Abstract
Background:
Mutations in the CACNA1C-encoded L-type calcium channel have been associated with Timothy syndrome (TS) with severe QT prolongation, syndactyly, facial dysmorphisms, developmental delay, and sudden death. Recently, patients hosting CACNA1C mutations with only long QT syndrome (LQTS) have been described. We sought to identify novel variants in CACNA1C associated with either TS or LQTS, and to determine the impact of the mutation on channel function.
Methods/results:
Two probands were identified with mutations in CACNA1C, one with a TS-associated mutation, G406R, and a second with genotype-negative LQTS. Illumina HiSeq 2000 whole exome sequencing on the genotype-negative LQTS proband revealed a novel variant, CACNA1C-L762F, that co-segregated within a multi-generational family. The missense mutation localized to the DII/DIII intracellular interlinker segment of the channel in a highly conserved region in close proximity to the 6th transmembrane segment of domain II (DIIS6). Whole cell patch clamp of heterologously expressed CACNA1C-L762F in TSA201 cells demonstrated slower inactivation tau and increased sustained and window current. Comprehensive review and topological mapping of all described CACNA1C mutations revealed TS-specific hotspots localizing to the cytoplasmic aspect of 6th transmembrane segment of respective domains. Probands hosting TS mutations were associated with elevated QTc, higher prevalence of 2:1 AV block, and a younger age at presentation compared to LQTS.
Conclusions:
The CACNA1C-L762F mutation is associated with development of LQTS through slower channel inactivation and increased sustained and window current. TS-associated mutations localize to specific areas of CACNA1C and are associated with a younger age at presentation, higher QTc, and 2:1 AV block than isolated LQTS-associated mutations.
Keywords: CACNA1C, Long QT syndrome, L-type calcium channel, Mutation, Timothy syndrome
1. Introduction
L-type calcium channel (LTCC, Cav1.2) is a macromolecular complex which plays a key role in cardiomyocyte calcium (Ca2+) cellular influx, the cardiocyte action potential, and excitation–contraction coupling [1]. The channel complex is comprised of three subunits including α1c, β2, and α2δ, whereby the CACNA1C-encoded α1c α-subunit (CACNA1C) is the channel pore. CACNA1C is comprised of 4 homologous domains (DI through DIV) that are connected by intracellular linker regions (I–II, II–III, and III–IV loops) and 6 transmembrane segments (S1 through S6) [2]. Mutations in CACNA1C have been associated with a number of human diseases that have cardiac manifestations.
Timothy syndrome (TS) is a multi-organ system genetic syndrome which can manifest as extreme QT prolongation, syndactyly, neurodevelopmental delay, and a significant risk of sudden cardiac death (SCD) in an autosomal dominant pattern [3,4]. Canonically associated with mutations in CACNA1C, these pathogenic defects induce gain-of-function alterations in the LTCC channel which result in impaired channel inactivation [5,6]. Recently, a handful of CACNA1C mutations have been identified in patients with QT prolongation in isolation and in the absence of syndactyly or other non-cardiac manifestations of TS. Congenital long QT syndrome (LQTS), with a prevalence as high as 1 in 2500 persons, comprises a distinct group of cardiac channelopathies characterized by delayed cardiac repolarization and increased risk for syncope, seizures, and SCD in the absence of underlying syndrome or structural heart disease [7]. To date, hundreds of mutations have been identified in 17 LQTS-susceptibility genes with approximately 75% of LQTS cases due to mutations in three genes: KCNQ1-encoded IKs potassium channel (Kv7.1, LQT type 1), KCNH2-encoded IKr potassium channel (Kv11.1, LQT2), and SCN5A-encoded INa sodium channel (NaV1.5, LQT3) [8]. These ion channels play key roles in the cardiac action potential and mutations in these channels delay repolarization leading to prolonged QT intervals in ECG. While findings of CACNA1C mutations associated with LQTS raises the possibility of a spectrum of phenotypic expression, this spectrum remains relatively unexplored. Further, recent identification of a CACNA1C-associated clinical entity that manifests with only cardiac abnormalities (QT prolongation, structural heart disease, and cardiomyopathy), without extracardiac abnormalities, so-called cardiac-only Timothy syndrome (COTS), offers additional phenotypic and clinical variation [9].
To this end, we have identified a novel CACNA1C mutation in a family with multiple generations of isolated QT prolongation and sudden death without other systemic or cardiac sequelae associated with TS. This mutation results in a gain-of function defect of the heterologously expressed LTCC with slower inactivation tau and increased sustained and window current. Comprehensive analysis of all reported CACNA1C variants demonstrated TS-associated mutations “hot spot” with a distinct clinical repolarization phenotype compared to LQTS and COTS-associated mutations.
2. Methods
2.1. Clinical evaluation and study enrollment
In this IRB-approved study, subjects presenting to the Texas Children’s Heart Center within the Division of Cardiology, Department of Pediatrics at the Baylor College of Medicine with SCD-predisposing disease were recruited for genetic testing. Subjects were evaluated by a pediatric electrophysiologist and a history, physical exam, echocardiogram, electrocardiogram, and ancillary testing were performed. QTc intervals were calculated using the Bazett formula. Following written receipt of informed consent, DNA was obtained from subjects and appropriate kindred.
2.2. Genetic analysis
Genomic DNA was isolated using Gentra Puregene Blood Kit (Qiagen, Valencia, CA)-based extraction from whole blood obtained from peripheral venipuncture. Whole exome sequencing was conducted as previously described [10]. Briefly, the Illumina HiSeq 2000 platform was coupled with a uniquely designed exome capture platform with an average of 100× per exome coverage. Exome sequences were processed via an in-house developed bioinformatic pipeline including the Burrows-Wheeler Alignment algorithm for mapping and the Genome Analysis Toolkit (Broad Institute) [11] and Atlas2 v1.4.3 r171 [12] platforms for variant calling. Variants were further annotated using an ANNOVAR-based in-house developed annotation program [13]. Candidate mutations were then confirmed by Sanger sequencing.
To confirm absence of putatively pathogenic mutations in ostensibly healthy individuals, presence of the mutation was analyzed in 1360 reference alleles (N = 680 subjects). Control genomic DNA was obtained from the European Collection of Cell Cultures (HPA Culture Collections, UK), the Human Genetic Cell Repository sponsored by the National Institute of General Medical Sciences, and the Coriell Institute for Medical Research (Camden, NJ). Further, absence of the mutation in all publically-held databases including the 1000 Genome Project (N = 1094 subjects) comprised of 381 whites, 246 blacks, 286 Asians, and 181 Hispanics [14]; the National Heart, Lung, and Blood Institute GO Exome Sequencing Project (N = 5379 subjects) comprise of 3510 whites and 1869 blacks [15]; the 12,000 Exome Chip (N = 12,000 subjects) [15]; and the ExAC database (N = 60,706) [16]. In total, putatively pathogenic variants were absent in approximately 99,006 subjects and 198,012 reference alleles.
