et al., 2012; Swingle et al., 2014, 2015; Lang et al., 2015b; Antony et al., 2016b). Image registration, which has received much less attention, has been used to align scans from different subjects (Chen et al., 2014), for longitudinal intra-subject registration (Niemeijer et al., 2012; Wu et al., 2014), and in applications of registration including voxel based morphometry (Antony et al., 2016a).
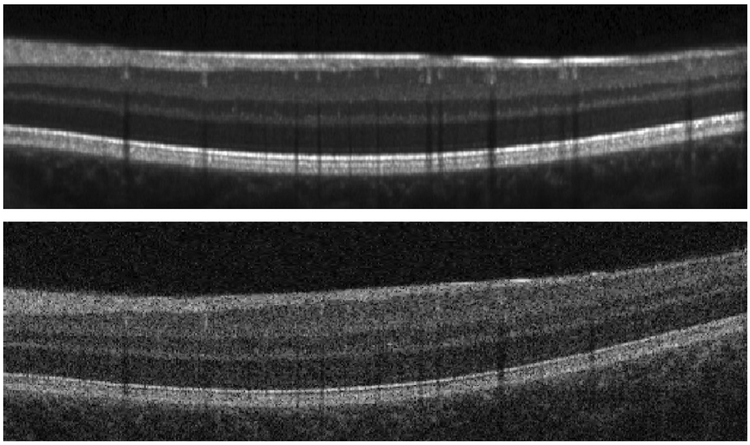
Such automated algorithms rely on the consistency of intensities within images and across subjects to perform optimally. Unfortunately, OCT images often have different intensity values between subjects and scanners. Though some segmentation methods (Novosel et al., 2017) have been designed specifically to only assume that layers are locally homogeneous. Figure 1 shows an example of intensity differences between images acquired on the same patient on different scanners. In general, differences in intensity are attributed to differences in scanner settings and protocols, in the opacity of the ocular media, and possibly in tissue properties like attenuation, which can change due to disease (Vermeer et al., 2012; Varga et al., 2015)
Figure 1:
B-scan images acquired on the same subject of (approximately) the same location on two different scanners demonstrating the variability in the intensity profile. The images were acquired on (top) a Heidelberg Spectralis scanner and (bottom) a Zeiss Cirrus scanner.
OCT images also suffer from intensity inhomogeneity problems, leading to variability within a single scan of the same subject. Intensity inhomogeneity occurs for a variety of reasons: off-axis acquisition resulting in signal loss (Arevalo, 2009), tissue attenuation (Vermeer et al., 2014), orientation of the cellular structure (Lujan et al., 2011), vignetting due to the iris (Drexler and Fujimoto, 2008), material inhomogeneity in the eye’s lens, cornea, and vitreous fluid (Arevalo, 2009), misalignment when averaging multiple images to improve image quality, and even dirt on the scanner eyepiece. Since the sources of intensity inhomogeneity are not necessarily consistent with each other, a systematic approach to correcting for it is not as straightforward as in magnetic resonance imaging (MRI), where the inhomogeneity is well understood (Sled et al., 1998). Basic methods for correcting OCT data from well-characterized physical sources like attenuation (Vermeer et al., 2014; Girard et al., 2011) may not correct for other sources of inhomogeneity. We note that the models used within attenuation coefficient correction approaches (Girard et al., 2011; Vermeer et al., 2014) represent the inhomogeneity as a multiplicative gain field model. Such a model has been explored in depth within the neuroimaging community, with examples including N3 (Sled et al., 1998), N4 (Tustison et al., 2010), and others (Roy et al., 2011).
Correction of intensity inhomogeneities in OCT images is expected to be beneficial for automated algorithms such as image segmentation and registration, which typically depend on consistent intensities for optimal performance. Since intensity variations may well be indicative of pathology, the use of inhomogeneity correction for diagnostic purposes may be ill-advised. Therefore, we specifically seek a correction method that will work well as a preprocessing approach in automated OCT image analysis.
A few methods have been developed to correct inhomogeneity in retinal OCT data. In (Kraus et al., 2014), the projected intensity pattern in the en-face plane was used for illumination correction of the data; however, the correction did not vary with depth. Novosel et al. (Novosel et al., 2015) use the attenuation correction method in (Vermeer et al., 2014) as a pre-processing step prior to running their layer segmentation method. The N4 algorithm (Tustison et al., 2010), originally developed for MRI data, was used by Kaba et al. (2015), but it was not able to remove all inhomogeneity as shown in their presented figures.
A common method to normalize the intensity range of OCT data is to rescale the intensity values, either across the volume or in individual B-scans, to a fixed range (Ishikawa et al., 2005; Ghorbel et al., 2011; Wilkins et al., 2012; Lang et al., 2013). These methods do not use the intensity range of specific layers when normalizing the data and thus inconsistencies still arise. In contrast, the work of Chen et al. (2015) used histogram matching to normalize the intensities of different scans, and showed improved stability of layer segmentation across a range of images with different quality. However, matching histograms may not be robust, especially in the presence of inhomogeneity, which will blur the peaks of the histograms.
In this paper, we propose a method for both inhomogeneity correction and normalization of macular OCT data that we call N3 for OCT (N3O). At the core of our method is the N3 algorithm, which was developed for inhomogeneity correction of brain MR data (Sled et al., 1998) and has been shown to be competitive with other state-of-the-art algorithms (Arnold et al., 2001; Hou, 2006). Since direct application of N3 does not produce satisfactory results on OCT data, we adapted the method, making several changes to improve both performance and efficiency. Some of this work was previously presented as a conference paper (Lang et al., 2016). In this paper, we have added several improvements to the method, significantly expanded the algorithmic details, and provided a more extensive validation, which includes the use of synthetic data and analysis of the effects of correction on image registration.
2. Methods
2.1. Overview
A goal of this work is to correct macular OCT data so that the pixel intensities of a layer at any region within that layer should be similar—that is have a small standard deviation relative to the standard deviation of intensities within the whole image—moreover they should be consistent with scans from other subjects. This would give us a subject independent standardized intensity scale for macular OCT data, which would be beneficial in creating normalized population atlases. In other words, the intensity values of a layer at one region of a volume should have similar values to those in another region of the same volume, as well as to scans from another subject. Our approach proceeds in three steps. First, we convert each B-scan into a computational domain that we denote macular flatspace (MFS)—in this domain, the second step of inhomogeneity correction is improved. This correction step estimates and then removes a smoothly varying gain field. Finally, the data is normalized so that the intensity values of the corrected data lie in a predefined range consistent across subjects.
2.2. Macular Flatspace
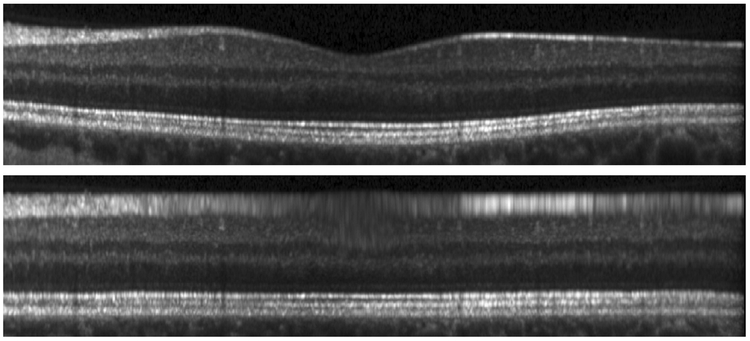
We transform an OCT image into MFS to create an image in which all of the layers appear flat; see Fig. 2 for an example. In the converted image, thin layers are stretched and thicker layers are compressed to achieve the goal of the layers being approximately flat. The transformation itself is applied only in the A-scan direction (vertically), and can therefore be thought of as applying a 1D deformation to each A-scan. However, knowledge of the 3D spatial position of the A-scans is necessary to determine which 1D transformation to apply.
Figure 2:
A B-scan image in (top) native space and (bottom) MFS.
MFS acts as a standardized computational space allowing for consistent processing across subjects. It also removes the curvature of the retina—which can vary significantly across acquisitions—allowing for different regions of the volume to be treated in the same manner. We also note that the coordinate system in MFS becomes meaningful relative to the “coordinate system” of the retina; traversing the x and y axes in MFS corresponds to movement within and between layers, respectively. As a result, operations like smoothing can be easily adapted to restrict smoothing to within or between certain layers.
Previously, the MFS transformation of OCT data has been used for layer segmentation where the conversion allowed for simplified constraints on the shape of the retina (Lang et al., 2014; Carass et al., 2014). Three distinct changes have been made to the MFS we use in this work: 1) a quadratic regression model replaces a linear model to better fit the retinal shape; 2) added regularization is used to enforce spatial smoothness; 3) cubic interpolation of the transformation is used to reduce artifacts.