2.3. In silico mutation pathogenicity and sequencing conservation
In silico mutation pathogenicity prediction was done using SIFT [17], PolyPhen-2 [18], and PhD-SNP [19]. In addition, PredictSNP, a compilation of multiple pathogenicity prediction tools was queried [20]. SIFT and PolyPhen-2 designation of damaging or probably damaging, PhD-SNP designation of deleterious, and PredictSNP designation of disease-associated were considered pathogenic. While respective designations of tolerated, benign, or neutral were considered benign. Sequence conservation analysis was done utilizing Multalin alignment algorithms [21]. Variants were mapped to the primary sequence of the CACNA1C [22].
2.4. CACNA1C expression vectors
The human wild-type (WT) CACNA1C cDNA with an N-terminal enhanced yellow fluorescence protein (EYFP) tag [(EYFP) Nα1c, 77] in the pcDNA vector, and the cDNA of the CACNA2D1 gene cloned in pcDNA3.1 vector was a generous gift from Dr. Antzelevitch. This included cDNA sequencing containing exon 8 A (accession No. Z34815) [23]. The cDNA of the CACNB2b gene was subcloned into the bicistronic pIRES2-dsRED2 vector (Clontech, Mountain View, CA). The L762F-CACNA1C missense mutation was engineered into pcDNA3-CACNA1C-WT–EYFP vector using the Quikchange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA).
2.5. Cell culture and transfection
TSA 201 cells were cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), 1.0% L-glutamine, and 1.2% penicillin/streptomycin solution in a 5% CO2 incubator at 37 °C. Heterologous expression of L-type Ca2+ channel, Cav1.2 was accomplished by co-transfecting 1 μg CACNA1C-EYFP WT or mutant (CACNA1C-L762F-EYFP) cDNA with 1 μg CACNB2b-pIRES2-dsRed2, 1 μg CACNA2D1-pcDNA3.1 and 0.25 μg Green Fluorescence Protein (GFP) cDNA with the use of 8 μl Lipofectamine 2000. The media were replaced with fresh OPTI-MEM after 4–6 h. Transfected TSA 201 cells were cultured in OPTI-MEM and incubated for 48 h before electrophysiological experiments.
2.6. Whole cell patch clamp
Standard whole-cell patch clamp technique was used to measure Cav1.2 CACNA1C wild type and mutant calcium currents at room temperature (22–24 °C) with the use of an Axopatch 200B amplifier, Digidata 1440A and pclamp version 10.2 software (Axon Instruments, Sunnyvale, CA). The extracellular (bath) solution contained (mmol/L): 2 CaCl2, 1 MgCl2, 150 TEA-Cl and 10 HEPES, pH adjusted to 7.35 with CsOH. The intracellular (pipette) solution contained (mmol/L): 110 CsCl, 0.1 CaCl2, 10 HEPES, 10 EGTA, 2 MgATP and 10 TEA-Cl, pH adjusted to 7.30 with CsOH as previously described [24]. Microelectrodes were pulled on a P-97 puller (Sutter Instruments, Novato, CA) and fire polished to a final resistance of 2–3 MΩ. Series resistance was compensated by 80–85%. Currents were filtered at 1 kHz and digitized at 5 kHz with an eight-pole Bessel filter. The voltage dependence of activation and inactivation was determined using voltage-clamp protocols described in the relevant figure legends. Data were analyzed using Clampfit (Axon Instruments, Sunnyvale, CA), Excel (Microsoft, Redmond, WA), and fitted with Origin 9.1 (OriginLab Corporation, Northampton, MA) software.
The voltage-dependence of activation curve was fitted with a Boltzmann function: GCa/GCa max = {1 + exp[(V − V1/2) / k]}−1, where V1/2 and k are the half-maximal voltage of activation and the slope factor respectively. The steady-state inactivation curve was fitted with a Boltzmann function: ICa/ICa max = {1 + exp[(V − V1/2) / k]}−1, where V1/2 and k are the half-maximal voltage of inactivation and the slope factor respectively. ICa decay was fitted with a two-exponential function: y = y0 + {1 − [Af exp(−t / τf)] + [As exp(−t / τs)]}, where Af and As represent the amplitudes of the fast and the slow inactivating components respectively and τf and τs represent the fast and slow time constants of inactivation respectively. Sustained Cav1.2 current was measured at the end of 500 ms long depolarization of +20 mV.
2.7. CACNA1C mutation compendium of clinically described probands
Comprehensive review of the literature identified all CACNA1C mutations associated with TS, COTS, or LQTS. Probands for whom QT interval and basic clinical demographic information was available were included. Probands with normal QT intervals who hosted variants that had no functional impact on the Cav1.2 channel on in vitro patch clamp analysis or were predicted to be benign in all in silico models were excluded. Probands with clinical findings not consistent with TS or COTS were excluded, and family members of affected probands were excluded. Clinical history was extracted and positive family history was considered positive in first-degree relatives.
2.8. Statistical analysis
For electrophysiology findings, all data points are shown as the mean value and bars represent the standard error of the mean. A Student’s t-test or Fisher’s exact test was performed to determine statistical significance between two groups as appropriate. A P value < 0.05 was considered to be significant.
3. Results
3.1. Clinical evaluation of CACNA1C-positive kindred
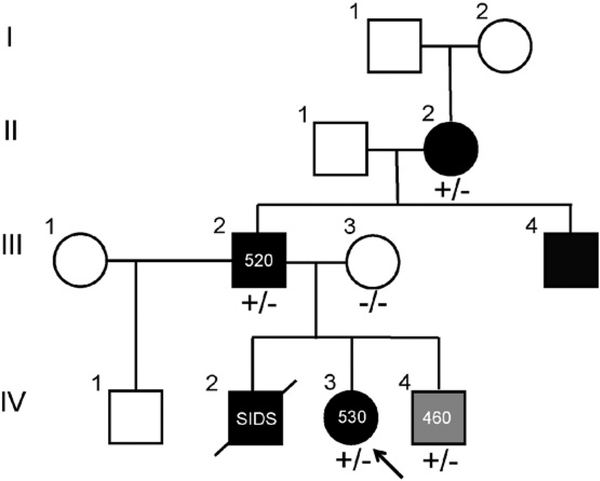
The proband identified was a Hispanic/Latino 6-year-old female who was otherwise healthy until presentation following a syncopal episode at school while sitting at a desk. She spontaneously regained consciousness. Subsequent clinical evaluation demonstrated a QTc of 530 ms and prolonged PR interval at rest in the setting of a structurally and functionally normal heart by echocardiogram (Fig. 1). She was started on beta blocker therapy, received an implantable cardioverter defibrillator, and demonstrated no defibrillator discharges or documented arrhythmia on serial evaluation. Her family history was positive for a brother who died of sudden infant death syndrome (SIDS) at 3 months of age, and a maternal miscarriage in the late first trimester of an otherwise healthy pregnancy. Subsequent clinical evaluation of the kindred revealed a 3-year-old brother with borderline QT prolongation (QTc of 460 ms), father with a QTc of 520 ms, as well as a paternal grandmother and a paternal uncle with diagnosed LQTS. The paternal uncle also carried a diagnosis of epilepsy since childhood. The proband’s mother had a normal QTc. The family’s pedigree is depicted in Fig. 2 with a summary of demographic, genotypic, and clinical findings in Table 1.
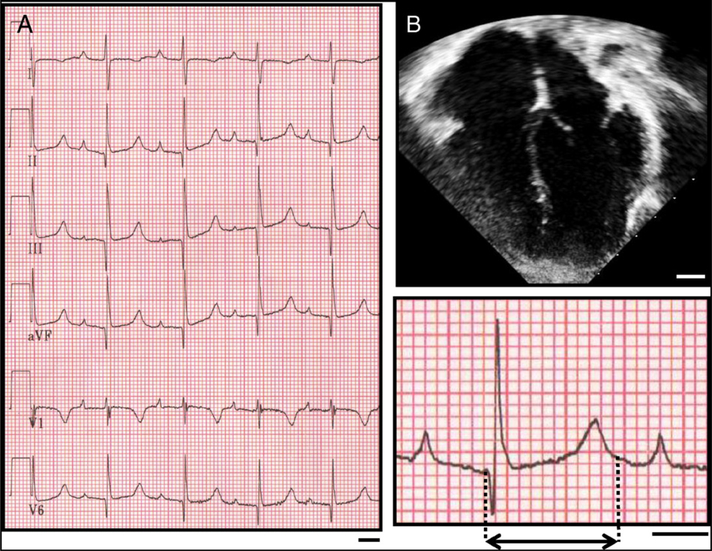
Fig. 1.
The proband hosting the CACNA1C-L762F mutation has QT prolongation and normal cardiac anatomy. A) Representative ECG obtained from proband demonstrating significantly prolonged QT interval reflected in a QTc of 530. An enhanced representative ECG tracing is shown in the inset. Dashed lines demarcate the QT interval. Bar denotes 200 ms. B) Representative apical 4-chamber echocardiographic window of the proband demonstrating normal cardiac structure. Bar denotes 1 cm.
Fig. 2.
CACNA1C-L762F co-segregates in a pedigree of LQTS. Pedigree of the CACNA1C mutation-positive family. Arrow denotes the proband (IV.3). Circles denote female and squares denote male. Black fill denotes diagnosis of LQTS. Gray fill denotes diagnosis of borderline QT prolongation. White fill denotes no cardiac diagnoses either following clinical evaluation or by reported history. +/− denotes heterozygosity for CACNA1C-L262F mutation while −/− denotes wild-type. Diagonal line denotes deceased.
Table 1.
Summary of clinical and genotype information for CACNA1C-mutation positive family.
| No. | Subject | Status | Age(y) | Sex | CACNA1C-L262F | KCNH2-K897T | SCN5A-H558R | KCNE1-G38S | Diagnosis | QTc | Clinical |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I.1 | Alive | 86 | M | NA | NA | NA | NA | None | NA | NA |
| 2 | I.2 | Alive | 86 | F | NA | NA | NA | NA | None | NA | NA |
| 3 | II.1 | Alive | 68 | M | NA | NA | NA | NA | None | NA | NA |
| 4 | II.2 | Alive | 66 | F | +/− | −/− | −/− | −/− | LQTS | NA | Diagnosed at 63 yo |
| 5 | III.1 | Alive | 44 | F | NA | NA | NA | NA | None | NA | NA |
| 6 | III.2 | Alive | 47 | M | +/− | +/− | +/− | +/− | LQTS | 520 | Diagnosed at 44 yo |
| 7 | III.3 | Alive | 41 | F | −/− | +/− | +/− | +/− | None | NA | Normal ECG and echocardiogram |
| 8 | III.4 | Alive | 49 | M | NA | NA | NA | NA | LQTS, Epilepsy | NA | Diagnosed with LQTS at 46 yo, diagnosed with epilepsy at 16 yo |
| 9 | IV.1 | Alive | 21 | M | NA | NA | NA | NA | None | NA | Normal ECG and echocardiogram |
| 10 | IV.2 | Deceased | 0.25 | M | NA | NA | NA | NA | SIDS | NA | Autopsy negative sudden infant death |
| 11 | IV.3 | Alive | 6 | F | +/− | +/− | +/+ | +/− | LQTS | 530 | Diagnosed at 3 yo |
| 12 | IV.4 | Alive | 10 | M | +/− | +/− | +/− | −/− | Borderline LQTS | 460 | Diagnosed at 7 yo |
M, male; F, female; NA, not applicable or not available; +/− heterozygous for variant; −/− homozygous wild-type.
A second, unrelated CACNA1C-mutation positive case was identified. Demonstrating classic features of TS, this proband is a Hispanic female who presented at birth following observance of fetal bradycardia and post-natal ECG demonstrating 2:1 AV block with a symptomatic ventricular rate of 60 bpm and a QTc of 655 ms. Echocardiogram demonstrated small ventricular septal defect, patent ductus arteriosus (PDA), mild right ventricular hypertrophy, moderate left ventricular hypertrophy with prominent left ventricular free wall trabeculations which did not meet criteria for non-compaction and diastolic dysfunction. She demonstrated dysmorphic craniofacial features and cutaneous syndactyly of the left hand and foot. Clinical genetic testing for CACNA1C demonstrated a heterozygous G to A substitution at nucleotide 1216 (c. G1216A), resulting in a glycine to arginine missense mutation at residue number 406 (CACNA1C-G406R). Given identification of a known TS-associated mutation, no further genetic testing was conducted.
3.2. Genetic analysis
Among the LQTS family, 5 kindred were analyzed by whole exome analysis which identified 832 variants in the proband following bioinformatic filtering. 130 variants were common to affected individuals in the family and had an allele frequency less than 2%. Following quality analysis and in silico pathogenicity analysis, a single variant was identified that co-segregated with disease in the family – a heterozygous C to T substitution at nucleotide 2284 (c. C2284T), resulting in a leucine to phenylalanine missense mutation at residue number 762 (CACNA1C-L762F, Fig. 3A). No other mutations were identified in CACNA1C. Comprehensive review of other LQTS-associated genes including KCNQ1, KCNH2, SCN5A, KCNE1, KCNE2, KCNJ2, CAV3, SCN4B, AKAP9, and SNTA1 were negative for rare variants that co-segregated with affected individuals. Three common functionally pro-arrhythmic genetic variants were identified in the proband including heterozygous KCNH2-K897T (2690A>C), homozygous SCN5A-H558R (1673A>G), and heterozygous KCNE1-G38S (112G>A) [25–27]. The paternal grandmother, who was diagnosed with LQTS late in life, was positive for the putative mutation. She did not host any of the three common functional polymorphisms. Further, the proband’s brother hosted two of the three variants, in addition to CACNA1C-L762F, and had a borderline QTc of 460 ms. In this way, these common polymorphisms did not co-segregate with incidence of disease nor did presence of the variants appear to dramatically modify QT intervals within the kindred. The paternal uncle was unable to be recruited for genetic analysis. A post-mortem autopsy tissue sample was not available for the sibling who died of SIDS to allow for genotyping. Genotypes are summarized in Table 1.
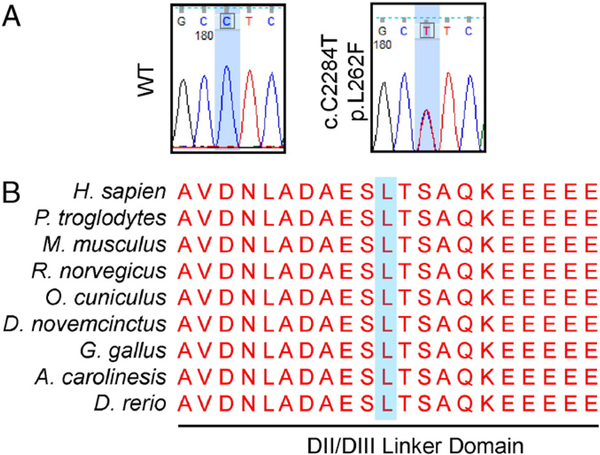
Fig. 3.