In order to transform the image to MFS, where the layers appear flat, we require an estimate of the boundary positions. In the case that a set of layer boundaries are available, perhaps as output by a layer segmentation algorithm, then these segmentation boundaries can be used directly to transform the data to MFS. However, N3O is intended to be used as a pre-processing step for such algorithms, and we therefore do not assume that a layer segmentation is available. Instead, we estimate each layer’s position based only on the positions of the top and bottom retinal boundaries, the inner limiting membrane (ILM) and Bruch’s membrane (BrM), respectively, which can be done very quickly and accurately due to their large gradient response.1
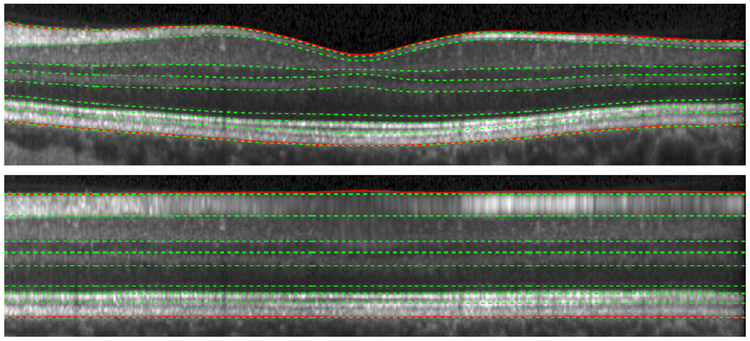
These two boundaries are then used to predict where the interior boundaries will be. For this prediction, we use separate regression models to find each boundary within each column, or A-scan, of an image. Figure 3 shows an example image with estimated boundaries as dashed green lines, which are computed based only on the solid red outer retina boundaries. Given the boundary positions, we construct a transformation going from the regression boundaries to a flattened position defined by the average position of each boundary in the native space. Definitions of the various layers can be seen in Lang et al. (2013) Fig. 1.
Figure 3:
A B-scan image in native space and MFS with retinal boundaries overlaid as solid lines in red and regression estimated boundaries overlaid as dashed lines in green.
2.2.1. Boundary estimation
We estimate the boundary positions within an A-scan assuming that the thick ness, ti(x), of layer i (i ∈ {1,…,8}) can be predicted given the total retina thickness, t(x) = b2(x) − b1(x), where b1 and b2 are the initial estimates of the ILM and BrM boundaries, respectively, and x indexes the A-scans. Specifically, we use a quadratic model expressed as
| (1) |
where αi,j are the regression coefficients. A similar regression model estimates t0(x), the distance from b1(x) to the true ILM boundary, correcting for any bias in the b1 estimate compared to ground truth data. Given an input set of retina boundaries and the result of each regression, the estimated boundary positions are given as
| (2) |
where j ∈ [1,…, 9} indicate the boundaries in order from the ILM to the BrM.
The regression model in (1) is trained using data from manually segmented macular OCT scans. Since this model is spatially varying over A-scans, we first align each scan to the fovea, providing a central reference point. Since the scans were acquired having a consistent orientation, no further alignment was done aside from flipping right eyes to appear as left eyes. Any remaining variability is expected to be captured by the regression. Given T manually segmented volumes, each having a fixed size of L × M × N voxels2, there are MN A-scans and 3MN coefficients to estimate for the quadratic regression of a single layer. We solve for the coefficients using a regularized least squares system of equations formulated as
| (3) |
where ti is a T MN × 1 vector of layer thicknesses derived from the manually de lineated boundary points, A is a 3MN × T MN block diagonal matrix of the form
| (4) |
with Vandermonde matrices Vk on the diagonal with the form
| (5) |
where each row uses thickness values from a different training subject. The regularization matrix Γ penalizes differences in the coefficients of neighboring A-scans (i.e. the first order difference in the two orthogonal directions of the data). This problem can be solved efficiently as a sparse system of equations using the QR decomposition (e.g. as implemented using the function in MATLAB).
2.2.2. MFS Transformation
Given an A-scan, the transformation from native space to MFS is constructed by mapping the regression boundaries so that they are flat. Specifically, given a set of points in native space {ri, i = 1,…, 9} and corresponding points in flat space {fi, i = 1,…, 9}, we need to find a smoothly varying and monotone transformation r = T(f) such that ri = T(fi) for all i.3 The monotone requirement preserves layer ordering and prevents folding in the transformation. Since a piecewise cubic Hermite interpolating spline (Fritsch and Carlson, 1980) fits these requirements, we use this method for interpolating the transformation between boundaries. (For further details see the MATLAB function pchip.) In previous work, this transformation was defined using linear interpolation (Lang et al., 2014; Carass et al., 2014), which can produce discontinuity artifacts at boundaries.
For the values of fi, we use the average value of ri over all A-scans (i.e. the native space boundaries are mapped to their average position), as such layers maintain their proportional thickness between native space and MFS (that is thin layers in native space are thin in MFS). Since the interpolation of the data into MFS is done on a regular lattice grid, we need to define the size of the interpolated MFS result. The width of the data is unchanged so we only need to choose the number of rows. Specifically, we do this by setting the pixel size in MFS such that the average height of each pixel in a layer is equal to 4 μm, which is close to the digital resolution of the data. Note that the true physical pixel size at every location varies laterally depending on the amount of compression or stretching in each A-scan. Padding is also added to the ILM and BrM at a fixed distance of 60 μm. The resulting MFS data has approximately 130 pixels per A-scan, depending on the subject.
2.3. N3 Inhomogeneity Correction
We briefly describe the details of N3 Sled et al. (1998) here, before detailing our modification for OCT data. The inhomogeneity model is assumed to be multiplicative with the intensity of an image v at position x given by v(x) = u(x)b(x) + n(x), where u is the corrected/underlying image, b is a smoothly varying gain field, and n is normally distributed noise. By taking a logarithm of the data, an additive model is created, leading to the model log . The additive field is assumed to be smoothly varying following a zero-mean Gaussian distribution. Note that the noise term is assumed to be zero in this transformed model, as detailed in the original work Sled et al. (1998). The derivation of the N3 algorithm operates under this assumption, despite it being incorrect. To partially counteract this problem, an iterative correction strategy is used by N3, incrementally estimating the inhomogeneity field. In practice, the method was found to work well on real data with varying levels of noise, and therefore it can be assumed that the noise-free assumption and iterative refinement strategy are appropriate.
The algorithm iterates over three steps of estimating û from given in the previous iteration, sharpening the distribution of û using the assumed normal distribution of , and smoothing the resulting estimate of from the sharpened û by fitting a cubic B-spline surface to the data. Iterations continue until either the field estimate converges to within a given threshold or the number of iterations reaches a specified limit.
To adapt N3 to work for OCT data, we incorporate three significant modifications to the original algorithm. First, instead of initializing the gain field to unity as in Sled et al. (1998), we use the value of the image divided by an average MFS image as the initialization, which improves the convergence of the algorithm. Second, while we maintain the smoothing step of the original algorithm at every iteration, we use a slightly different and more efficient B-spline smoothing model. This new model produces similar results to the original method at a fraction of the computational cost. Finally, we run N3 independently on every 2D image in a 3D volume as opposed to running it fully in 3D was as done in the original work. In practice, we have found that intensity inhomogeneity in OCT data can vary a lot between images due to serial acquisition of the data and a full 3D model does not work as well.
2.3.1. Initialization
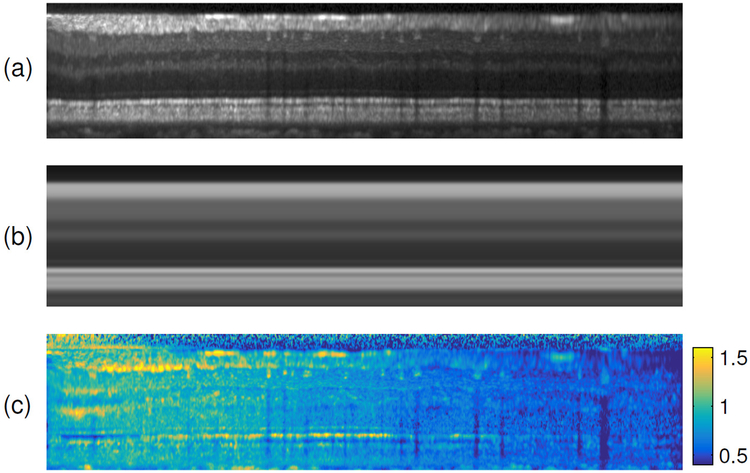
By averaging over all A-scans in an MFS-converted OCT volume, we create an average A-scan profile that is then replicated back to the size of the original data to produce a template MFS image. This template image serves as a guide to what the original data should look like without any inhomogeneity. We can then compute an initial estimate of the gain field by dividing the input image by the template image. An example of these three images is shown in Fig. 4. This initialization has artifacts since the boundaries in the initialization are not perfectly flat. However, the iterative refinements of the algorithm allow for convergence to an accurate estimate of the true gain field, which produces corrected intensities similar to those of the template image.