L762F localizes to a highly conserved interlinker segment of the CACNA1C. A) Representative sequencing chromatograms for wild-type genotype and the CACNA1C-L262F heterozygous genotype. B) Primary sequence alignment from multiple divergent species. The mutation localizes to the DII/DIII interlinker segment of the CACNA1C-encoded α1c α-subunit of the LTCC which is completely conserved across species. The mutated residue is denoted with a blue background.
The CACNA1C mutation localized to the DII/DIII intracellular linker segment of the channel pore within close proximity to the 6th transmembrane segment of domain II (DIIS6), 8 residues distal to the C-terminal aspect of the transmembrane segment. This residue and the surrounding region is conserved completely across all species queried (Fig. 3B). Further, this mutation was predicted to be highly damaging/pathogenic by SIFT, PolyPhen-2, PhD-SNP, and PredictSNP in silico modeling. The divergent clinical phenotype of these two CACNA1C mutations prompted additional exploration into TS- and LQTS-associated mutations.
3.3. Patch clamp analysis of CCNA1C-L762F
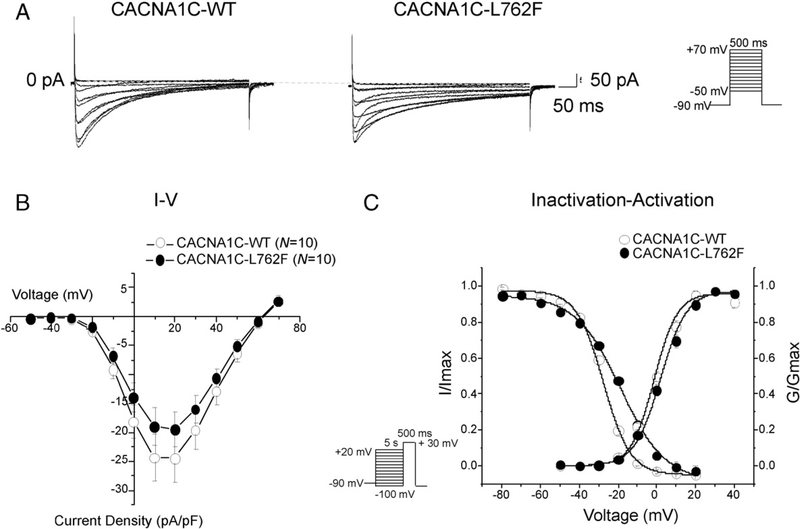
To evaluate the effect of CACNA1C-L762F on the Cav1.2 channel, we utilized whole cell patch clamp to determine whether there are electro-physiological differences between the mutant and wild-type Cav1.2 (WT) channels in transfected TSA201 cells. Typical Cav1.2 tracings of voltage-dependent activation were obtained from CACNA1C-WT and CACNA1C-L762F transfected cells with a holding potential at −90 mV. Analysis of the current–voltage relationship demonstrated no significant difference in current density in WT compared to CACNA1C-L762F (Fig. 4A and B). Analysis of channel activation curves demonstrated no significant difference in the V1/2 of activation between WT with −1.3 ± 1.4 mV (N = 10) and CACNA1C-L762F with 2.1 ± 1.2 mV (N = 10). The slope factor (k) remained unchanged as well with WT of 6.0 ± 0.6 (N = 10) and CACNA1C-L762F of 6.8 ± 0.6 (N = 10, Fig. 4C). Steady-state inactivation was assessed by a standard 2 pulse voltage-clamp protocol and inactivation curves were plotted. The CACNA1C-L762F mutation was associated with a V1/2 of inactivation shift toward more depolarized potential. Specifically, mutant cells demonstrated a V1/2 of inactivation of −18.7 ± 0.9 (N = 8) compared to −27.5 ± 0.8 mV among WT cells (N = 10) which was a shift of +8.8 mV (P < 0.05). Further, the slope factor demonstrated a significant difference with the WT of 6.9 ± 0.6 (N = 10) and the mutant of 11.6 ± 0.8 (N = 8, P < 0.05). When voltage-dependent activation and steady-state inactivation were plotted together, a large increase in the window current can be observed (Fig. 4C).
Fig. 4.
CACNA1C-L762F increases Cav1.2 window current in heterologous TSA 201 cells. (A) Whole cell Cav1.2 current representative tracings from TSA201 cells expressing CACNA1C-WT or L762F determined from a holding potential of −90 mV to testing potential of +70 mV in 10 mV increments with 500 ms duration. (B) Current–voltage relationship for WT (N = 10) and L762F (N = 10). All values represent mean ± SEM. (C) Inactivation curves of WT (N = 10) and L762F (N = 8), determined from a holding potential of −90 mV to pre-pulse of 20 mV in 10 mV increments with 5 s duration followed by a test pulse of 30 mV with 500 ms duration. I/Imax represent normalized calcium current fitted with a Boltzmann function. Activation curves of WT (N = 10) and CACNA1C-L762F (N = 10). G/Gmax represents normalized conductance fitted with a Boltzmann function.
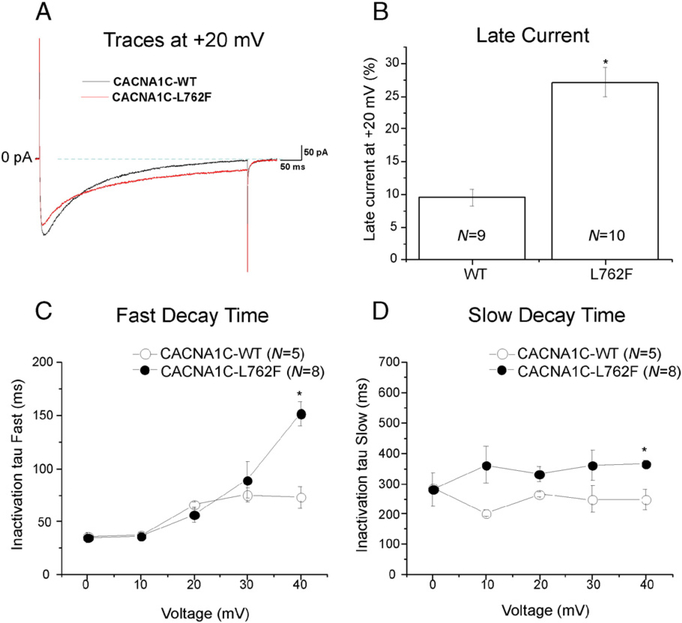
Next, we examined sustained current of the L-type calcium channel between WT and mutant cells which was normalized to the peak current at +20 mV and compared as the percent of each group. The CACNA1C-L762F mutation conferred an increased sustained current by 1.9 fold from 9.5% ± 1.3 in the WT (N = 9) to 27.1% ± 2.2 in the mutant (N = 10, P < 0.05). These results are summarized in Fig. 5A and B. We then examined channel inactivation tau (τ) of the L-type calcium channel by measuring Cav1.2 current decay after 90% of peak and identifying a best fit model with two exponentials with two τ values representing fast and slow inactivation. At +40 mV, CACNA1C-L762F revealed a slower inactivation τ in the fast and slow component of the decay time compared to WT (P < 0.05, Fig. 5C and D).
Fig. 5.
CACNA1C-L762F missense mutation increased Cav1.2 sustained current and inactivation tau (τ). A) Representative tracings of sustained (late) current representing CACNA1C-WT and CACNA1C-L762F measured at the end of 500 ms long depolarization of +20 mV. (B) Normalized sustained current to peak current at +20 mV shown as percentages in a bar graph for WT and cL762F. All values represent mean ± SEM. * P < 0.05 vs. CACNA1C-WT. (C) Inactivation time constants (τ) for the fast phase of Cav1.2 decay time of WT (N = 5) and mutant (N = 8) as a function of voltage. Time constants for each voltage step were determined by fitting a biexponential function to current decay. *P < 0.05 vs. CACNA1C-WT. (D) Inactivation time constants (τ) for the slow phase of Cav1.2 decay time of WT (N = 5) and CACNA1C-L762F (N = 8) as a function of voltage. *P < 0.05 vs. WT.
3.4. Compendium of Timothy syndrome, cardiac-only Timothy syndrome, and long QT syndrome-associated CACNA1C mutations
To gain an understanding of the possible genetic and phenotypic differences between TS-, LQTS-, and COTS-associated CACNA1C mutations, a comprehensive review of the literature was conducted to compile all known CACNA1C mutations associated with prolonged cardiac repolarization. A total of 28 independent probands were identified in the literature with CACNA1C mutations for which clinical information including QT interval could be obtained. All probands hosted single, heterozygous, missense mutations. Probands included 9 with TS or a TS-like clinical phenotype, and 16 with non-syndromic delayed cardiac repolarization consistent with LQTS, and 3 COTS probands. A comprehensive summary of all CACNA1C probands is provided in Table 2.
Table 2.
Compendium of CACNA1C mutations in the literature associated with prolonged cardiac repolarization disease or arrhythmia.
| No | Residue | Disease | Mutation | Age (y) | Sex | Race/Nationality | QTc | AVB | Arrhythmia | SCD | Fam Hx | Fam Hx SCD | Extracardiac | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 28 | LQTS, borderline | A28 T | 12 | F | NA | 450 | N | N | Y | N | – | Wemhoner K, 2015 | |
| 2 | 381 | LQTS | P381S | 8 | F | Japan | 480 | N | N | Y | N | – | Fukuyama M, 2014 | |
| 3 | 402 | TS | G402S | 4 | M | Caucasian | 620 | N | A fib, VT, TdP | Y | N | N | HCM | Splawski I, 2005 |
| 4 | 402 | TS, atypical | G402S | 8 | F | Lebanon | 468–547 | N | TdP | Y | N | N | Syndactyly, precocious puberty, GDD, epilepsy | Frohler S, 2014 |
| 5 | 402 | LQTS | G402S | 13 | F | European | 480 | N | VF | Y | N | N | – | Hiipala A, 2015 |
| 6 | 406 | TS | G406R | 1.4 | M | Caucasian | 697 | Y | N | Y | N | Syndactyly, small teeth | Dufendach K, 2013 | |
| 7 | 406 | TS | G406R | 2.5 | M | Korean | 580–600 | Y | VF | Y | N | N | Syndactyly, small teeth, hypoglycemia | An H, 2013 |
| 8 | 406 | TS | G406R | 0.1 | F | Caucasian | 730 | N | VT, TdP | Y | N | N | Syndactyly, GDD, cavities, facial anomalies | Splawski I, 2005 |
| 9 | 406 | TS | G406R | 0.25 | F | Mexico | 655 | Y | N | N | N | Syndactyly, facial anomalies | ||
| 10 | 518 | COTS | R518C | 25 | F | Caucasian | 500 | Y | N | Y | Y | – | Boczek N, 2015 | |
| 11 | 518 | COTS | R518C | 10 | F | Caucasian | 500 | N | Y | Y | N | – | Boczek N, 2015 | |
| 12 | 518 | COTS | R518H | 25 | F | Caucasian | 480 | N | SA node | N | Y | N | – | Boczek N, 2015 |
| 13 | 582 | LQTS | A582D | 12 | F | Japan | 597 | N | N | N | N | – | Fukuyama M, 2014 | |
| 14 | 762 | LQTS | L762F | 3 | F | Central American | 530 | N | N | Y | Y | None | ||
| 15 | 834 | LQTS | K834E | 15 | F | Caucasian | 475 | N | N | N | N | – | Boczek N, 2013 | |
| 16 | 857 | LQTS | P857R | 27 | F | Caucasian | 498 | N | N | N | N | – | Boczek N, 2013 | |
| 17 | 857 | LQTS | P857L | 15 | M | Caucasian | 514 | N | N | Y | Y | – | Boczek N, 2013 | |
| 18 | 858 | LQTS | R858H | 54 | F | Japan | 435 | N | VF | Y | Y | Y | – | Fukuyama M, 2014 |
| 19 | 858 | LQTS | R858H | 7 | M | Japan | 476 | N | N | N | N | – | Fukuyama M, 2014 | |
| 20 | 858 | LQTS | R858H | 15 | M | Japan | 420 | N | N | N | N | – | Fukuyama M, 2014 | |
| 21 | 860 | LQTS | R860G | 30 | M | NA | 498 | N | Y | N | N | – | Wemhoner K, 2015 | |
| 22 | 1166 | TS | I1166T | 0.1 | M | Caucasian | 595–812 | Y | N | N | N | Clinodactyly, osteopenia, seizures, small teeth | Boczek N, 2015 | |
| 23 | 1166 | TS | I1166T | 1 | F | NA | 550 | Y | Y | N | N | Syndactyly, GDD | Wemhoner K, 2015 | |
| 24 | 1166 | LQTS, borderline | I1166V | 15 | F | NA | 452 | N | N | Y | N | – | Wemhoner K, 2015 | |
| 25 | 1473 | TS | A1473G | 2 | M | Caucasian | 640 | Y | VT | N | N | N | Syndactyly, joint contractures, GDD, seizures, hypoglycemia | Gillis J, 2012 |
| 26 | 1475 | LQTS | I1475M | 14 | F | NA | 473 | N | Y | Y | N | – | Wemhoner K, 2015 | |
| 27 | 1496 | LQTS | E1496K | 26 | F | NA | 480 | N | Y | N | N | – | Wemhoner K, 2015 | |
| 28 | 1906 | LQTS | R1906Q | 39 | F | Caucasian | 513 | N | N | N | N | – | Boczek N, 2013 |
M, male; F, female; SCD, personal history of aborted sudden cardiac death; Fam Hx, family history of disease in a 1st degree relative; Fam Hx SCD, family history of sudden cardiac death in a 1st degree relative; A fib, atrial fibrillation; AVB, 2:1 atrioventricular nodal block; VT, ventricular tachycardia; VF, ventricular fibrillation; TdP, torsades de pointes; HCM, hypertrophic car diomyopathy; PDA, patent ductus arteriosus; NA, not available.