Figure 4:
The data (a) in MFS is divided by (b) the template image to generate (c) the initial gain field.
2.3.2. B-spline smoothing model
In N3, the estimated gain field is smoothed by fitting a tensor cubic B-spline surface to the data. In two dimensions, this B-spline function is written as
| (6) |
where Bi(x) is a 1D B-spline along the x dimension, centered at control point i, with a similar definition for Bj(y), and αij are weights of the 2D tensor B-spline function centered at the control point indexed by i and j. Control points are equally spaced over the data with the spacing between points in each of the two directions given as algorithm parameters. Smoothness of the fit is enforced both by increasing the distance between the control points, and by adding a regularization term to the least squares fitting problem.
The B-spline model is fit to the data by solving for the B-spline coefficients that minimize an energy function as
| (7) |
where E(α) measures the average squared error of the B-spline fit, R(α) measures the roughness of the fit, and β is a balance parameter. In Sled et al. (1998) the data term was defined as
| (8) |
where In is the measurement of the intensity at coordinate xn = (xn, yn), and the roughness term is defined by the thin plate bending energy as
| (9) |
where the integral is over the region C containing the data, which has area A Sled et al. (1998), and N is the number of pixels in the image.
We use the same form of the data energy E(α), but a different form for R(α), following the work of Eilers et al. (2006). Specifically, instead of minimizing the energy over the entire B-spline surface μ, we minimize over only the B-spline coefficients α. The regularization term used for this problem has the form (using the notation of the original paper)
| (10) |
where D2 is a second order difference matrix and αi• is a row vector containing values of αij over all j, with a similar definition for α•j as a column vector. Assuming that the number of control points is much smaller than the number of pixels in the image, this form of regularization constructs a far smaller matrix allowing the problem to be solved in an efficient way Eilers et al. (2006). While this model produces slightly different results than the original N3 one, we have empirically found that a similar result can be produced by tuning the regularization parameter β. When tuned to provide similar results using the same control point grid, the fit using the new model is about 150 times faster than running the fit using the old model (0.05 vs. 0.8 seconds per B-scan).4
2.4. Intensity Normalization
Since N3 only acts to sharpen the peaks of a histogram, the intensity ranges of corrected images are not necessarily consistent across scans. As we do not assume any knowledge of the segmentation of the layers, other than the retinal boundaries used in our MFS step, we cannot simply scale the intensity values within each layer to a predefined value. Instead, we scale the intensities based on the peak values found in the histograms of the vitreous region above the ILM and in the layer found above the BrM, known as the retinal pigment epithelium (RPE).
Since the peak value in the vitreous histogram is consistent and easy to find, we use the maximum value in the histogram directly for scaling; let this value be I1. While the RPE generally contains bright intensities, there are often two peaks in its histogram since darker intensity values also appear due to its proximity to the choroid, the appearance of blood vessel shadows, and the dark bands of the photoreceptor layers. For clarity, we define the choroid region to be everything within 40 μm of the BrM boundary. As a result, using the peak value of the histogram may produce a value not representative of the RPE band. We counteract this possibility in three ways: 1) Restrict the region we compute the histogram over to be from the BrM to 25 μm above it, which may not fully encompass the RPE layer; 2) Compute the histogram using a kernel density estimate in which the kernel is Gaussian with a relatively wide bandwidth of 0.05; 3) Find all peaks in the histogram and choose the one centered at the largest value, which we denote as I2. We normalize the data by contrast stretching, mapping values in the range [I1, I2] to the range [0.1, 0.65].
These values are arbitrary, however they produce images with an intensity range that is consistent with those in the native data.
3. Experiments
To evaluate the performance of our algorithm, N3O, we constructed three separate experiments. 1) We created synthetic OCT data free of any inhomogeneity and looked at the accuracy of using N3O to recover randomized gain fields having different characteristics, which are added to the data. 2) We looked at the variability of the intensities within each layer before and after running N3O to explore how the stability and consistency of the intensities changes in real data. 3) We ran an intensity based registration algorithm (Chen et al., 2013, 2014) on N3O processed data to show the improvement in performance offered by N3O. The first two experiments provide quantitative measures to show not only that N3O accurately corrects inhomogeneity, but that it does so more accurately than other methods. The third experiment provides a practical application of using N3O as a pre-processing step for an automated image analysis algorithm, in which we again compare its performance against other methods.
3.1. OCT Data
The data used in our experiments were acquired using either a Zeiss Cirrus scanner (Carl Zeiss Meditec, Dublin, CA, USA) or a Heidelberg Spectralis scanner (Heidelberg Engineering, Heidelberg, Germany). All images from the respective scanner were scanned using the same protocol. Both Cirrus and Spectralis data cover a 6 × 6 mm area of the macula centered at the fovea, with the Cirrus imaging to a depth of 2 mm and the Spectralis to a depth of 1.9 mm. B-scan images have a size of 1024 × 512 pixels for the Cirrus data with 128 equally spaced B-scans per volume. The Spectralis images have a size of 496 × 1024 pixels with 49 equally spaced B-scans per volume. The Spectralis scanner also used the automatic realtime (ART) setting where a minimum of 12 B-scan images of the same location were averaged to reduce noise. We also use default parameters within the scanner for selecting the focal plane, beam shape and signal roll-off.
3.2. Algorithm details
Training for the MFS was done using manually segmented Spectralis data from 41 subjects. Each boundary has one pixel per A-scan and therefore has 496 × 49 points over the volume. Rather than modifying the regularization in (3) to account for the anisotropy of the data, we resized the boundary maps to have a size of 224×224 pixels before computing the regression. We set λ= 4 in (3), based on a ten fold cross validation by minimizing the average standard deviation of the fitting error in the subjects left out of a fold.
We used default parameters from the original N3 algorithm for the number of iterations (50) and the convergence threshold (0.001) and used a FWHM of 0.1 for the Gaussian distribution used for sharpening. Downsample factors of 2 were used for each dimension in Cirrus B-scans and a factor of 4 horizontally in Spectralis B-scans (no vertical downsampling). For the experiment of running N3 on native space data, the region of the retina between the estimated ILM and BrM boundaries was used as a mask for running the algorithm.
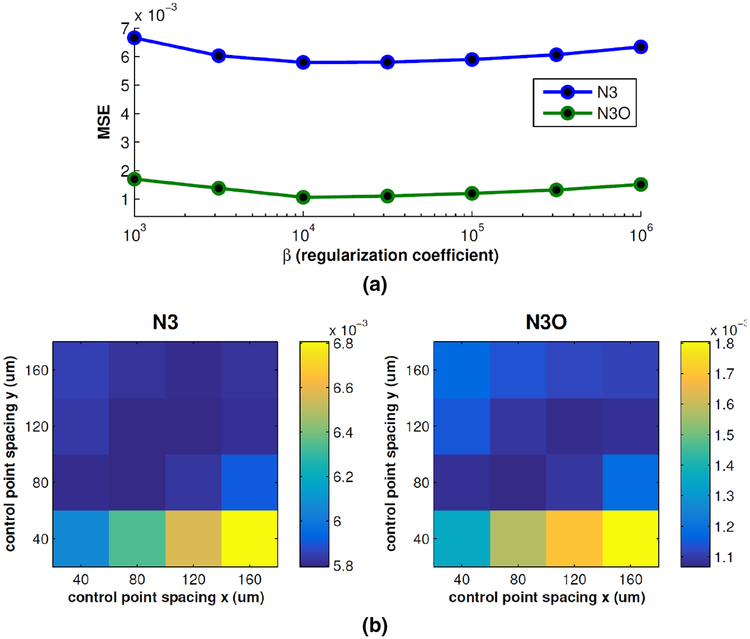
Finally, the values of the B-spline control point spacing in the x and y directions and the regularization parameter β in (7) were chosen by tuning the parameters over a range of values and choosing those with the best performance. Evaluation was done on an independent set of simulated OCT data, generated as described for experiments in Section 3.3. This set consisted of six synthetic OCT volumes (three Spectralis and three Cirrus), with five gain fields randomly generated using each of the four models described in Sec. 3.3.2 added to each volume. Thus, for a given set of parameters, the mean squared error between the true and estimated gain fields were averaged over all 120 data sets. The results of fixing the control point spacing to 80 μm in each direction and searching over different values of β, as well as fixing β = 104 and searching over the 2D space of control point values is show in Fig. 5. The values that worked best for both N3 and N3O were control point spacing in both directions of 80 μm and β = 104.
Figure 5:
Plots of the mean squared error (MSE) of the gain fields recovered using N3 and N3O while (a) varying and fixing the control point spacing, and (b) varying the control point spacing in the x and y directions while fixing.