Among the patients with TS or TS-like clinical phenotype, the average age at diagnosis was 2.2 ± 0.9 years while LQTS was significantly older at 19.1 ± 3.3 years (P = 0.001). A higher proportion of TS probands were male (56%) in comparison to LQTS probands (25%) and COTS probands (0%) although this did not reach statistical significance. QTc was significantly higher in TS probands (650 ± 29 ms) vs LQTS (486 ± 10 ms, P < 0.0001) and there was a high prevalence of AV block (55%) in TS which was absent in LQTS probands (0%, P = 0.001). These findings are summarized in Table 3.
Table 3.
Comparison of Timothy syndrome, cardiac-only Timothy syndrome, and long QT syndrome probands in the literature.
| TS | COTS | LQTS | Total | P-valuea | |
|---|---|---|---|---|---|
| Probands | 9 | 3 | 16 | 28 | |
| Mutations | 4 | 2 | 14 | 20 | |
| Age (y) | 2.15 ± 0.85 | 20.0 ± 5.0 | 19.1 ± 3.3 | 13.7 ± 2.5 | 0.001 |
| % female | 44% | 100% | 75% | 68% | 0.2 |
| QTc | 650 ± 29 | 493 ± 7 | 486 ± 10 | 539 ± 18 | <0.0001 |
| AV block | 66% | 0% | 0% | 25% | 0.0024 |
| SCD | 55% | 33% | 31% | 39% | 0.12 |
| Fam Hx | 11% | 100% | 44% | 39% | 0.39 |
| Fam Hx SCD | 0% | 33% | 19% | 14% | 0.28 |
TS vs LQTS cohorts; COTS, cardiac-only Timothy syndrome, SCD, personal history of aborted sudden cardiac death; Fam Hx, family history of disease in a 1st degree relative; Fam Hx SCD, family history of sudden cardiac death in a 1st degree relative.
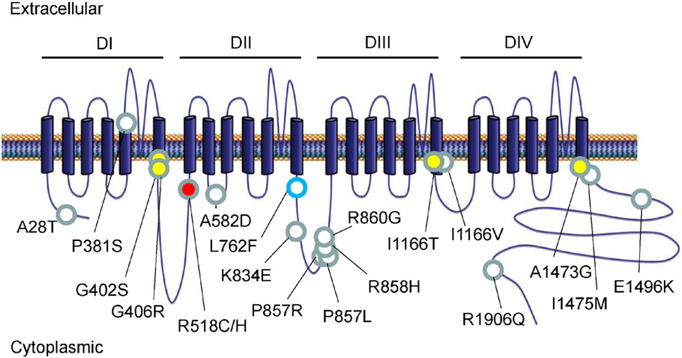
Topological mapping of all variants along the linear protein topology of CACNA1C demonstrated disease-specific loci. Specifically, TS-associated mutations exclusively localized to 6th transmembrane segments across the homologous domains. Conversely, LQTS-associated mutations localize diffusely across the cytoplasmic N- and C-terminal segments as well as the transmembrane linker regions (Fig. 6). To evaluate whether mutation location or disease association predicted the biophysical impact of the mutation on CACNA1C, available patch clamp data for each mutation was compiled. A variety of alterations to Cav1.2 channel kinetics were noted; however, there was no clear association between observed changes and either the location of the mutation or disease association. These results are summarized in Table 4.
Fig. 6.
Topological map of all known TS, COTS, and LQTS-associated CACNA1C mutations. 4 homologous domains are designated (DI-DIV) comprised of transmembrane segments (columns), cytoplasmic segments, and transmembrane linker segments. TS- (yellow fill), COTS- (red fill), and LQTS-associated (white fill) mutations are overlaid the linear topology of the CACNA1C. The CACNA1C-L762F mutation is noted in blue outline.
Table 4.
Compendium of Cav1.2 channel function alterations from in vitro patch clamp studies of known CACNA1C mutations.
| Mutation | Disease | Current density | Act shift | Inact shift | Inact tau | Persistent current | Window current | Reference |
|---|---|---|---|---|---|---|---|---|
| A28T | LQTS, Borderline | Increase | No change | Positive | No change | No change | Wemhoner K, J Mol Cell Cardiol, 2015 | |
| P381S | LQTS | No change | No change | No change | No change | Fukuyama M, Europace, 2014 | ||
| G402S | TS | No change | Much slower | Splawski I, PNAS 2005 | ||||
| G406R | TS | No change | Negative | Much slower | Splawski I, PNAS 2005 | |||
| M456I | LQTS | No change | No change | No change | No change | Fukuyama M, Europace, 2014 | ||
| R518C | COTS | Decrease | No change | Positive | Slower | Increase | Increase | Boczek N, Circ Arrhythm Electrophysiol, 2015 |
| R518H | COTS | Decrease | No change | Positive | Slower | Increase | Increase | Boczek N, Circ Arrhythm Electrophysiol, 2015 |
| A582D | LQTS | No change | No change | No change | Slower | Fukuyama M, Europace, 2014 | ||
| L762F | LQTS | No change | No change | Positive | Slower | Increase | Increase | |
| P857L | LQTS | Increase | Boczek N, Circ Cardiovasc Genetics, 2013 | |||||
| R858H | LQTS | Increase | No change | No change | No change | Fukuyama M, Europace, 2014 | ||
| R860G | LQTS | No change | No change | Positive | No change | Increase | Wemhoner K, J Mol Cell Cardiol, 2015 | |
| I1166T | TS | Decrease | Negative | No change | Increase | Boczek N, Heart Rhythm, 2014 | ||
| I1166T | TS | Increase | Negative | No change | No change | Increase | Wemhoner K, J Mol Cell Cardiol, 2015 | |
| I1166V | LQTS, Borderline | Increase | No change | No change | No change | No change | Wemhoner K, J Mol Cell Cardiol, 2015 | |
| I1475M | LQTS | No change | Negative | No change | No change | Increase | Wemhoner K, J Mol Cell Cardiol, 2015 | |
| E1496K | LQTS | No change | Negative | No change | Slower | Increase | Wemhoner K, J Mol Cell Cardiol, 2015 | |
| G1783C | LQTS | No change | No change | No change | No change | Fukuyama M, Europace, 2014 |
Act, activation; Inact, inactivation; VF, ventricular fibrillation.
4. Discussion
The identification of CACNA1C mutations in a variety of cardiovascular and syndromic diseases offers a microcosm of the challenge of linking genetic changes to clinical disease. In addition to disease with impaired cardiac repolarization, such as TS and LQTS, mutations in CACNA1C have been associated with other arrhythmias, such as Brugada syndrome and early repolarization syndrome [23]. In addition, the recent discovery of TS without extracardiac manifestations, so-called COTS, highlights the widening degree of variable expressivity that mutations in a single ion channel can convey [9,28]. As studies into the genetic etiologies of diseases advances, a paradigm has emerged in which mutations within a single ion channel, and indeed identical residues, result in a variety of phenotypic changes. For example, mutations in JPH2-encoded junctophilin type 2 have been linked with cardiomyopathy, heart failure, and atrial tachyarrhythmias [29–32]. Further, cardiac ion channels have an established history of variable expressivity when functionally perturbed. The SCN5A-encoded cardiac sodium channel (Nav1.5) has been associated with a number of cardiac arrhythmias and defective electrical properties including LQTS, Brugada syndrome (BrS), as well as conduction perturbations [33]. Classically, gain-of-function mutations in Nav1.5, such as window current or late current, have been associated with LQT3 while loss-of-function mutations result in BrS1. Yet despite this traditional dichotomy, there is marked overlap in the clinical presentation of patients [34]. The complexity of Nav1.5-associated diseases continues to increase as recent evidence suggestions that loss of function SCN5A mutations have been associated with sinus arrhythmia and poor pacemaker capture [35].