3.3. Gain Field Recovery from Synthetic Data
For the first experiment, we created several sets of synthetic OCT data free of inhomogeneity and therefore useful for estimating the performance of N3O by applying artificially generated gain fields and comparing the recovered field to the true field. The artificial gain fields were generated randomly by following one of two different inhomogeneity models, one having decreased intensities near the edges of the data field-of-view, simulating effects of curvature and vignetting, and one having decreased intensities over different regions of the data, with the size and number of regions varying randomly. In addition to these two global inhomogeneity models, we included an additional inhomogeneity field independently to each B-scan to simulate effects due to scan averaging, raster-scan acquisition, and eye movement.
3.3.1. Synthetic Data
To create synthetic OCT data, we followed a similar process to how the template images were created for the N3 initialization step, as described in Sec. 2.3. Specifically, an input volume was converted to MFS where all of the A-scans were then averaged to create a 1D template A-scan. This template was then replicated back to the size of the original volume and transformed back to native space so as to match the shape of the ILM and BrM boundaries in the original image.
Noise was added to each image according to the OCT speckle model described by Serranho et al. (2011). Details of the algorithm are left to the referred paper. The algorithm has several parameters, and we used a different set for each scanner’s data. For the Cirrus data, we used the same parameters described in the paper but amplified the scale of the additive noise by 25%. For the Spectralis data, we changed algorithm parameters β1 = 0.1 and β3 = 0.6. We also decreased the amplitude of the additive noise by 50% and smoothed the final noise field with an isotropic Gaussian kernel (α = 2.5 μm). These changes are heuristic in nature, designed only to produce visually similar images to those we acquire on living persons.
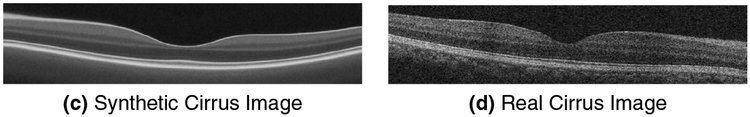
Example synthetic B-scans from both scanners are shown in Fig. 6. We note that blood vessels are removed by this process. Blood vessels could be added back into the data, however we did not do this so as to assess the performance purely based on layer intensities.
Figure 6:
Examples of synthetic OCT data generated from real scans acquired by the (a) Spectralis and (c) Cirrus scanners next to their corresponding real images (b, d).
3.3.2. Artificial Gain Fields
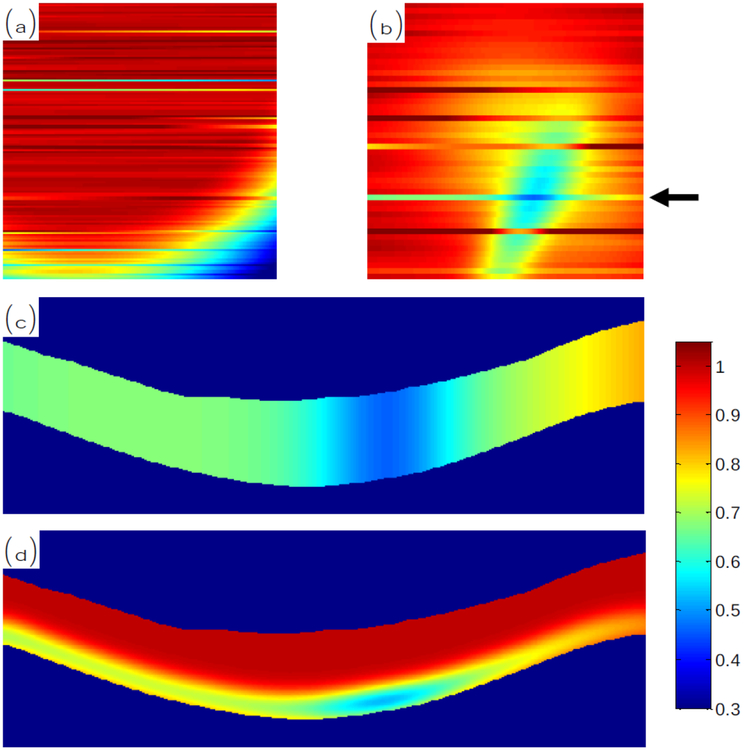
The gain fields are randomly generated as 2D patterns on the top-down en-face plane of the data. The pattern is then projected down through the data such that either the gain has the same value throughout the entire A-scan, or it has the same value only within the RPE region. When restricted to the RPE region, we also smooth the resulting gain field so that it is not discontinuous at the boundaries. An example of this process is shown in Fig. 7 where we show the gain field pattern in 2D and then projected through the volume.
Figure 7:
En-face plane views of an example (a) Type 1 and (b) Type 2 gain field patterns, each having added variation within each B-scan. In (c) and (d), we show the gain field at the location pointed by the arrow in (b) projected through the volume to cover the whole retina or only the RPE region, respectively.
Two different types of randomized gain fields, which we will denote as Type 1 and Type 2, were added to the synthetic data. While these two types of fields are modeled after realistic patterns found in OCT data, such as those that may be found as a result of retinal pathology, however, they are exaggerated to be relatively extreme cases. The Type 1 pattern reduces the intensities around the edge of the field-of-view of the data. Specifically, the multiplicative gain field has a unity value within a circle of radius mm (thus, the circle can circumscribe the square 3 × 3 mm area of the data). The center of this circle is then randomly placed between 2 and 3 mm of the center of the scan. The gain field decays in a Gaussian shape outwardly from the edge of the circle with a variance such that the smallest value over the entire image is scaled equal to 0.2.
The Type 2 gain field pattern includes random areas of decreasing intensity simulating local areas of inhomogeneity, which commonly arise in the data. Specifically, we randomly include between 1 and 4 spots, centered randomly within the central 5 mm area of the data. The spots are modeled as anisotropic Gaussian functions with a randomly chosen standard deviation value between 0.25 and 2 in each direction, as well as being oriented in a random direction, and having a peak magnitude between 0.2 and 0.5.
Finally, to model inhomogeneity patterns that differ between B-scans, we linearly vary fields across each B-scan. Specifically, the gain values on the left and right edge of each image are chosen randomly from a normal distribution with a mean value of 1 and a standard deviation of 0.02 for 85% of the images, and with a standard deviation of 0.2 for 15% of the images. Thus, the added inhomogeneity pattern is smaller for most of the data, which is an effect commonly seen in data acquired from the Spectralis scanner.
3.3.3. Experiments
We generated ten synthetic OCT scans of healthy controls, with five coming from each of the Spectralis and Cirrus scanners. For each synthetic scan, we randomly generated ten inhomogeneity fields using each of the two models. We further restricted each of the fields to be applied either to the entire retina or to only the RPE region. Restricting only to the RPE mimics changes in the intensity of only a single layer or region, which is similar to attenuation differences over varying thicknesses. Thus, a total of 400 synthetic data sets were created. Note that noise was added to the synthetic images after applying the respective gain field.5
To evaluate the algorithm performance, we compute the average RMS error across all scans as
| (11) |
where the summation is over the n pixels in the retina masked region M, bgt,i and best,i are the ground truth and estimated gain fields, respectively, indexed by pixel i, and ω is a normalization factor accounting for a scale difference between the two fields. The value of ω is computed in closed form by minimizing the sum-of-squared-differences Chua et al. (2009). Finally, we evaluate the performance using the simulated data after running N3O versus running N3 on the native space data with initialization using a unity gain field and the modified smoothing model described in Section 2.3 (thus we do not compare against the original N3 algorithm, but a version modified to work on OCT data).
3.4. Consistency and Contrast of Layer Intensities
For the second experiment, we measure the variability of intensities within each layer before and after running N3O. While not a direct measure of inhomogeneity correction, measuring this variability provides a surrogate measure of algorithm performance since a stated goal was to increase the consistency of intensities within each layer.
To measure the intensity variability, we computed the coefficient of variation (CV) of the intensity values within each layer, with lower CV values indicating better performance (more stability). CV measures the ratio of the standard deviation to the mean value and is independent of the absolute scaling of the estimated gain field and thus is directly comparable between algorithms and data sets. Layers were defined based on the results of using a validated automated segmentation algorithm on the data (Lang et al., 2013). To reduce the effect of boundary errors, the resulting segmentation labels for each layer were eroded by one pixel. Using this type of conservative segmentation has also been shown to be more correlated with the accuracy of estimated gain fields in MR data (Chua et al., 2009).
An image having a uniform intensity everywhere would be best according to CV, so we therefore also look at the contrast between adjacent layers to show that we maintain the differences between layer intensities after running N3O. The contrast between adjacent layers i and j is defined as where denotes the average intensity within layer i. Larger values of Cij indicate increased levels of contrast. Since contrast is not invariant to linear transformations of the intensity, we rescaled the intensity range within each image so that the vitreous region has a value of 0.2 and the RPE region has a value of 0.65. We did not use histograms for normalization as in Sec. 2.4, since the histogram is less reliable in the presence of inhomogeneity.
In our experiments, we analyzed 80 scans from each of the Spectralis and Cirrus scanners—160 subjects in total. The 80 scans consisted of 40 healthy control subjects and 40 multiple sclerosis (MS) subjects. While there was some overlap in subjects scanned on both scanners, many were separate. We compared the results using N3O with those on the original data, using only the intensity normalization strategy described in Section 2.4, and an attenuation correction method (Girard et al., 2011). While the work of Girard et al. (2011) does not explicitly correct for inhomogeneity, it does aim to standardize the intensities across scans by estimating the attenuation coefficient of each pixel as part of a multiplicative model.
3.5. Registration and Segmentation
As a final experiment, we assess the performance of segmenting OCT data by label transfer using image registration. Label transfer segmentation, sometimes called atlas-based segmentation, produces a segmentation of an unlabeled image by registration to an image that has a manual segmentation. After registration, the segmentation of the labeled data is transferred through the registration to the unlabeled subject. Note that this type of segmentation methodology is applicable to volumetric, intensity-based registration methods rather than surface-based methods, which requires a segmentation as a part of the registration process.
Looking at retinal OCT data, atlas-based segmentation has only recently began to be used for finding the retinal layers (Zheng et al., 2013; Chen et al., 2013, 2014), showing inferior accuracy to current state-of-the-art segmentation methods. Despite its inaccuracies image registration enables a full volumetric analysis that is not accessible using other segmentation methods. Specifically, through registration to a normalized space, like an average atlas, spatial differences within layers can be extracted and compared across populations of subjects. Such an analysis is sometimes called voxel based morphometry (VBM). Recently, Antony et al. (2016a) used VBM to show localized statistically significant differences between populations of healthy controls and patients with multiple sclerosis. Considering its success and popularity in other imaging modalities like CT and MRI Isgum et al. (2009); Cabezas et al. (2011), registration will likely become a more common technique for analyzing retinal OCT data in the future.
To look at the effect of N3O on registration, we use a deformable registration method developed by Chen et al. (2013, 2014), which, after an affine alignment step to align the outer retinal boundaries, uses one-dimensional radial basis functions to model deformations along each A-scan. Since a full registration is carried out separately on each A-scan, a regularization term is used to ensure that the resulting deformations are smoothly varying between A-scans. Further details of the full registration algorithm can be found in Chen et al. (2013, 2014).
One additional change was made to the presented method where we included an additional regularization term to the energy minimization problem to add smoothness of the deformation within each A-scan. Specifically, given the deformation field dm(x) of an A-scan indexed by m, we encourage the smoothness of the deformation by penalizing the gradient magnitude ||∇dm(x)||2. While the registration was shown to work well using the original formulation, this added term helps to improve the registration in areas where the intensity is uniform (e.g. within the layers).
To evaluate the results of the label transfer segmentation on the OCT data, we registered a set of 5 randomly chosen subjects to each of a separate set of 10 subjects, with the subjects chosen from the same cohort of data used in Sec. 3.4. Since the registration algorithm has not yet been validated using Cirrus data, this experiment was restricted to only Spectralis data. Evaluation was done by looking at the average unsigned boundary error between the registered data and the ground truth segmentations produced over all 50 registrations.
4. Results
In Table 1, we show the results of running N3 and N3O on the synthetic OCT data. We see that N3O performs better than N3 in every case with statistical significance shown for every comparison (p < 10−10 using paired two-tailed t-tests). Looking within specific experiments, recovery of the whole scan field had a smaller error than recovery of the field restricted to the RPE for N3O. This is because the restricted RPE field had an abrupt change in the gain field which could not be recovered as accurately due to the gain field smoothing step of N3. We also see similar performance for recovery of the second type of gain field as compared to the first type. While the second type appeared to be locally more difficult where the gain field changes in smaller regions, looking globally averaged away these differences.
Table 1:
Mean RMS error in the recovered gain field. Results for each field type and method are averaged over 100 trials. All results show significant improvement using N3O over N3 (p < 10−10 using a paired two-tailed t-test).
| Whole Scan | RPE | ||||
|---|---|---|---|---|---|
| Type 1 | Type 2 | Type 1 | Type 2 | Mean | |
| Spectralis | |||||
| N3 | 0.095 | 0.076 | 0.047 | 0.049 | 0.067 |
| N3O | 0.015 | 0.014 | 0.036 | 0.042 | 0.027 |
| Cirrus | |||||
| N3 | 0.094 | 0.078 | 0.046 | 0.048 | 0.067 |
| N3O | 0.010 | 0.009 | 0.030 | 0.039 | 0.022 |
We observe from Table 1, that the gain field is more accurately recovered on synthetic Cirrus data as opposed to Spectralis data. This, on the surface, is counterintuitive as Cirrus data is noisier than Spectralis data. We believe that this result is due to the Spectralis and Cirrus data having different histogram profiles as well as different contrast between each of the layers. Since N3 acts on the histogram of the data, differences between the histograms from these two scanners produce different results. When the Spectralis noise model was applied to simulated Cirrus data, and vice versa, we see the expected result with the smaller Spectralis noise performing better than with the Cirrus noise.
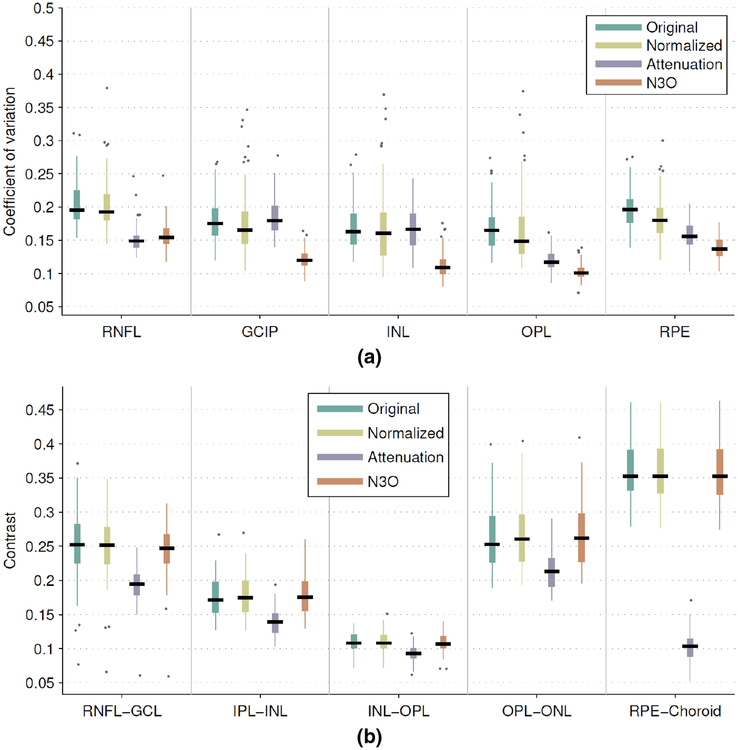
Figure 8(a) shows the result of computing both the layer intensities on the original data and the data after intensity normalization, after attenuation correction, and after running N3O. The presented results are only shown for the Spectralis data on a restricted set of layers, with results for all of the layers, and for the Cirrus data provided in the Supplementary Material. Overall, we see that N3O has significantly better CV values than the first two methods for all layers (p < 10−9), and significantly better than the attenuation correction for all layers except the RNFL, OPL, and ISOS layers (p < 10−6, see Supp. Fig. S1 for ISOS result), where the attenuation correction was significantly better (p < 0.01). The Cirrus data showed similar results, with added significance over attenuation correction for all layers (p < 0.01). Interestingly, the normalization only result on the Cirrus had worse CV values than in the original data (see Supp. Fig. S1), which is likely due to noise in the data affecting the histogram peak estimation. Since the histogram peaks are sharpened after running N3, N3O did not have this problem. Thus, intensity normalization by peak finding is not a recommended strategy for uncorrected OCT data.
Figure 8:
Box and whisker plots of (a) the coefficient of variation of the intensities within a select set of layers and (b) the contrast between successive layers. For each box, the median value is indicated by the central black line with the width of the boxes extending to the 25th and 75th percentiles, the whiskers extending to 1.5 times the interquartile range, and single points representing outliers beyond the whiskers.
Figure 8(b) also shows that the contrast of the data is not negatively affected by running N3O. While some layers showed significant differences when compared to the original data, the contrast values were consistent with each other to within a median value of 0.011 for every layer. We also see that while attenuation correction helps to remove inhomogeneity in the data by normalizing to attenuation values (thus improving the CV), some layers end up with less contrast, for instance, the RPE to choroid interface.
In Fig. 9, we show an example result on a Spectralis and a Cirrus image. Shown are the input images, the N3O result images, and the estimated gain fields. Note that the result image is the input image divided by the gain field as a result of the multiplicative model used by both N3 and N3O. To emphasize the gain field correction in the region where the data is converted to MFS, we have masked out the background in the gain field images.