Identification of the novel CACNA1C-L762F mutation associated with a markedly prolonged QT interval in a multigenerational family otherwise negative for canonical LQTS-associated mutations suggests that this mutation may be associated with deranged cardiac repolarization. Multiple in silico modeling programs predict a highly deleterious effect on the native protein which is supported by the absence of this mutation in over 99,000 subjects. Further, whole cell patch clamp of heterologously expressed mutant CACNA1C in TSA201 cells demonstrate markedly slower inactivation tau and increased sustained and window current compared to wild-type channel. Comparison of TS-and LQTS-associated mutations, as well as mutations localizing to common areas of Cav1.2, do not appear to have consistent biophysical signatures when autologously expressed. This phenotypic complexity is also reflected within the genotype of the CACNA1C-L762F kindred. While all individuals in the family hosting the L762F mutation, three potentially modifying genetic variants were identified including KCNH2-K897T, SCN5A-H558R, and KCNE1-G38S. Each of these variants has been reported in the literature to modify cardiac repolarization; however, are frequent in the population and are unlikely to be a sole genetic etiology of QT prolongation. While it is possible that high genetic burden of these variants, in the context of the CACNA1C-L762F variant, may cause a more severe repolarization delay, additional genetic and functional analyses are needed to explore this possibility.
Following compilation and analysis of all TS-, COTS-, LQTS-associated mutations, disease-specific mutation locations begin to emerge. Classically, TS is associated with mutations CACNA1C-G402S and G406R [5,6]. Indeed, we have identified a Hispanic female with classic features of TS including marked QT prolongation with a QTc of 655 ms, fetal bradycardia secondary to 2:1 AV block, dysmorphic facial features, and syndactyly. To our knowledge, this is the first TS proband described from Central and/or South America. Recently, additional TS-associated mutations have been identified, including CACNA1C-I1166T and -A1473G [28,36]. While these mutated residues are found on divergent domains of CACNA1C, each localizes to the respective C-terminal aspect of the 6th transmembrane segment near the interlinker region [28].
Conversely, mutations associated with LQTS, without the extracardiac syndromic manifestations of TS, localize across the linear topology of the protein. Interestingly, recent identification of the CACNA1C-I116V mutation in a patient with LQTS offers a genetic enigma as it localizes to the identical residue as the CACNA1C-I1166T mutation which is associated with TS [37]. There is currently no mechanistic explanation for this divergence in clinical disease given nearly identical mutations. It is possible that the divergent biochemical properties of the mutant valine, a non-polar small residue, compared to the mutant threonine, a polarized residue susceptible to a number of post-translational modifications, may account for this difference; however, independent mechanistic experimentation will be needed to verify this. Further, it is notable that all three probands identified with COTS host mutations that localize to an apparent disease-specific hotspot at the C-terminal interlinker between DI and DII [9]. Certainly, additional mechanistic studies are required to explain the disease specific mechanisms leading to the TS clinical phenotype versus LQTS in the face of overlapping mutation locations and varied biophysical changes to CACNA1C.
Future research directions should be targeted toward identifying these underlying mechanistic differences, particularly in more physiologic cellular backgrounds, including primary cardiac myocytes. The recent availability of human induced pluripotent cells offers a potentially robust model from which to test patient-specific mechanisms of genetic diseases and allow dissection of multi-factorial etiologies of disease expression.
Supplementary Material
Acknowledgements
This work was supported in part by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (M.J.A.).
Abbreviations:
- CACNA1C
the CACNA1C-encoded α1c α-subunit of the L-type calcium channel
- COTS
cardiac-only Timothy syndrome
- LQTS
long QT syndrome
- LTCC
L-type calcium channel
- SCD
sudden cardiac death
- SIDS
sudden infant death syndrome
- TS
Timothy syndrome
Footnotes
Disclosures
M.J.A. is a consultant for Boston Scientific, Gilead Sciences, Medtronic, and St. Jude Medical. M.J.A. and Mayo Clinic receive sales based royalties from Transgenomic with respect to their FAMILION-LQTS and FAMILION-CPVT genetic tests. No financial support was provided by any of these entities for this study.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ijcard.2016.06.081
References
- [1].Bers DM, Cardiac excitation–contraction coupling, Nature 415 (2002) 198–205. [DOI] [PubMed] [Google Scholar]
- [2].Napolitano C, Antzelevitch C, Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac voltage-dependent L-type calcium channel, Circ. Res 108 (2011) 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Reichenbach H, Meister EM, Theile H, The heart-hand syndrome. A new variant of disorders of heart conduction and syndactylia including osseous changes in hands and feet, Kinderarztl. Prax 60 (1992) 54–56. [PubMed] [Google Scholar]
- [4].Marks ML, Trippel DL, Keating MT, Long QT syndrome associated with syndactyly identified in females, Am. J. Cardiol 76 (1995) 744–745. [DOI] [PubMed] [Google Scholar]
- [5].Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT, Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations, Proc. Natl. Acad. Sci. U.S.A 102 (2005) 8089–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT, CaV1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism, Cell 119 (2004) 19–31. [DOI] [PubMed] [Google Scholar]
- [7].Landstrom AP, Tester DJ, Ackerman MJ, Role of genetic testing in athletes in sports, in: Lawless C (Ed.), Cardiology Essentials: Evaluation, Management and Case Studies New York, Springer Science, New York, 2011. [Google Scholar]
- [8].Tester D, Will M, Haglund C, Ackerman M, Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing, Heart Rhythm. 2 (2005) 507–517. [DOI] [PubMed] [Google Scholar]