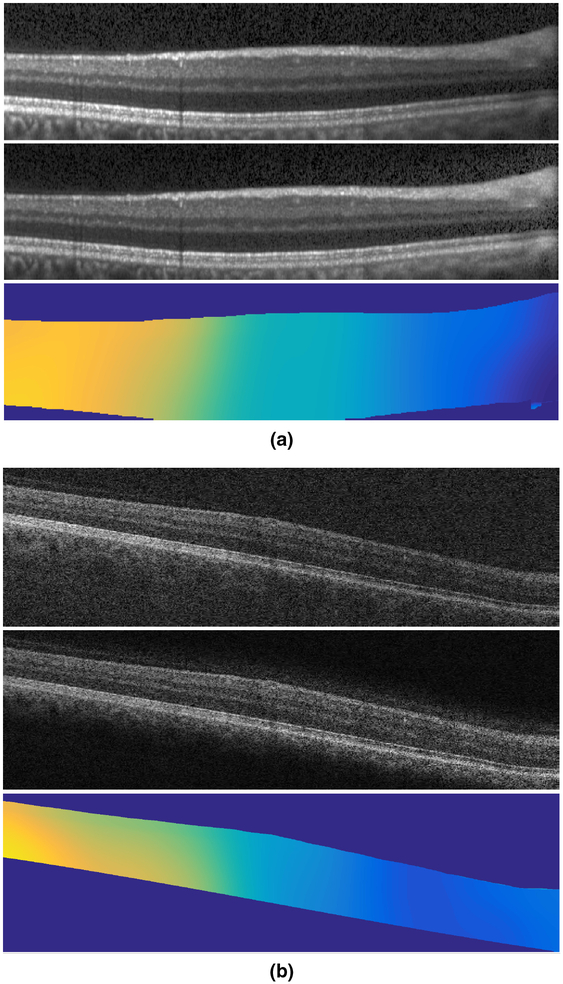
Figure 9:
Example results from (a) a Spectralis scanner and (b) a Cirrus scanner. For each example, the input image is shown on top, the N3O result in the middle, and the estimated gain field on the bottom. Darker colors in the gain field images indicate smaller gain values.
Finally, we show the results of the label transfer segmentation experiment in Table 2. Here, we see that N3O improves the performance of image registration of OCT data on average when compared to using each of the other three methods, with significantly better results for six of the nine boundaries at the p < 0.001 level. Similarly, we see that running N3 without conversion to flatspace is more accurate than running intensity normalization alone, which is in turn better than no normalization of the data at all.
Table 2:
Mean absolute errors (μm) in boundary positions between the ground truth segmentation and the segmentation results after registration. Standard deviation across subjects are in parentheses. Significance testing done using a paired two-tailed t-test.
| ILM | RNFL-GCL | IPL-INL | |
| Original | 3.90 (±0.68) | 9.84 (±2.83) | 11.95 (±5.07) |
| Norm. | 3.72 (±0.46) | 9.09 (±2.52) | 9.86 (±2.98) |
| N3 | 3.71 (±0.54) | 8.29 (±2.56)*† | 7.77 (±3.02)*† |
| N3O | 3.65 (±0.51) | 7.78 (±2.51)*†‡ | 7.22 (±3.30)*† |
| INL-OPL | OPL-ONL | ELM | |
| Original | 10.77 (±4.49) | 9.11 (±4.09) | 5.86 (±1.47) |
| Norm. | 8.58 (±2.02)* | 7.18 (±1.62) | 5.32 (±1.12)* |
| N3 | 7.11 (±1.69)*† | 6.18 (±1.23)*† | 5.00 (±1.07)*† |
| N3O | 6.63 (±1.73)*†‡ | 6.00 (±1.21)*† | 4.84 (±1.00)*†‡ |
| IS-OS | OS-RPE | BrM | |
| Original | 4.29 (±1.43) | 6.37 (±1.53) | 4.94 (±2.04) |
| Norm. | 3.87 (±1.34)* | 6.24 (±1.59) | 4.13 (±1.16)* |
| N3 | 3.65 (±1.60)* | 6.07 (±1.63)* | 2.98 (±0.51)*† |
| N3O | 3.48 (±1.47)*†‡ | 5.91 (±1.53)*†‡ | 2.83 (±0.53)*†‡ |
Significant at the p < 0.001 level compared to the Original.
Significant at the p < 0.001 level compared to Norm.
Significant at the p < 0.001 level compared to N3.
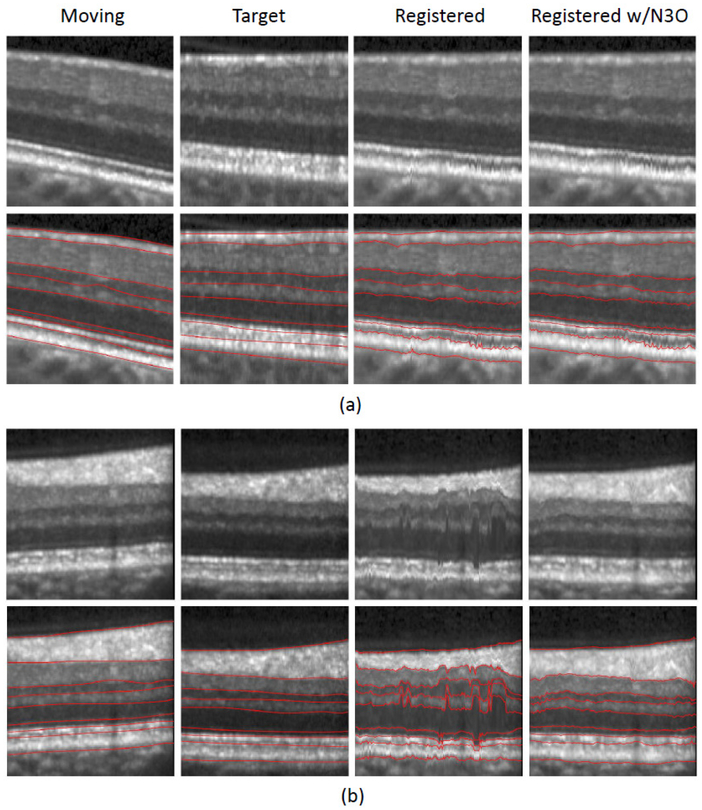
Figure 10 shows an enlarged portion of two sample registration results, displaying both the subject and target as well as the results of the registration using the original intensity image and the N3O corrected image. We see that when the intensity values between the moving and target images are close, the unprocessed result performs as well as the N3O result, however, N3O performs much better when the moving and target images have different intensity ranges. Note that while N3O improves the registration result, there is still room for improvement, as indicated by some of the larger values in Table 2. This problem is mainly due to the variability inherent in the registration. The algorithm parameters were not tuned to optimize the performance of the segmentation, and better results would likely be possible if they were.
Figure 10:
Cropped registration results of two separate examples are shown in (a) and (b). Shown are the moving image, the target image, and the registration results both without intensity correction and after running N3O on each image prior to registration. Overlaid segmentation boundaries are shown in the second row of (a) and (b). Manual segmentations are shown for the moving and target images, while the manual labels from the moving image are overlaid after registration on the respective registration images.
5. Discussion and Conclusion
We have proposed a method for inhomogeneity correction and intensity normalization of macular OCT data. While the previously developed N3 method is used for correction, it required adaptation for OCT by converting the data to a macular flat space before running it. In addition to the use of MFS, we improved the performance of N3 by modifying the model used to smooth the gain field at every iteration. This modification produces similar results to the original algorithm with improved efficiency.
The importance of converting the data to MFS before running N3 was shown in our first and third experiments. Specifically, conversion to flatspace prior to running N3 produced more accurate recovery of gain fields and also enabled a more accurate registration of intensity corrected data. We believe there are several reasons for these results. First, MFS allows the estimated gain field to vary smoothly within layers thereby improving the consistency of the resulting intensities in each layer. Second, it also allows us to initialize the gain field in a meaningful way by creating a template image to estimate the initial field. By initializing the algorithm closer to the solution, the N3 iterations are better able to converge to an accurate solution. Note however that there is some robustness built into the original N3 algorithm, since it was originally developed for brain MR data, which can have a lot of variability. This was shown in Table 2 where the N3 result outperformed our previously developed intensity normalization method when looking at image registration.
Experiments showed that N3O can accurately recover gain fields applied to synthetic data. While the process used to create the synthetic data did not follow physical principles of how OCT is generated, realistic images were created with the performance of the algorithm further validated by looking at the intensity consistency as well as the results of registration. Since simulation of OCT was not a research aim of this work, we believe the model used in our experiments provides sufficient evidence for the performance of N3O. In the future, validation based on imaging a phantom could be used. However, questions about how realistic the phantom is with respect to the shape, texture, and number of layers will likely still arise.
Additionally, the assumption that the gain field is a multiplicative model may not be appropriate, but the resulting images showed improved consistency according to our initial goal of reducing the variability of intensities within each layer. There is also evidence to show that a multiplicative model might be correct in some sense since the attenuation correction of OCT data as modeled in Vermeer et al. (2014) and Girard et al. (2011) is multiplicative. It is prudent to point out that our model is multiplicative within the processed image intensities—log transformed—as is typically available from the scanner, whereas the attenuation correction models are multiplicative on the unprocessed image intensities—prior to being log transformed. We note that the changes in contrast in attenuation correction methods may be due to the depth based correction of the intensities. Attenuation correction methods depend on the integrated intensity along an A-scan to correct the data. This means that for adjacent layers with large intensity differences (like those in the RPE and choroid), the correction factor can vary significantly between layers. In some cases, such as the RPE and choroid interface, the correction brings the intensities of the two layers closer together, thus reducing their contrast.
To further validate our method based on the performance of automated methods, we used the application of image registration, which depends entirely on the intensity of the data. While alternative cost functions, like mutual information and cross correlation, may be less sensitive to intensity variations, the method we used was validated using the sum of square differences measure and we wanted to maintain consistency in our comparison to previous work (Chen et al., 2013, 2014).
When initially undertaking this research, we hypothesized that N3O would improve the accuracy of our retinal layer segmentation algorithm (Lang et al., 2013). However, our experiments (not described here) have shown no significant differences in the results. Since this algorithm uses a classifier to learn boundary positions based on a variety of features, including both intensity and spatially varying contextual features, there is a learned robustness to intensity differences. We believe that this accounts for the lack of statistical significance. Other work has shown that intensity normalization is important for generating consistent results using other segmentation algorithms Chen et al. (2015), and therefore, we believe N3O would be an important pre-processing step for such methods. Though some segmentation methods (Novosel et al., 2017) have been designed specifically to only assume that layers are locally homogeneous and may not benefit from N3O based correction.
One limitation to running N3O is that it is only applicable to data without severe pathologies like macular edema, macular degeneration, and retinal detachments. There are several reasons why the method may not perform well on such data. First, the outer boundaries of the retina must be very accurately detected in the first step of the algorithm. The regression model relies on these boundaries to estimate the position of each layer. Any disruption will mean that the layers will no longer be flat after transformation to MFS. Since we showed that N3 works better when the layers are flat, this will have an adverse effect on the intensity correction. Second, the transformation to MFS uses a regression model that was trained on healthy data, meaning that when applying the method to new data, the shape of the retina should have a similar shape/structure to what a healthy retina looks like for the boundaries to be accurately estimated. Clinical data that maintains a relatively normal shape, like in patients with multiple sclerosis and glaucoma, where macular thinning in the inner retina is the predominant pathology, should work with N3O provided the thinning is not too severe.
To adapt N3O to work for a wider range of pathologies, both the outer retinal boundary estimation step and the regression model for estimating the inner boundaries would need to be refined. For the regression model, training data containing pathological cases can be included. Additionally, a non-linear regression model could be used to better capture the retinal shape variability. The presence of fluid in 675 the retina presents another difficult challenge, since the fluid can severely alter the position of the boundaries. One possibility for handling these cases is to segment the fluid and exclude those regions in the regression model. This method, however, would rely on the layers being relatively normal around the fluid.
In the future, we hope to extend the algorithm to work fully on 3D data, instead of running independently on 2D B-scan images. While we showed good performance when running the algorithm in 2D, we expect improvements in the consistency of the results between adjacent images to improve after incorporating an added 3D component to our model The difficulty of implementing this idea is that the gain field will need two components, one varying in 2D within each image independently, and one varying smoothly between images in 3D, since we see both types of problems in OCT data, depending on the source of the inhomogeneity. Additionally, our approach drusen. Given that there are existing methodologies for providing segmentation of the pathological regions. We believe it is feasible that N3O could be used in an iterative framework to help reduce inhomogeneity, which in turn turn can provide an improved segmentation. With this process iterating until the segmentations stabilize.
Finally, we note the computational performance of our algorithm. The method takes on average 0.06 seconds per B-scan image for the Spectralis data and 0.12 seconds for the Cirrus data, with code written in MATLAB (MathWorks, Natick, MA, USA). Approximately 40% of this time is spent on converting the data to and from MFS (e.g. interpolation), with the remaining time spent on the N3 inhomogeneity correction and intensity normalization. Performance was measured on a 1.73 GHz quad core computer running Windows 7 and ultimately can be improved by both conversion to a low level programming language and through parallelization for each B-scan. The code for N3O may be downloaded from http://www.nitrc.org/projects/aura_tools/.
Supplementary Material
Highlights.
Optical coherence tomography (OCT) intensity inhomogeneity correction based on image characteristics, which outperforms the physics based models.
Introduction of macular flat space as a unique computational domain for processing retinal images.
Improved the performance of the N3 inhomogeneity correction software, which is critically important in neuroimaging.
A framework for applying synthetic gainfields to OCT images for validating inhomogeneity correction.
First OCT demonstration of improved registration performance after application of inhomogeneity correction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
To estimate the ILM and BrM, we use a previously described method (Lang et al., 2013) where, in brief, the boundaries are found by looking for the largest positive and negative gradients in each column of the data with some added outlier removal and smoothing.
All of our training data had the same size. For testing data acquired with a different number of A- or B-scans, we can resize the coefficient maps accordingly.
We define the transformation from MFS to native space since the mapping of the volume into MFS uses a pull-back transformation at each pixel defined in the opposite direction. However, since the mapping is invertible due to its monotonicity, we can compute the transformation in both directions.
Run times come from comparing our implementation of N3 incorporating Eilers et al. (2006) in MATLAB with the implementation of Sled et al. (1998) available at https://github.com/BIC-MNI/N3.
While speckle can be viewed as a tissue property affected by inhomogeneity, the noise in real images appears amplified after inhomogeneity correction, and thus, this model may be appropriate
References
- Antony BJ, Chen M, Carass A, Jedynak BM, Al-Louzi O, Solomon SD, Saidha S, Calabresi PA, Prince JL, 2016a. Voxel based morphometry in optical coherence tomography: Validation & core findings. In: Proc. SPIE Med. Imag p. 97880P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony BJ, Lang A, Swingle EK, Al-Louzi O, Carass A, Solomon SD, Calabresi PA, Saidha S, Prince JL, 2016b. Simultaneous Segmentation of Retinal Surfaces and Microcystic Macular Edema in SDOCT Volumes. Proceedings of SPIE Medical Imaging (SPIE-MI 2016), San Diego, CA, February 27-March 3, 2016 9784, 97841C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo JF (Ed.), 2009. Retinal Angiography and Optical Coherence Tomography. Springer-Verlag; New York. [Google Scholar]
- Arnold JB, Liow J-S, Schaper KA, Stern JJ, Sled JG, Shattuck DW, Worth AJ, Cohen MS, Leahy RM, Mazziotta JC, Rottenberg DA, 2001. Qualitative and quantitative evaluation of six algorithms for correcting intensity nonuniformity effects. NeuroImage 13 (5), 931–943. [DOI] [PubMed] [Google Scholar]
- Cabezas M, Oliver A, Lladó X, Freixenet J, Cuadra MB, 2011. A review of atlas-based segmentation for magnetic resonance brain images. Comput. Methods Programs Biomed 104 (3), e158–e177. [DOI] [PubMed] [Google Scholar]