- [9].Boczek NJ, Ye D, Jin F, Tester DJ, Huseby A, Bos JM, Johnson AJ, Kanter R, Ackerman MJ, Identification and functional characterization of a novel CACNA1Cmediated cardiac disorder characterized by prolonged QT intervals with hypertrophic cardiomyopathy, congenital heart defects, and sudden cardiac death, Circ. Arrhythm. Electrophysiol. 8 (2015) 1122–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yamamoto S, Jaiswal M, Charng W-L, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman Nele A, Xiong B, Zhang K, Bayat V, David G, Li T, Chen K, Gala U, Harel T, Pehlivan D, Penney S, Vissers Lisenka ELM, de Ligt J, Jhangiani SN, Xie Y, Tsang Stephen H, Parman Y, Sivaci M, Battaloglu E, Muzny D, Y-W W, Liu Z, Lin-Moore Alexander T, Clark Robin D, Curry CJ, Link N, Schulze KL, Boerwinkle E, Dobyns WB, Allikmets R, Gibbs RA, Chen R, Lupski JR, Wangler MF, Bellen HJ, A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases, Cell 159 (2014) 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data, Genome Res. 20 (2010) 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Challis D, Yu J, Evani U, Jackson A, Paithankar S, Coarfa C, Milosavljevic A, Gibbs R, Yu F, An integrative variant analysis suite for whole exome nextgeneration sequencing data, BMC Bioinforma. 13 (2012) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang K, Li M, Hakonarson H, ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data, Nucleic Acids Res. 38 (2010), e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Consortium TGP, An integrated map of genetic variation from 1092 human genomes, Nature 491 (2012) 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Project NES, Akey JM, Analysis of 6515 exomes reveals the recent origin of most human protein-coding variants, Nature 493 (2013) 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, O’DonnellLuria A, Ware J, Hill A, Cummings B, Tukiainen T, Birnbaum D, Kosmicki J, Duncan L, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Cooper D, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki M, Levy Moonshine A, Natarajan P, Orozco L, Peloso G, Poplin R, Rivas M, Ruano-Rubio V, Ruderfer D, Shakir K, Stenson P, Stevens C, Thomas B, Tiao G, Tusie-Luna M, Weisburd B, Won H-H, Yu D, Altshuler D, Ardissino D, Boehnke M, Danesh J, Roberto E, Florez J, Gabriel S, Getz G, Hultman C, Kathiresan S, Laakso M, McCarroll S, McCarthy M, McGovern D, McPherson R, Neale B, Palotie A, Purcell S, Saleheen D, Scharf J, Sklar P, Patrick S, Tuomilehto J, Watkins H, Wilson J, Daly M, MacArthur D, Analysis of protein-coding genetic variation in 60,706 humans, bioRxiv (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ng PC, Henikoff S, Predicting deleterious amino acid substitutions, Genome Res. 11 (2001) 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR, A method and server for predicting damaging missense mutations, Nat. Methods 7 (2010) 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Capriotti E, Calabrese R, Casadio R, Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information, Bioinformatics 22 (2006) 2729–2734. [DOI] [PubMed] [Google Scholar]
- [20].Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J, Brezovsky J, Damborsky J, PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations, PLoS Comput. Biol 10 (2014), e1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Corpet F, Multiple sequence alignment with hierarchical clustering, Nucleic Acids Res. 16 (1988) 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tanabe T, Takeshima H, Mikami A, Flockerzi V, Takahashi H, Kangawa K, Kojima M, Matsuo H, Hirose T, Numa S, Primary structure of the receptor for calcium channel blockers from skeletal muscle, Nature 328 (1987) 313–318. [DOI] [PubMed] [Google Scholar]
- [23].Antzelevitch C, Pollevick GD, Cordeiro JM, Casis O, Sanguinetti MC, Aizawa Y, Guerchicoff A, Pfeiffer R, Oliva A, Wollnik B, Gelber P, Bonaros EP, Burashnikov E, Wu Y, Sargent JD, Schickel S, Oberheiden R, Bhatia A, Hsu L-F, Haïssaguerre M, Schimpf R, Borggrefe M, Wolpert C, Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death, Circulation 115 (2007) 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hu D, Barajas-Martinez H, Nesterenko VV, Pfeiffer R, Guerchicoff A, Cordeiro JM, Curtis AB, Pollevick GD, Wu Y, Burashnikov E, Antzelevitch C, Dual variation in SCN5A and CACNB2b underlies the development of cardiac conduction disease without Brugada syndrome, Pacing Clin. Electrophysiol 33 (2010) 274–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nof E, Cordeiro JM, Pérez GJ, Scornik FS, Calloe K, Love B, Burashnikov E, Caceres G, Gunsburg M, Antzelevitch C, A common single nucleotide polymorphism can exacerbate long QT type 2 syndrome leading to sudden infant death, Circ. Cardiovasc. Genet 3 (2010) 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shinlapawittayatorn K, Du XX, Liu H, Ficker E, Kaufman ES, Deschênes I, A common SCN5A polymorphism modulates the biophysical defects of SCN5A mutations, Heart Rhythm. 8 (2011) 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ehrlich JR, Zicha S, Coutu P, Hébert TE, Nattel S, Atrial fibrillation-associated minK38G/S polymorphism modulates delayed rectifier current and membrane localization, Cardiovasc. Res. 67 (2005) 520–528. [DOI] [PubMed] [Google Scholar]
- [28].Boczek NJ, Miller EM, Ye D, Nesterenko VV, Tester DJ, Antzelevitch C, Czosek RJ, Ackerman MJ, Ware SM, Novel Timothy syndrome mutation leading to increase in CACNA1C window current, Heart Rhythm. 12 (2015) 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Landstrom AP, Beavers DL, Wehrens XHT, The junctophilin family of proteins: from bench to bedside, Trends Mol. Med 20 (2014) 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Landstrom AP, Weisleder N, Batalden KB, Martijn Bos J, Tester DJ, Ommen SR, Wehrens XHT, Claycomb WC, Ko J-K, Hwang M, Pan Z, Ma J, Ackerman MJ, Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans, J. Mol. Cell. Cardiol 42 (2007) 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].van Oort R, Garbino A, Wang W, Dixit S, Landstrom A, Gaur N, De Almeida A, Skapura D, Rudy Y, Burns A, Ackerman M, Wehrens X, Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice, Circulation 123 (2011) 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Beavers DL, Wang W, Ather S, Voigt N, Garbino A, Dixit SS, Landstrom AP, Li N, Wang Q, Olivotto I, Dobrev D, Ackerman MJ, Wehrens XHT, Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization, J. Am. Coll. Cardiol 62 (2013) 2010–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Grant A, Carboni M, Neplioueva V, Starmer C, Memmi M, Napolitano C, Priori S, Long QT syndrome, Brugada syndrome, and conduction system disease are linked to a single sodium channel mutation, J. Clin. Invest 110 (2002) 1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Makita N, Behr E, Shimizu W, Horie M, Sunami A, Crotti L, Schulze-Bahr E, Fukuhara S, Mochizuki N, Makiyama T, Itoh H, Christiansen M, McKeown P, Miyamoto K, Kamakura S, Tsutsui H, Schwartz PJ, George AL, Roden DM, The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome, J. Clin. Invest 118 (2008) 2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chiang DY, Kim JJ, Valdes SO, de la Uz C, Fan Y, Orcutt J, Domino M, Smith M, Wehrens XHT, Miyake CY, Loss-of-function SCN5A mutations associated with sinus node dysfunction, atrial arrhythmias, and poor pacemaker capture, Circ. Arrhythm. Electrophysiol 8 (2015) 1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gillis J, Burashnikov E, Antzelevitch C, Blaser S, Gross G, Turner L, Babul-Hirji R, Chitayat D, Long QT, syndactyly, joint contractures, stroke and novel CACNA1C mutation: expanding the spectrum of Timothy syndrome, Am. J. Hum. Genet. A 158A (2012) 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wemhöner K, Friedrich C, Stallmeyer B, Coffey AJ, Grace A, Zumhagen S, Seebohm G, Ortiz-Bonnin B, Rinné S, Sachse FB, Schulze-Bahr E, Decher N , Gainof-function mutations in the calcium channel CACNA1C (Cav1.2) cause nonsyndromic long-QT but not Timothy syndrome, J. Mol. Cell. Cardiol 80 (2015) 186–195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.