- Carass A, Lang A, Hauser M, Calabresi PA, Ying HS, Prince JL, 2014. Multiple-object geometric deformable model for segmentation of macular OCT. Biomed. Opt. Express 5 (4), 1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-L, Ishikawa H, Wollstein G, Bilonick RA, Sigal IA, Kagemann L, Schuman JS, 2015. Histogram matching extends acceptable signal strength range on optical coherence tomography images. Invest. Ophthalmol. Vis. Sci 56 (6), 3810–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lang A, Sotirchos E, Ying HS, Calabresi PA, Prince JL, Carass A, 2013. Deformable registration of macular OCT using A-mode scan similarity In: 10th International Symposium on Biomedical Imaging (ISBI 2013). pp. 476–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lang A, Ying HS, Calabresi PA, Prince JL, Carass A, 2014. Analysis of macular OCT images using deformable registration. Biomed. Opt. Express 5 (7), 2196–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niemeijer M, Zhang L, Lee K, Abramoff MD, Sonka M, 2012. Three-dimensional segmentation of fluid-associated abnormalities in retinal OCT: Probability constrained graph-search-graph-cut. IEEE Trans. Med. Imag 31 (8), 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S, 2010. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt. Express 18 (18), 19413–19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua ZY, Zheng W, Chee MWL, Zagorodnov V, 2009. Evaluation of performance metrics for bias field correction in MR brain images. J. Magn. Reson. Imaging 29 (6), 1271–1279. [DOI] [PubMed] [Google Scholar]
- Drexler W, Fujimoto JG (Eds.), 2008. Introduction to Optical Coherence Tomography. Springer Berlin Heidelberg. [Google Scholar]
- Eilers PHC, Currie ID, Durbán M, 2006. Fast and compact smoothing on large multidimensional grids. Comput. Stat. Data Anal 50 (1), 61–76. [Google Scholar]
- Fritsch FN, Carlson RE, 1980. Monotone piecewise cubic interpolation. SIAM J. Numer. Anal 17, 238–246. [Google Scholar]
- Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M, 2009. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans. Med. Imag 28 (9), 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbel I, Rossant F, Bloch I, Tick S, Paques M, 2011. Automated segmentation of macular layers in OCT images and quantitative evaluation of performances. Pattern Recogn. 44 (8), 1590–1603. [Google Scholar]
- Girard MJA, Strouthidis NG, Ethier CR, Mari JM, 2011. Shadow removal and contrast enhancement in optical coherence tomography images of the human optic nerve head. Invest. Ophthalmol. Vis. Sci 52 (10), 7738–7748. [DOI] [PubMed] [Google Scholar]
- Hee MR, Izatt JA, Swanson EA, Huang D, Schuman JS, Lin CP, Puliafito CA, Fujimoto JG, 1995. Optical coherence tomography of the human retina. Arch. Ophthalmol 113 (3), 325–332. [DOI] [PubMed] [Google Scholar]
- Hou Z, 2006. A review on MR image intensity inhomogeneity correction. Int. J. Biomed. Imaging 2006, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgum I, Staring M, Rutten A, Prokop M, Viergever MA, van Ginneken B, 2009. Multi-atlas-based segmentation with local decision fusion — application to cardiac and aortic segmentation in CT scans. IEEE Trans. Med. Imag 28 (7), 1000–1010. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Stein DM, Wollstein G, Beaton S, Fujimoto JG, Schuman JS, 2005. Macular segmentation with optical coherence tomography. Invest. Ophthalmol. Vis. Sci 46 (6), 2012–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaba D, Wang Y, Wang C, Liu X, Zhu H, Salazar-Gonzalez AG, Li Y, 2015. Retina layer segmentation using kernel graph cuts and continuous max-flow. Opt. Express 23 (6), 7366–7384. [DOI] [PubMed] [Google Scholar]
- Kraus MF, Liu JJ, Schottenhamml J, Chen C-L, Budai A, Branchini L, Ko T, Ishikawa H, Wollstein G, Schuman J, Duker JS, Fujimoto JG, Hornegger J, 2014. Quantitative 3D-OCT motion correction with tilt and illumination correction, robust similarity measure and regularization. Biomed. Opt. Express 5 (8), 2591–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Carass A, Al-Louzi O, Bhargava P, Ying HS, Calabresi PA, Prince JL, 2015a. Longitudinal graph-based segmentation of macular OCT using fundus alignment. Proceedings of SPIE Medical Imaging (SPIE-MI 2015), Orlando, FL, February 21–26, 2015 9413, 94130M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Carass A, Bittner AK, Ying HS, Prince JL, 2017. Improving graph-based OCT segmentation for severe pathology in Retinitis Pigmentosa patients. Proceedings of SPIE Medical Imaging (SPIE-MI 2017), Orlando, FL, February 11 – 16, 2017 10137, 101371M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Carass A, Calabresi PA, Ying HS, Prince JL, 2014. An adaptive grid for graph-based segmentation in retinal OCT In: Proc. SPIE Med. Imag p. 903402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Carass A, Hauser M, Sotirchos ES, Calabresi PA, Ying HS, Prince JL, 2013. Retinal layer segmentation of macular OCT images using boundary classification. Biomed. Opt. Express 4 (7), 1133–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Carass A, Jedynak B, Solomon SD, Calabresi PA, Prince JL, 2016. Intensity inhomogeneity correction of macular OCT using N3 and retinal flatspace In: 12th International Symposium on Biomedical Imaging (ISBI 2015). pp. 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Carass A, Swingle EK, Al-Louzi O, Bhargava P, Saidha S, Ying HS, Calabresi PA, Prince JL, 2015b. Automatic segmentation of microcystic macular edema in OCT. Biomed. Opt. Express 6 (1), 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan BJ, Roorda A, Knighton RW, Carroll J, 2011. Revealing Henle’s fiber layer using spectral domain optical coherence tomography. Invest. Ophthalmol. Vis. Sci 52 (3), 1486–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros FA, Zangwill LM, Alencar LM, Bowd C, Sample PA, Susanna R Jr, Weinreb RN, 2009. Detection of glaucoma progression with stratus OCT retinal nerve fiber layer, optic nerve head, and macular thickness measurements. Invest. Ophthalmol. Vis. Sci 50 (12), 5741–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeijer M, Lee K, Garvin MK, Abràmoff MD, Sonka M, 2012. Registration of 3D spectral OCT volumes combining ICP with a graph-based approach. In: Proc. SPIE Med. Imag p. 83141A. [Google Scholar]
- Novosel J, Thepass G, Lemij HG, de Boer JF, Vermeer KA, van Vliet LJ, 2015. Loosely coupled level sets for simultaneous 3D retinal layer segmentation in optical coherence tomography. Med. Image Anal 26 (1), 146–158. [DOI] [PubMed] [Google Scholar]
- Novosel J, Vermeer KA, de Jong JH, Wang Z, van Vliet LJ, 2017. Joint Segmentation of Retinal Layers and Focal Lesions in 3-D OCT Data of Topologically Disrupted Retinas. IEEE Trans. Med. Imag 36 (6), 1276–1286. [DOI] [PubMed] [Google Scholar]
- Roy S, Carass A, Bazin P-L, Prince JL, 2011. Intensity Inhomogeneity Correction of Magnetic Resonance Images using Patches. In: Proceedings of SPIE Medical Imaging (SPIE-MI 2011), Orlando, FL, February 12–17, 2011. Vol. 7962 pp. 79621F–79621F–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidha S, Syc SB, Ibrahim MA, Eckstein C, Warner CV, Farrell SK, Oakley JD, Durbin MK, Meyer SA, Balcer LJ, Frohman EM, Rosenzweig JM, Newsome SD, Ratchford JN, Nguyen QD, Calabresi PA, 2011. Primary retinal pathology in multiple sclerosis as detected by optical coherence tomography. Brain 134 (2), 518–533. [DOI] [PubMed] [Google Scholar]
- Serranho P, Maduro C, Santos T, Cunha-Vaz J, Bernardes R, 2011. Synthetic OCT data for image processing performance testing. In: Proc. 18th IEEE Int. Conf. Image Process pp. 409–412. [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC, 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imag 17 (1), 87–97. [DOI] [PubMed] [Google Scholar]
- Swingle EK, Lang A, Carass A, Al-Louzi O, Prince JL, Calabresi PA, 2015. Segmentation of microcystic macular edema in macular Cirrus data. Proceedings of SPIE Medical Imaging (SPIE-MI 2015), Orlando, FL, February 21–26, 2015 9417, 94170P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle EK, Lang A, Carass A, Calabresi PA, Ying HS, Prince JL, 2014. Microcystic macular edema detection in retina OCT images. Proceedings of SPIE Medical Imaging (SPIE-MI 2014), San Diego, CA, February 15–20, 2014 9038, 90380O. [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imag 29 (6), 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga BE, Gao W, Laurik KL, Tátrai E, Simó M, Somfai GM, Cabrera DeBuc D, 2015. Investigating tissue optical properties and texture descriptors of the retina in patients with multiple sclerosis. PLoS ONE 10 (11), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer KA, Mo J, Weda JJA, Lemij HG, de Boer JF, 2014. Depthresolved model-based reconstruction of attenuation coefficients in optical coherence tomography. Biomed. Opt. Express 5 (1), 322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer KA, van der Schoot J, Lemij HG, de Boer JF, 2011. Automated segmentation by pixel classification of retinal layers in ophthalmic OCT images. Biomed. Opt. Express 2 (6), 1743–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer KA, van der Schoot J, Lemij HG, de Boer JF, 2012. RPE-normalized RNFL attenuation coefficient maps derived from volumetric OCT imaging for glaucoma assessment. Invest. Ophthalmol. Vis. Sci 53 (10), 6102–6108. [DOI] [PubMed] [Google Scholar]
- Wilkins GR, Houghton OM, Oldenburg AL, 2012. Automated segmentation of intraretinal cystoid fluid in optical coherence tomography. IEEE Trans. Biomed. Eng 59 (4), 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Gerendas BS, Waldstein SM, Langs G, Simader C, Schmidt-Erfurth U, 2014. Stable registration of pathological 3D SD-OCT scans using retinal vessels In: 16th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2014) the 1st Int. Workshop on Ophthalmic Medical Image Analysis. Boston, MA, pp. 1–8. [Google Scholar]
- Zheng Y, Xiao R, Wang Y, Gee JC, 2013. A generative model for OCT retinal layer segmentation by integrating graph-based multi-surface searching and image registration In: 16th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2013). Vol. 8149 of Lecture Notes in Computer Science. Springer; Berlin Heidelberg, pp. 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.